Introduction to Illumina Sequencing Day 1 Video 2


- Slides: 15

Introduction to Illumina Sequencing Day 1, Video 2 • Overview of Next-gen sequencing • Introduction to Illumina sequencing • Multiplexing • Sequencing run statistics

Next-Gen Sequencing • Millions of reactions performed in parallel • Shorter read lengths, higher error rate • Sample/library prep is required • Many different approaches • Illumina sequencing-by-synthesis (Solexa technology) • Roche 454 pyrosequencing • AB SOLID color-based sequencing by ligation • Ion Torrent semiconductor sequencing • Single-molecule sequencing (Pac. Bio, Min. ION, etc)

Some general terminology • SR: single-read sequencing, sequence from only one end • PE: paired-end sequencing, sequence from both ends • Adapters: DNA added to the ends of DNA/RNA fragments to be sequenced. The adapters allow the DNA/RNA to attach to the flowcell • Index/barcode: used interchangeable to indicate sequence identifier for multiplexing • Phi. X: commercially available genomic library of Phi. X bacteriophage DNA, commonly spiked into libraries

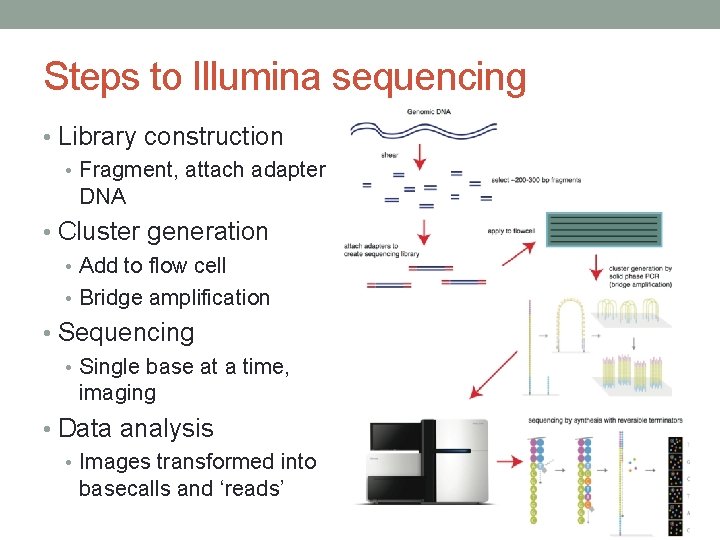
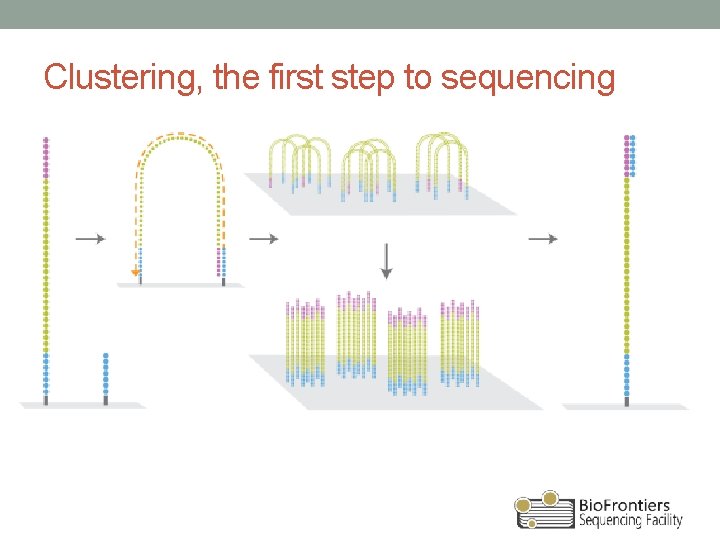
Steps to Illumina sequencing • Library construction • Fragment, attach adapter DNA • Cluster generation • Add to flow cell • Bridge amplification • Sequencing • Single base at a time, imaging • Data analysis • Images transformed into basecalls and ‘reads’

Illumina sequencing • SBS chemistry video • http: //www. illumina. com/technology/next-generationsequencing/sequencing-technology. html

Clustering, the first step to sequencing

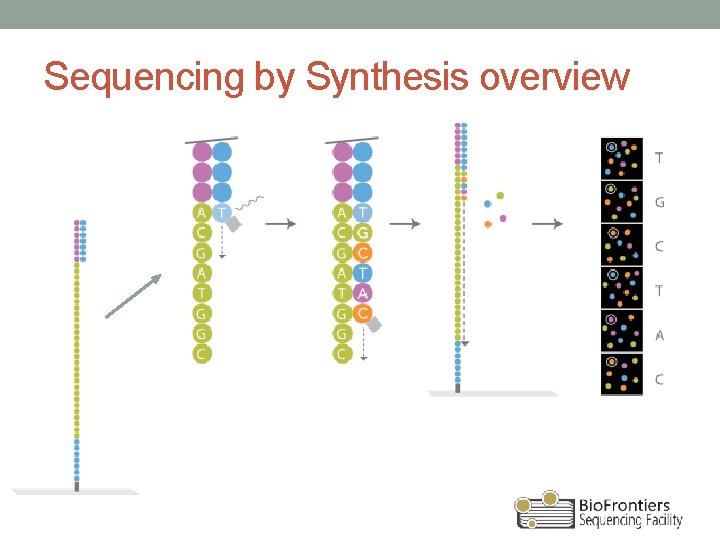
Sequencing by Synthesis overview

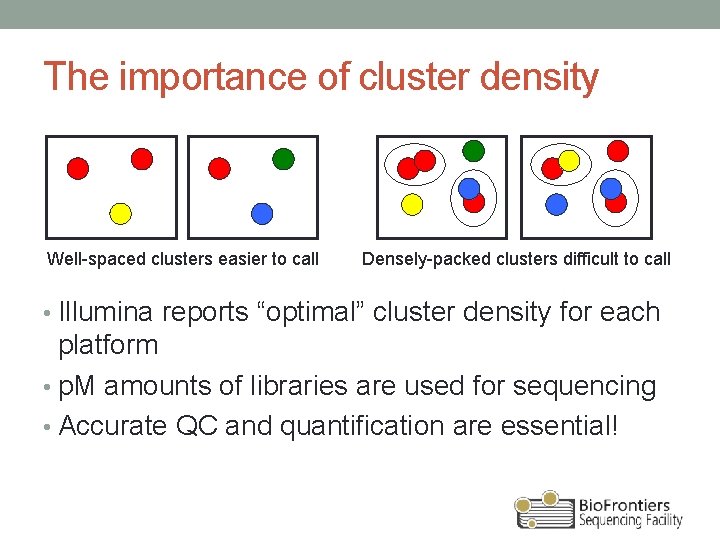
The importance of cluster density Well-spaced clusters easier to call Densely-packed clusters difficult to call • Illumina reports “optimal” cluster density for each platform • p. M amounts of libraries are used for sequencing • Accurate QC and quantification are essential!

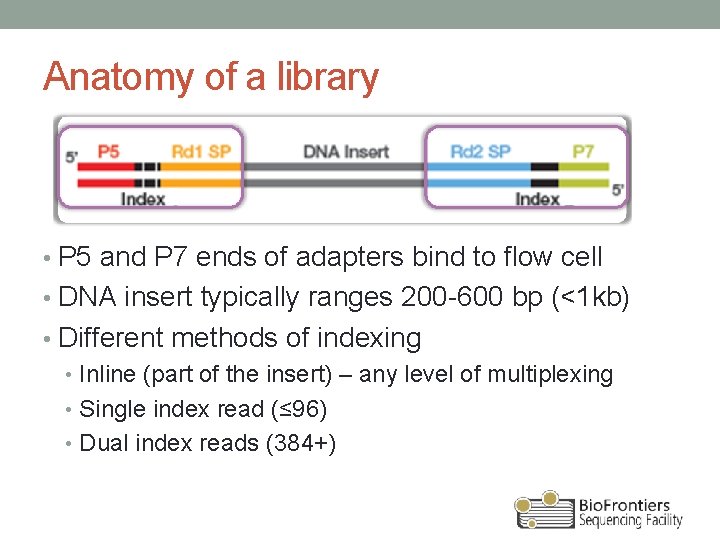
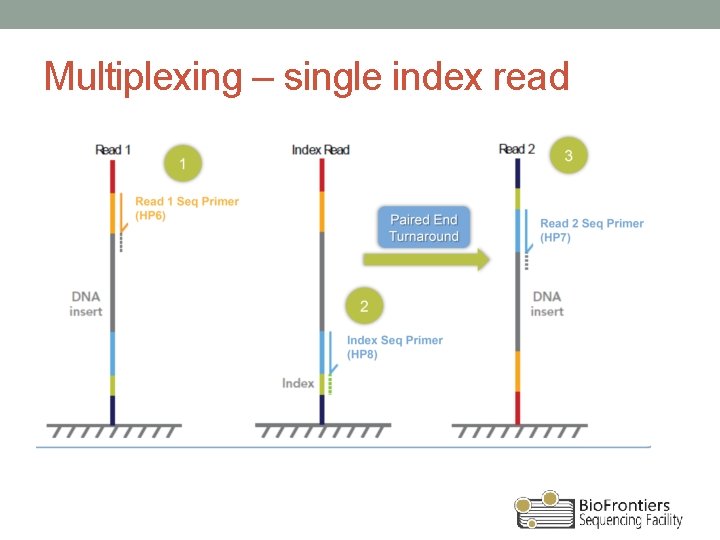
Anatomy of a library • P 5 and P 7 ends of adapters bind to flow cell • DNA insert typically ranges 200 -600 bp (<1 kb) • Different methods of indexing • Inline (part of the insert) – any level of multiplexing • Single index read (≤ 96) • Dual index reads (384+)

Multiplexing – single index read

Multiplexing – dual index reads • hf

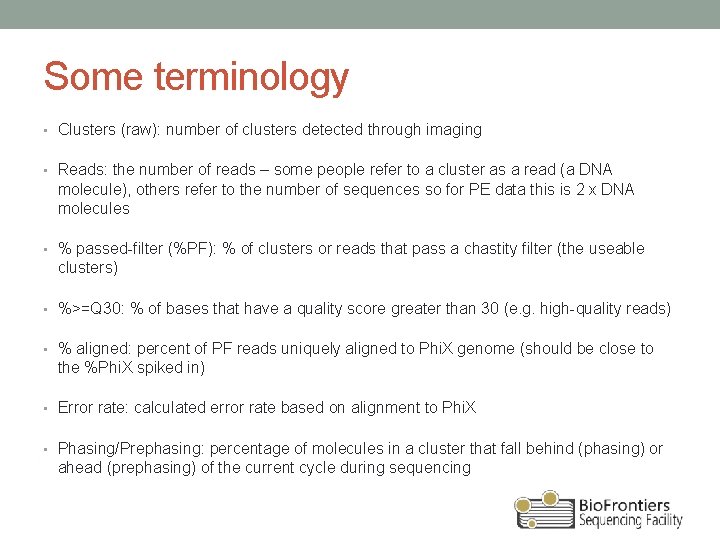
Some terminology • Clusters (raw): number of clusters detected through imaging • Reads: the number of reads – some people refer to a cluster as a read (a DNA molecule), others refer to the number of sequences so for PE data this is 2 x DNA molecules • % passed-filter (%PF): % of clusters or reads that pass a chastity filter (the useable clusters) • %>=Q 30: % of bases that have a quality score greater than 30 (e. g. high-quality reads) • % aligned: percent of PF reads uniquely aligned to Phi. X genome (should be close to the %Phi. X spiked in) • Error rate: calculated error rate based on alignment to Phi. X • Phasing/Prephasing: percentage of molecules in a cluster that fall behind (phasing) or ahead (prephasing) of the current cycle during sequencing

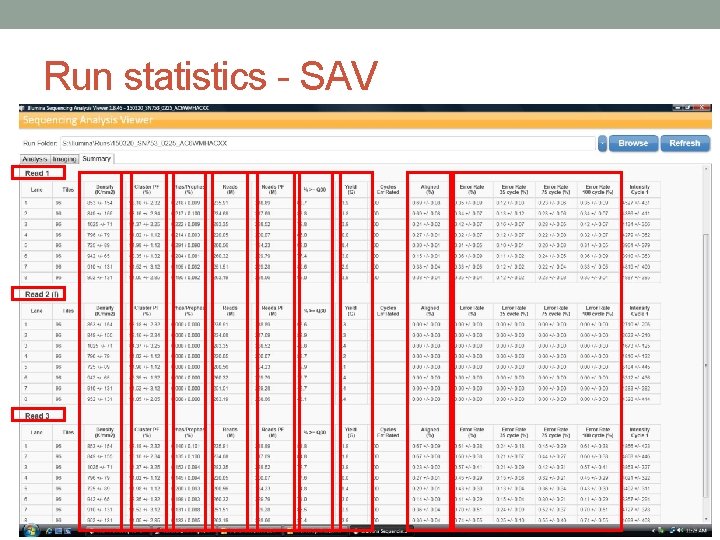
Run statistics - SAV • df

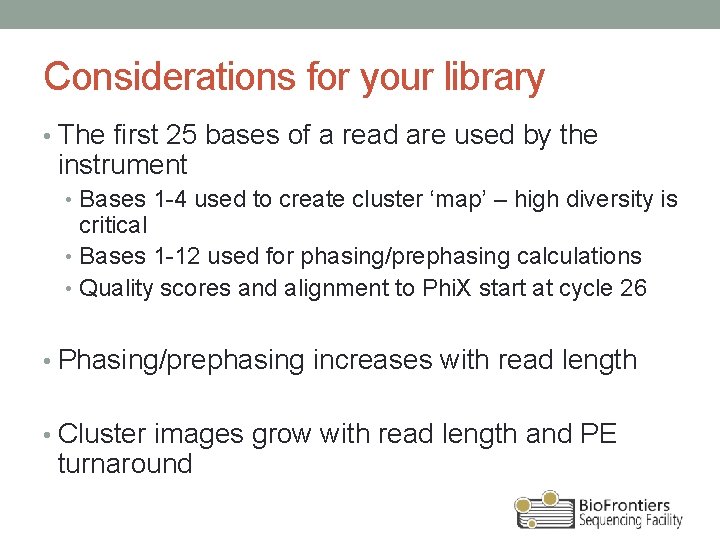
Considerations for your library • The first 25 bases of a read are used by the instrument • Bases 1 -4 used to create cluster ‘map’ – high diversity is critical • Bases 1 -12 used for phasing/prephasing calculations • Quality scores and alignment to Phi. X start at cycle 26 • Phasing/prephasing increases with read length • Cluster images grow with read length and PE turnaround

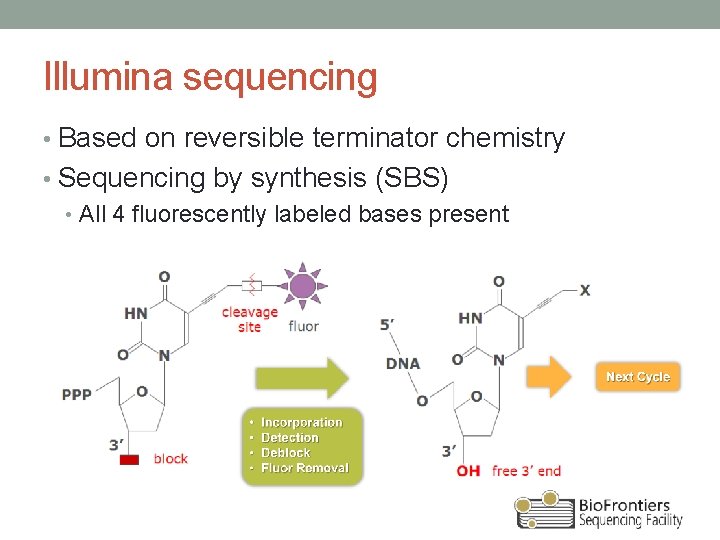
Illumina sequencing • Based on reversible terminator chemistry • Sequencing by synthesis (SBS) • All 4 fluorescently labeled bases present