The NHLBI Exome Sequencing Project Stephen S Rich

The NHLBI Exome Sequencing Project Stephen S. Rich, Ph. D September 30, 2013
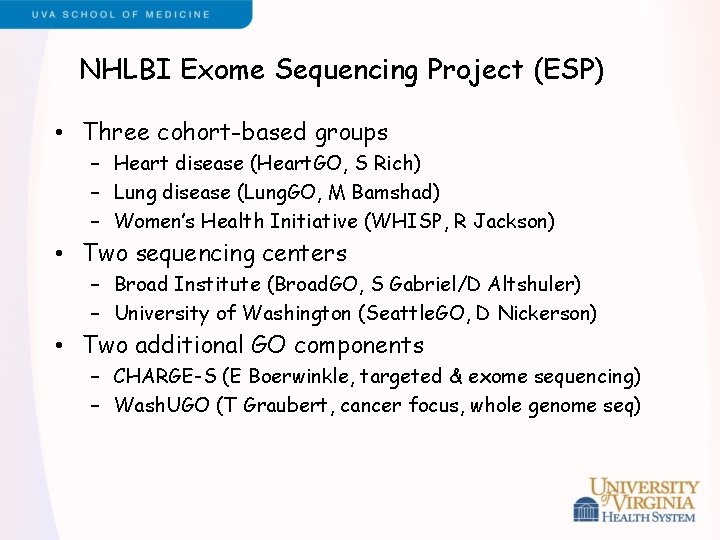
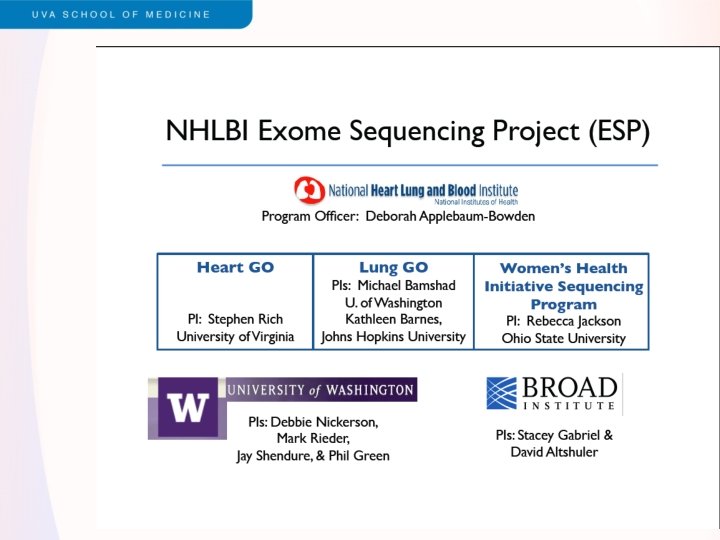
NHLBI Exome Sequencing Project (ESP) • Three cohort-based groups – Heart disease (Heart. GO, S Rich) – Lung disease (Lung. GO, M Bamshad) – Women’s Health Initiative (WHISP, R Jackson) • Two sequencing centers – Broad Institute (Broad. GO, S Gabriel/D Altshuler) – University of Washington (Seattle. GO, D Nickerson) • Two additional GO components – CHARGE-S (E Boerwinkle, targeted & exome sequencing) – Wash. UGO (T Graubert, cancer focus, whole genome seq)
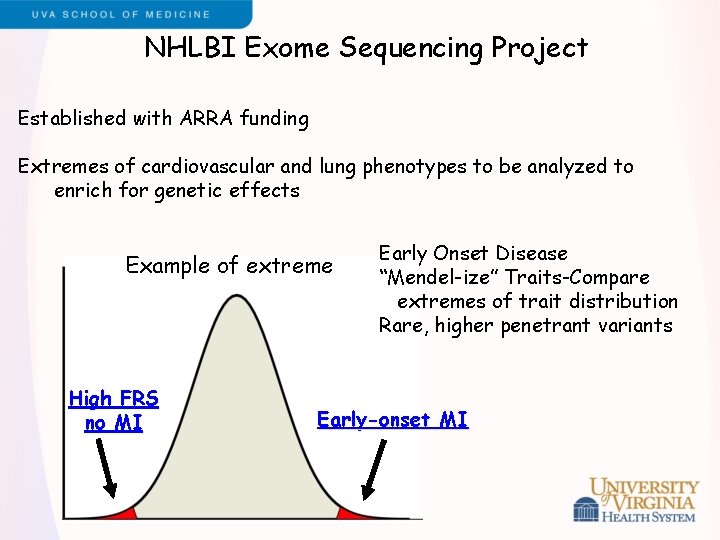
NHLBI Exome Sequencing Project Established with ARRA funding Extremes of cardiovascular and lung phenotypes to be analyzed to enrich for genetic effects Example of extreme High FRS no MI Early Onset Disease “Mendel-ize” Traits-Compare extremes of trait distribution Rare, higher penetrant variants Early-onset MI
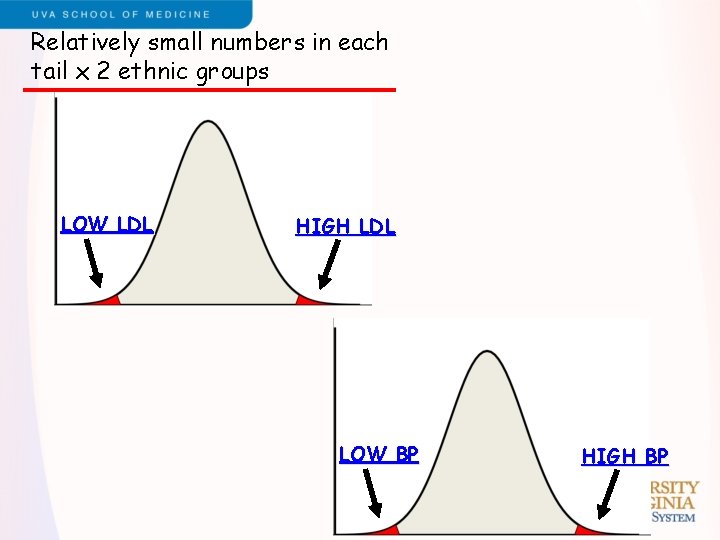
Relatively small numbers in each tail x 2 ethnic groups LOW LDL HIGH LDL LOW BP HIGH BP
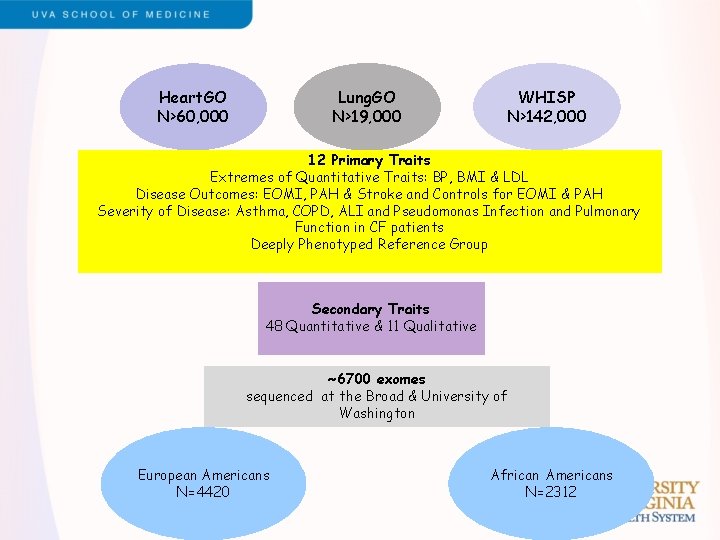
Lung. GO N>19, 000 Heart. GO N>60, 000 WHISP N>142, 000 12 Primary Traits Extremes of Quantitative Traits: BP, BMI & LDL Disease Outcomes: EOMI, PAH & Stroke and Controls for EOMI & PAH Severity of Disease: Asthma, COPD, ALI and Pseudomonas Infection and Pulmonary Function in CF patients Deeply Phenotyped Reference Group Secondary Traits 48 Quantitative & 11 Qualitative ~6700 exomes sequenced at the Broad & University of Washington European Americans N=4420 African Americans N=2312
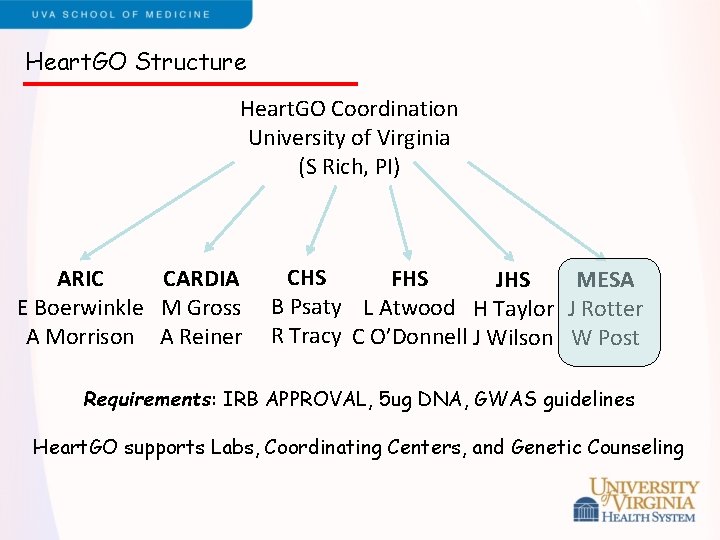
Heart. GO Structure Heart. GO Coordination University of Virginia (S Rich, PI) ARIC CARDIA E Boerwinkle M Gross A Morrison A Reiner CHS FHS JHS MESA B Psaty L Atwood H Taylor J Rotter R Tracy C O’Donnell J Wilson W Post Requirements: IRB APPROVAL, 5 ug DNA, GWAS guidelines Heart. GO supports Labs, Coordinating Centers, and Genetic Counseling
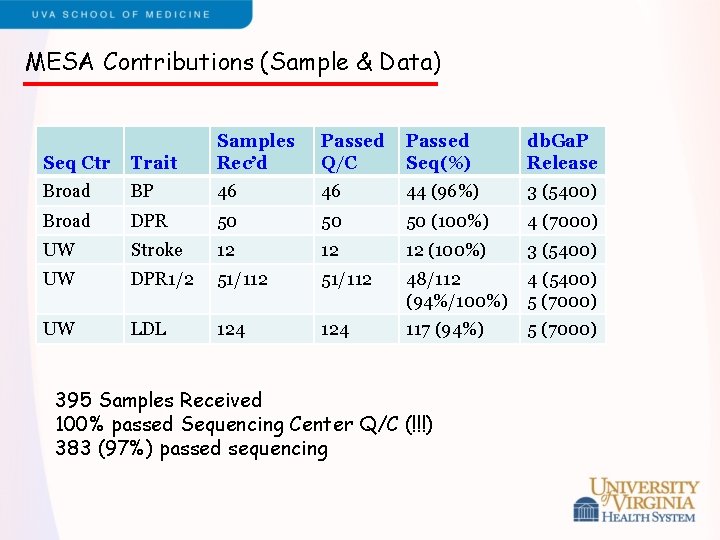
MESA Contributions (Sample & Data) Seq Ctr Trait Samples Rec’d Passed Q/C Passed Seq(%) db. Ga. P Release Broad BP 46 46 44 (96%) 3 (5400) Broad DPR 50 50 50 (100%) 4 (7000) UW Stroke 12 12 12 (100%) 3 (5400) UW DPR 1/2 51/112 48/112 (94%/100%) 4 (5400) 5 (7000) UW LDL 124 117 (94%) 5 (7000) 395 Samples Received 100% passed Sequencing Center Q/C (!!!) 383 (97%) passed sequencing

A Few Publications from ESP Tennessen JA, Bigham AW, O’Connor TD, Fu W, Kenny EE, Gravel S et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science 2012 Jul 6; 337(6090) 64 -9 Emond MJ, Louie T, Emerson J, Zhao W, Mathias RA, Knowles MR et al. Exome sequencing of extreme phenotypes identifies DCTN 4 as a modifier of chronic Pseudomonas aeruginosa infection in cystic fibrosis. Nat Genet 2012 Jul 8; 44(8): 886 -9 Auer PL, Johnsen JM, Johnson AD, Logsdon BA, Lange LA, Nalls MA et al. Imputation of exome sequence variants into population-based samples and blood-cell-trait-associated loci in African-Americans: NHLBI GO Exome Sequencing Project. Am J Hum Genet 2012 Nov 2; 91(5): 794 -808 Fu W, O’Connor TD, Jun G, Kang HM, Abecasis GR, Leal SM et al. Analysis of 6, 515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013 Jan 10; 493(7431): 216 -20 Johnsen JM, Auer PL, Morrison AC, Jiao S, Wei P, Haessler J et al. Common and rare von Willebrand factor (v. WF) coding variants, VWF levels, and factor VIII levels in African Americans: the NHLBI Exome Sequencing Project. Blood 2103 Jul 25; 122(4): 590 -7
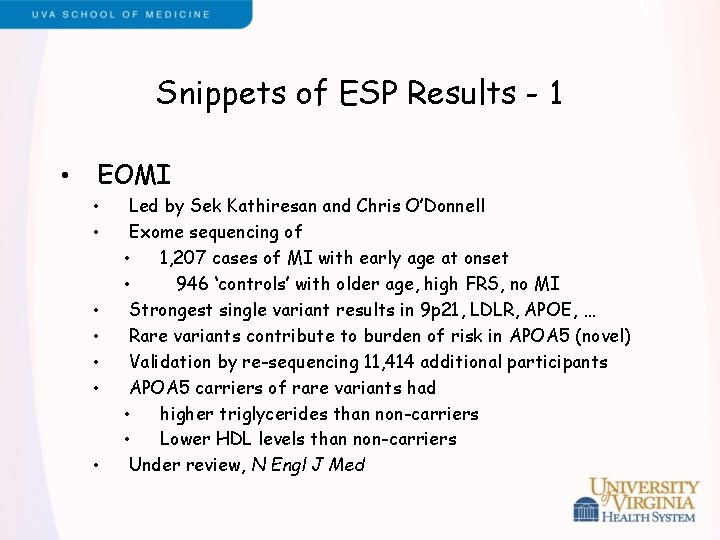
Snippets of ESP Results - 1 • EOMI • • Led by Sek Kathiresan and Chris O’Donnell Exome sequencing of • 1, 207 cases of MI with early age at onset • 946 ‘controls’ with older age, high FRS, no MI Strongest single variant results in 9 p 21, LDLR, APOE, … Rare variants contribute to burden of risk in APOA 5 (novel) Validation by re-sequencing 11, 414 additional participants APOA 5 carriers of rare variants had • higher triglycerides than non-carriers • Lower HDL levels than non-carriers Under review, N Engl J Med
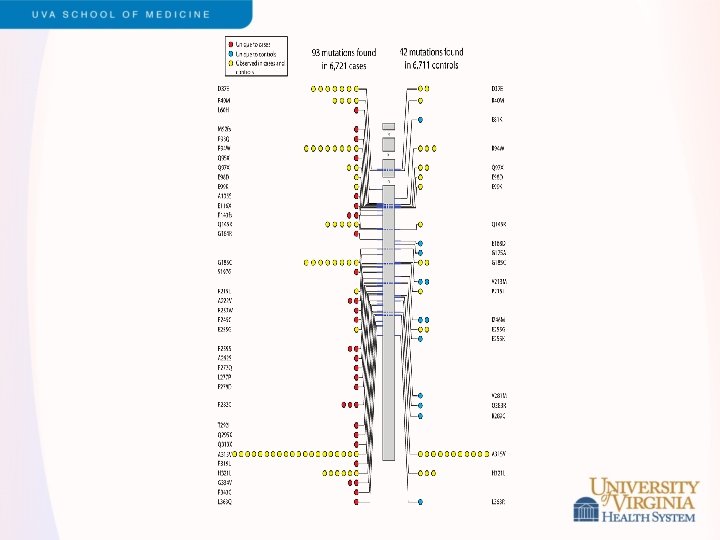
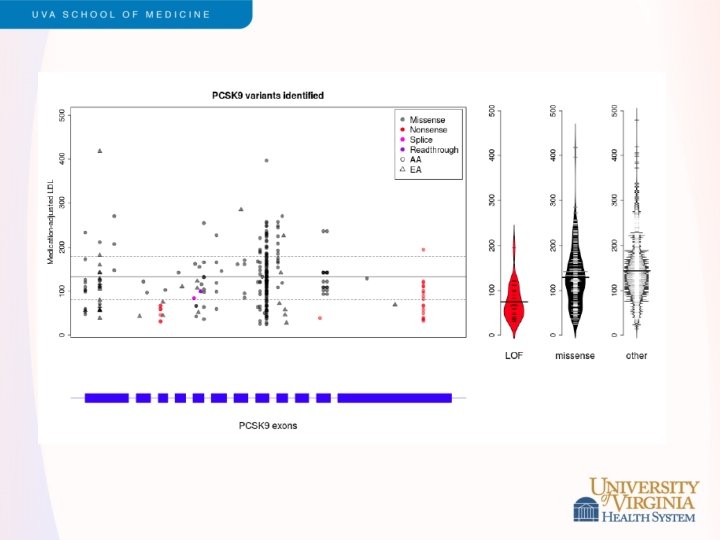
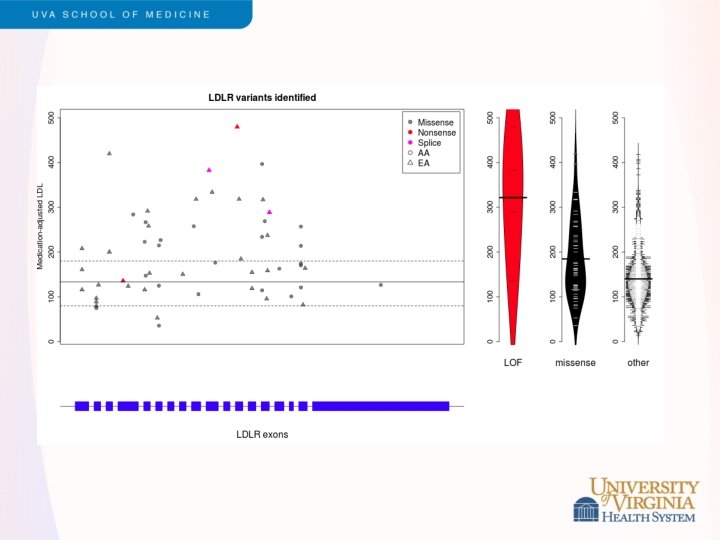
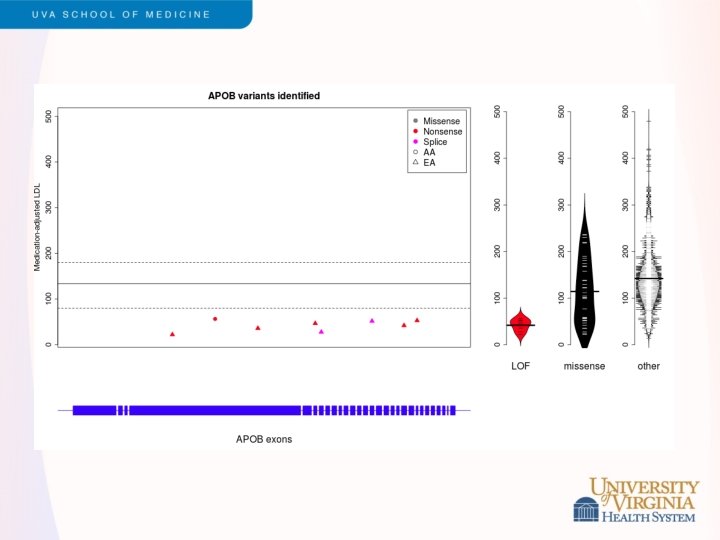
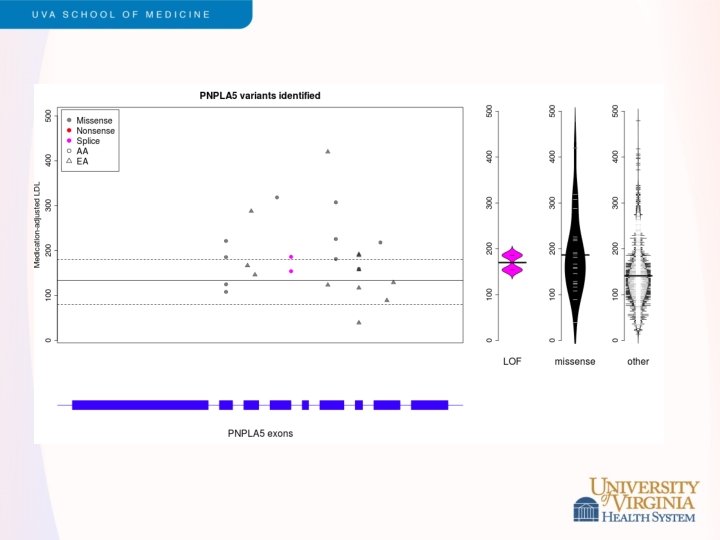
Snippets of ESP Results - 2 • LDL cholesterol levels • • Led by Cristen Willer and Leslie Lange Exome sequencing of 2, 005 participants, including 554 selected for extremes of LDL (>98 th or <2 nd percentile) Strongest single variant results in APOE Rare variants contribute to burden of risk in PCSK 9, LDLR, APOB and PNPLA 5 (novel) Validation in GOT 2 D sequencing study of 2, 084 participants Different genes exhibited different genetic architecture • PCSK 9 – loss of function variants (MAF < 5%) • LDLR – rare missense, loss of function variants (MAF<0. 1%) Under review, Am J Hum Genet (after a long tortuous path with Nat Genet)

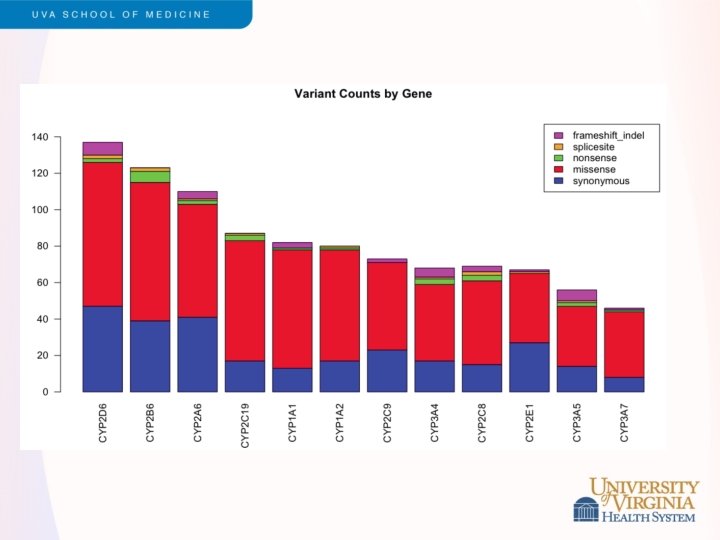
Snippets of ESP Results - 3 • Pharmacogenomics • • Led by Adam Gordon (Nickerson lab) Exome sequencing of 2, 203 African Americans and 4, 300 Caucasians Survey of coding variation in 12 CYP 450 genes Discovered 1, 006 unique variants (275 known, 731 novel); 486 missense variants and 42 nonsense/splice or frameshift CYP 2 A 6, CYP 2 B 6, CYP 2 D 6 contain most nonsynonymous variation (and are in the top 20% exome-wide) Predicted function -> drug metabolism? • 11. 7% AA and 7. 6% EA carry a predicted functional variant • 21. 8% AA and 14. 1% EA have variants in one of 12 CYP genes Under review, Hum Mol Genet
MESA ESP data in db. Ga. P

MESA ESP db. Ga. P access
The Exome. Chip Exome sequencing studies are likely well-powered to discover shared (MAF > 0. 1%) variants that contribute to diseases of interest Exome sequencing studies of (non-Mendelian) traits may be underpowered to demonstrate association to those variants Array-based genotyping is less expensive per sample than exome sequencing
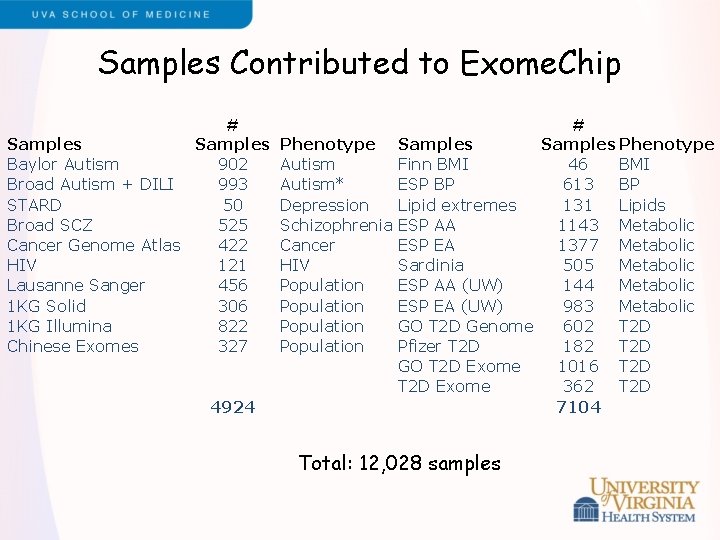
Samples Contributed to Exome. Chip # Samples Baylor Autism 902 Broad Autism + DILI 993 STARD 50 Broad SCZ 525 Cancer Genome Atlas 422 HIV 121 Lausanne Sanger 456 1 KG Solid 306 1 KG Illumina 822 Chinese Exomes 327 4924 # Phenotype Samples Phenotype Autism Finn BMI 46 BMI Autism* ESP BP 613 BP Depression Lipid extremes 131 Lipids Schizophrenia ESP AA 1143 Metabolic Cancer ESP EA 1377 Metabolic HIV Sardinia 505 Metabolic Population ESP AA (UW) 144 Metabolic Population ESP EA (UW) 983 Metabolic Population GO T 2 D Genome 602 T 2 D Population Pfizer T 2 D 182 T 2 D GO T 2 D Exome 1016 T 2 D Exome 362 T 2 D 7104 Total: 12, 028 samples
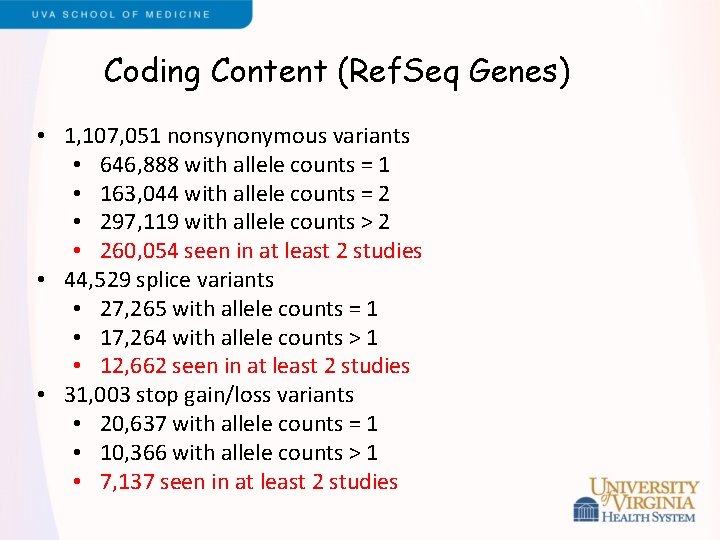
Coding Content (Ref. Seq Genes) • 1, 107, 051 nonsynonymous variants • 646, 888 with allele counts = 1 • 163, 044 with allele counts = 2 • 297, 119 with allele counts > 2 • 260, 054 seen in at least 2 studies • 44, 529 splice variants • 27, 265 with allele counts = 1 • 17, 264 with allele counts > 1 • 12, 662 seen in at least 2 studies • 31, 003 stop gain/loss variants • 20, 637 with allele counts = 1 • 10, 366 with allele counts > 1 • 7, 137 seen in at least 2 studies
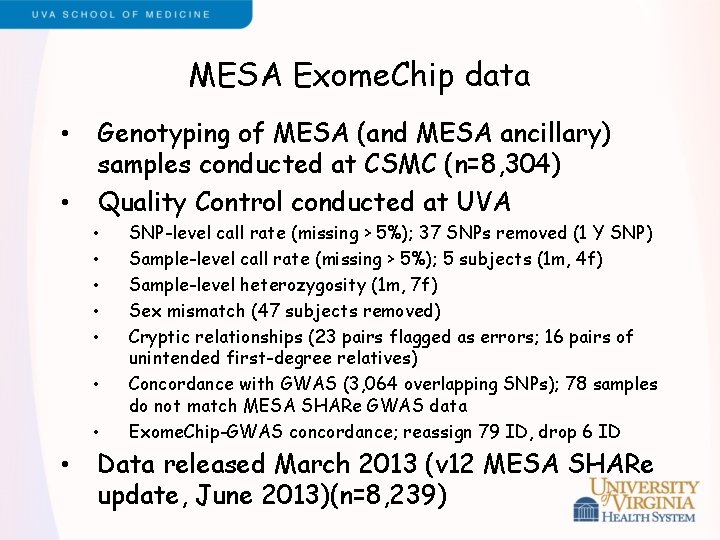
MESA Exome. Chip data • • Genotyping of MESA (and MESA ancillary) samples conducted at CSMC (n=8, 304) Quality Control conducted at UVA • • SNP-level call rate (missing > 5%); 37 SNPs removed (1 Y SNP) Sample-level call rate (missing > 5%); 5 subjects (1 m, 4 f) Sample-level heterozygosity (1 m, 7 f) Sex mismatch (47 subjects removed) Cryptic relationships (23 pairs flagged as errors; 16 pairs of unintended first-degree relatives) Concordance with GWAS (3, 064 overlapping SNPs); 78 samples do not match MESA SHARe GWAS data Exome. Chip-GWAS concordance; reassign 79 ID, drop 6 ID Data released March 2013 (v 12 MESA SHARe update, June 2013)(n=8, 239)

MESA Exome. Chip activity • • Take advantage of multi-ethnic structure and diverse phenotypes that would otherwise not be considered in large consortia Phenotypes • • Lipids (levels and subfractions) Lung (PFT and CT measures of volume, COPD, emphysema) Hematology and blood count factors Inflammatory and endothelial biomarkers Imaging of the heart (cardiac MRI, LV/RV parameters) Fatty acid and lipid metabolism Develop active collaboration with basic scientists
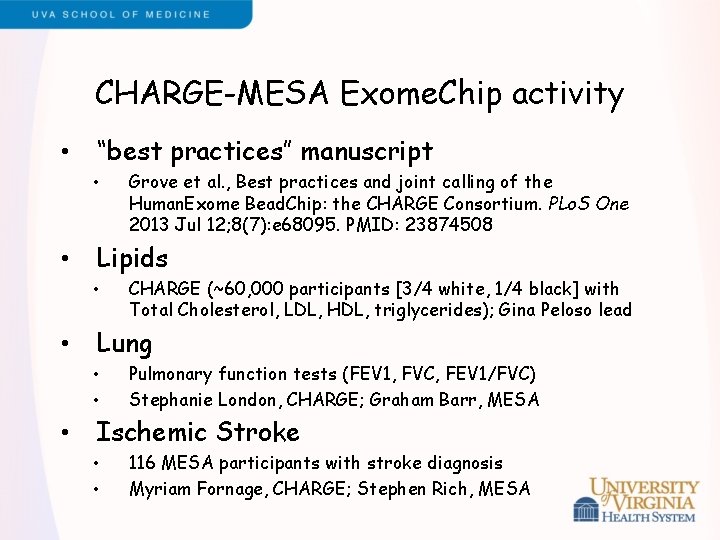
CHARGE-MESA Exome. Chip activity • “best practices” manuscript • • Lipids • • CHARGE (~60, 000 participants [3/4 white, 1/4 black] with Total Cholesterol, LDL, HDL, triglycerides); Gina Peloso lead Lung • • • Grove et al. , Best practices and joint calling of the Human. Exome Bead. Chip: the CHARGE Consortium. PLo. S One 2013 Jul 12; 8(7): e 68095. PMID: 23874508 Pulmonary function tests (FEV 1, FVC, FEV 1/FVC) Stephanie London, CHARGE; Graham Barr, MESA Ischemic Stroke • • 116 MESA participants with stroke diagnosis Myriam Fornage, CHARGE; Stephen Rich, MESA
Future Directions • Exome Sequence Project • • • Exome. Chip • • • MESA/Heart. GO ESP with CHARGE-S Translating candidate genes with burden of rare variants into mechanisms of pathology Leverage MESA ethnic-diversity and phenotypes into MESAcentric studies Continue MESA collaborations with CHARGE Initiate MESA as discovery cohort with PAGE (NHGRI) Utilizing MESA expression/methylation data with genomic data into Systems Genetics MESA serving as a foundation for functional studies with basic scientists
- Slides: 27