CH 339 K Proteins Amino Acids Primary Structure



- Slides: 55

CH 339 K Proteins: Amino Acids, Primary Structure, and Molecular Evolution

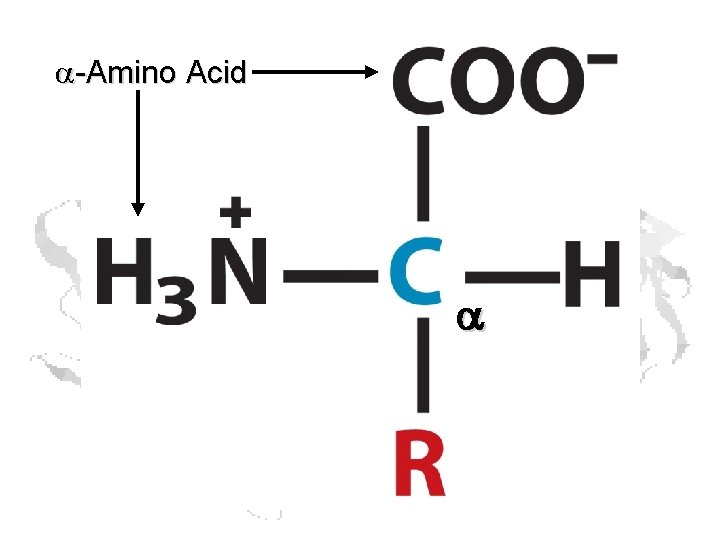
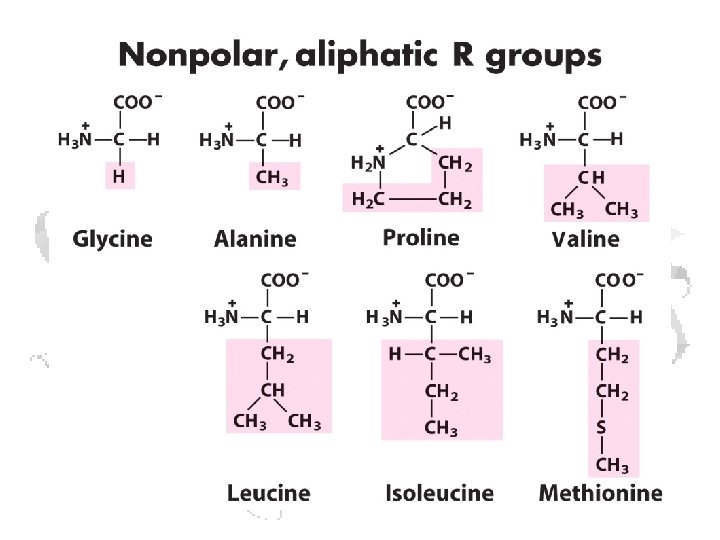
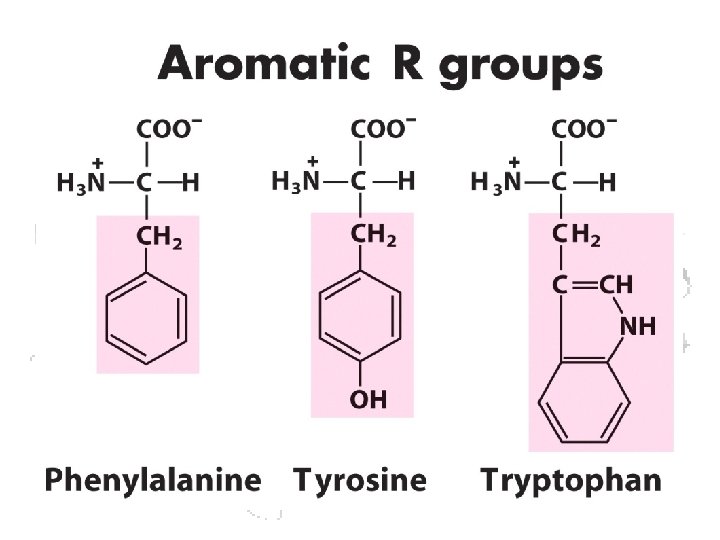
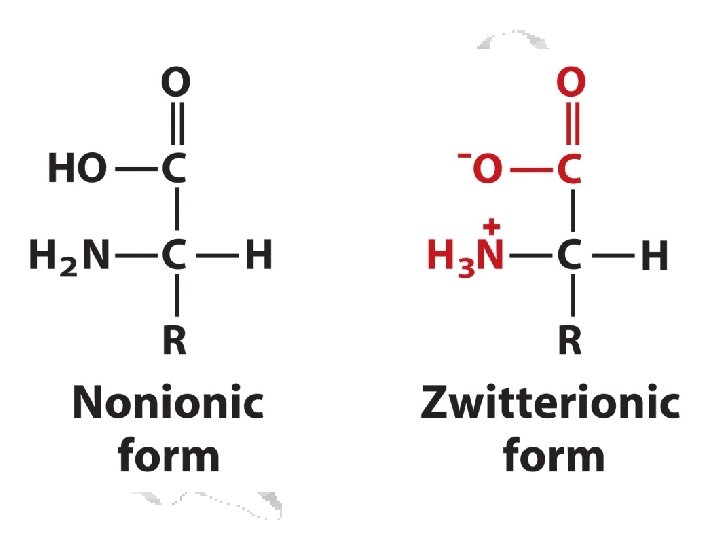
a-Amino Acid a

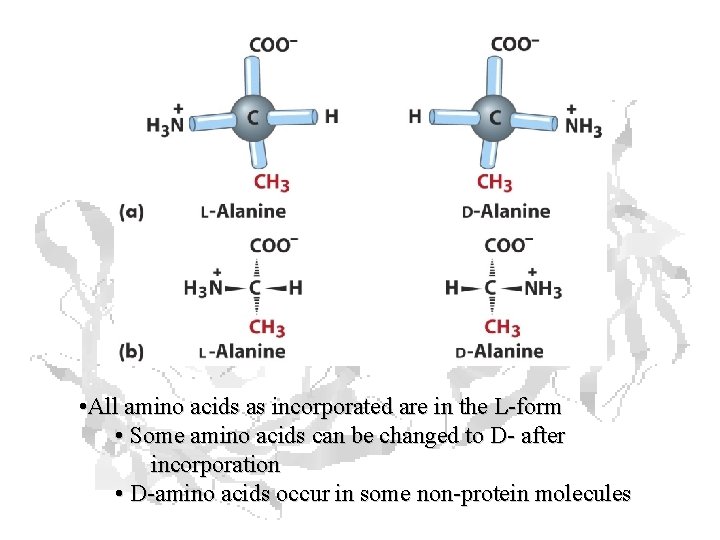
• All amino acids as incorporated are in the L-form • Some amino acids can be changed to D- after incorporation • D-amino acids occur in some non-protein molecules

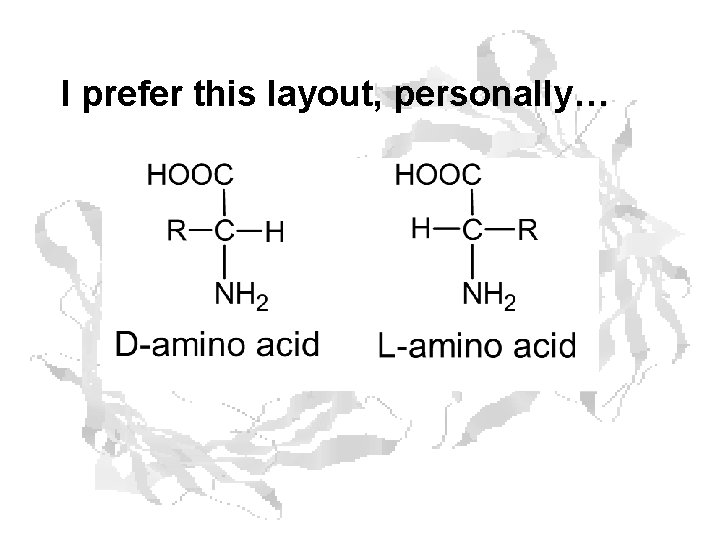
I prefer this layout, personally…




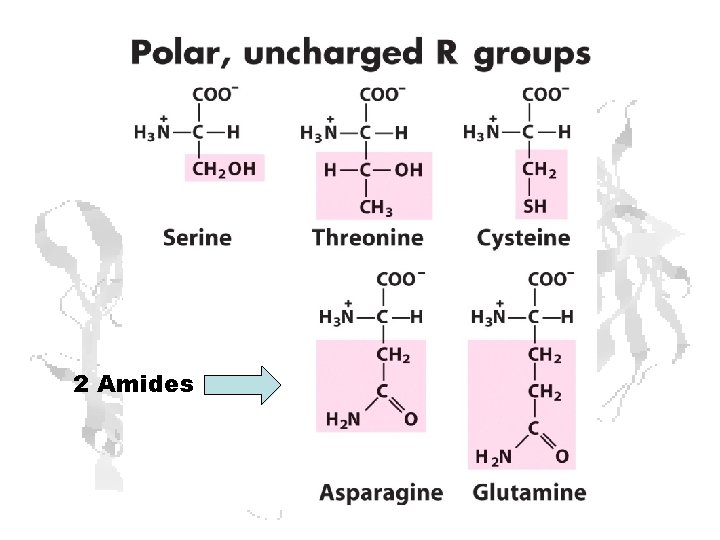
2 Amides

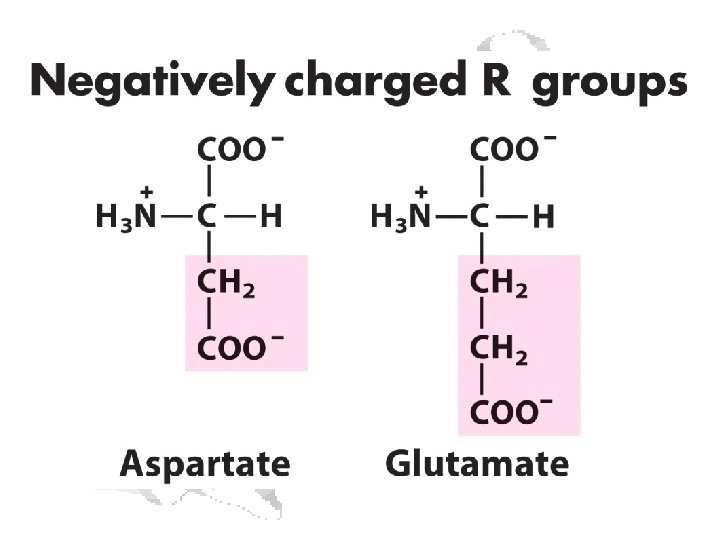
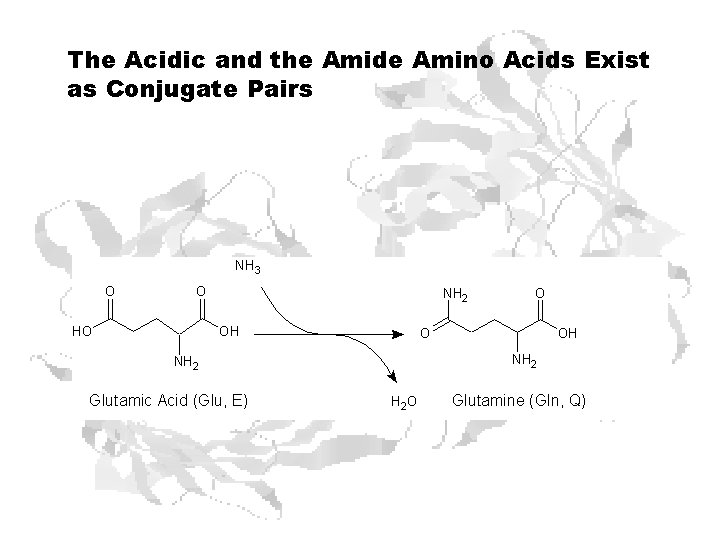
The Acidic and the Amide Amino Acids Exist as Conjugate Pairs




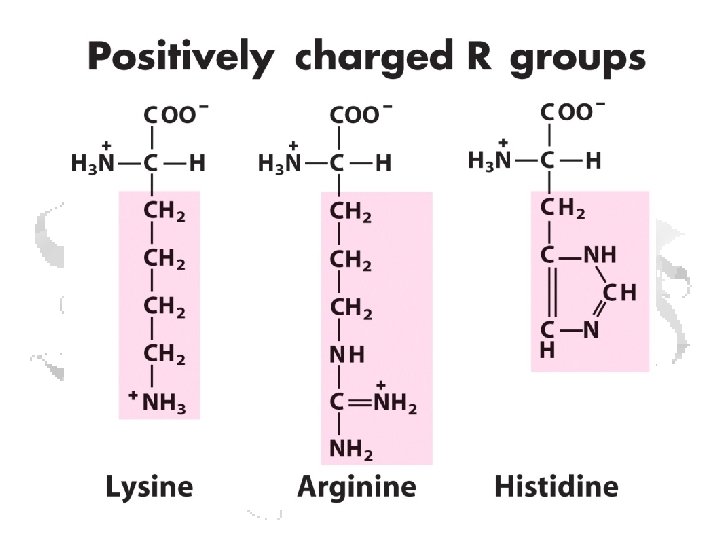
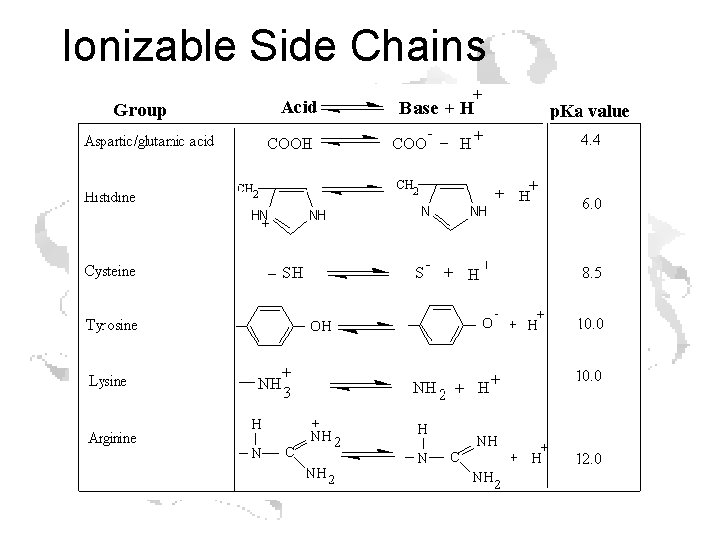
Ionizable Side Chains

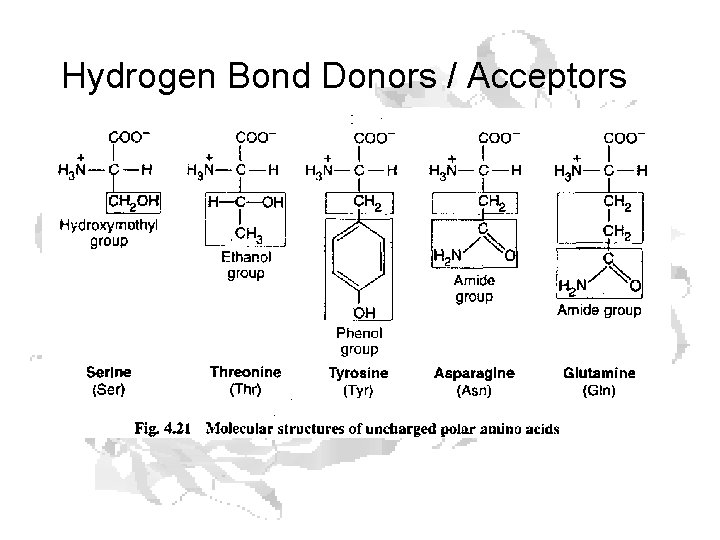
Hydrogen Bond Donors / Acceptors

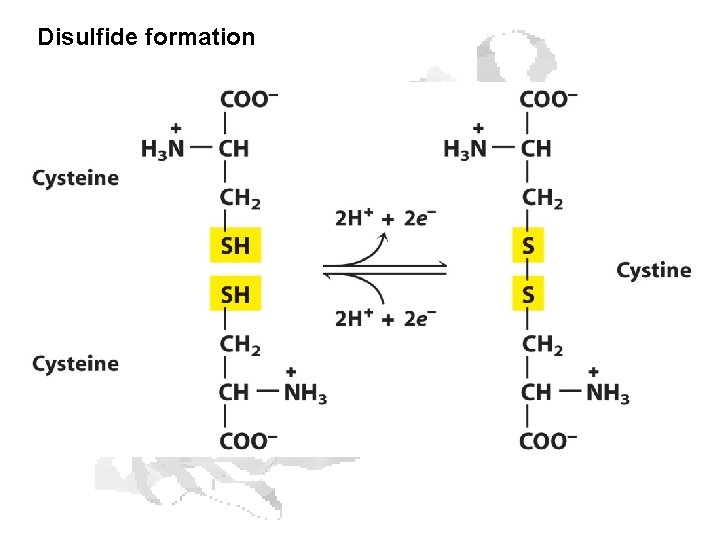
Disulfide formation

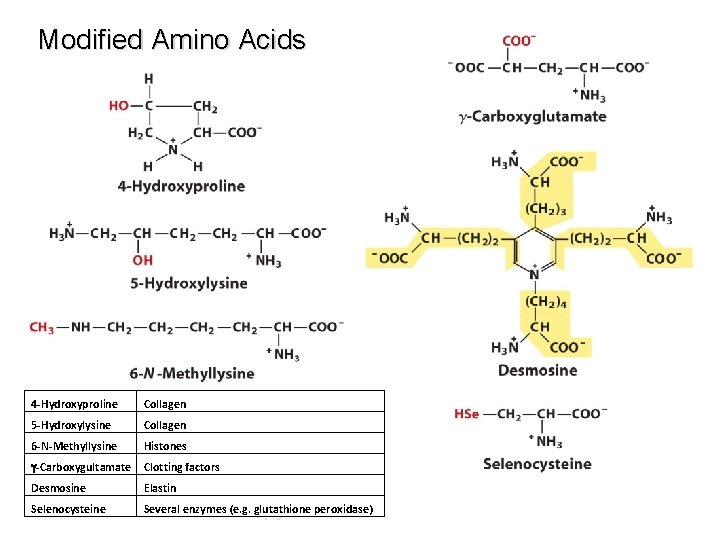
Modified Amino Acids 4 -Hydroxyproline Collagen 5 -Hydroxylysine Collagen 6 -N-Methyllysine Histones g-Carboxygultamate Clotting factors Desmosine Elastin Selenocysteine Several enzymes (e. g. glutathione peroxidase)

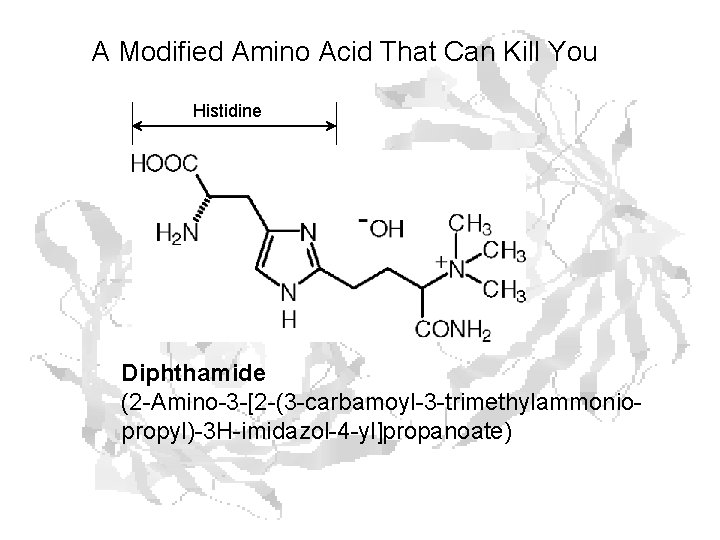
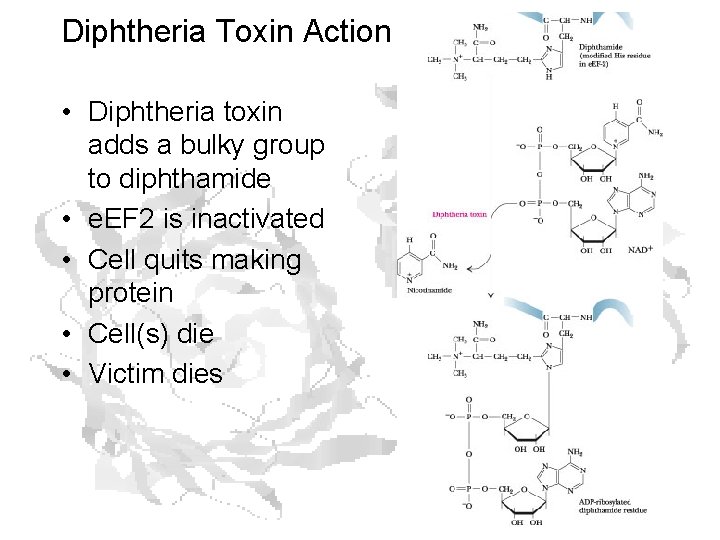
A Modified Amino Acid That Can Kill You Histidine Diphthamide (2 -Amino-3 -[2 -(3 -carbamoyl-3 -trimethylammoniopropyl)-3 H-imidazol-4 -yl]propanoate)

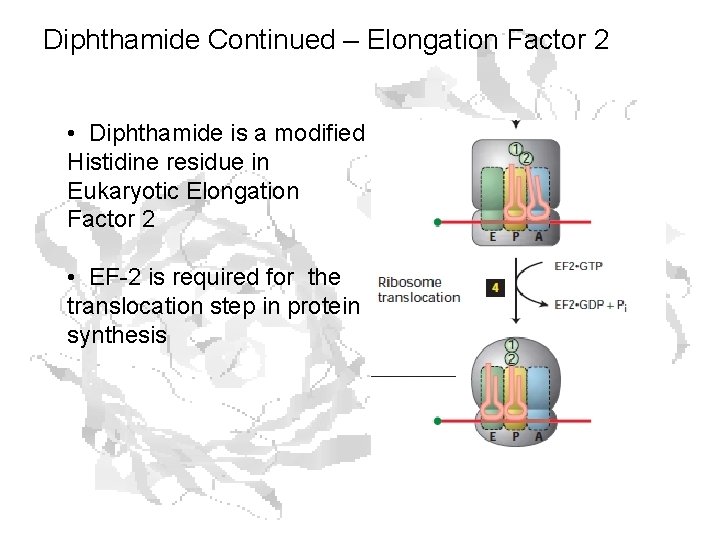
Diphthamide Continued – Elongation Factor 2 • Diphthamide is a modified Histidine residue in Eukaryotic Elongation Factor 2 • EF-2 is required for the translocation step in protein synthesis

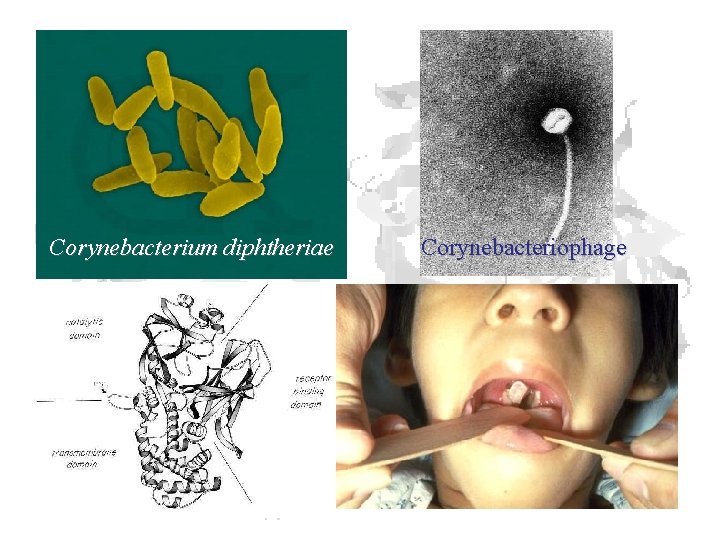
Corynebacterium diphtheriae Corynebacteriophage

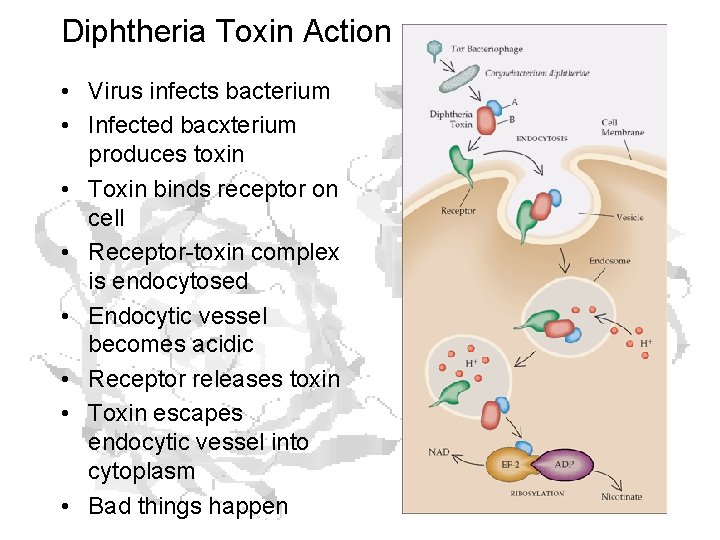
Diphtheria Toxin Action • Virus infects bacterium • Infected bacxterium produces toxin • Toxin binds receptor on cell • Receptor-toxin complex is endocytosed • Endocytic vessel becomes acidic • Receptor releases toxin • Toxin escapes endocytic vessel into cytoplasm • Bad things happen

Diphtheria Toxin Action • Diphtheria toxin adds a bulky group to diphthamide • e. EF 2 is inactivated • Cell quits making protein • Cell(s) die • Victim dies

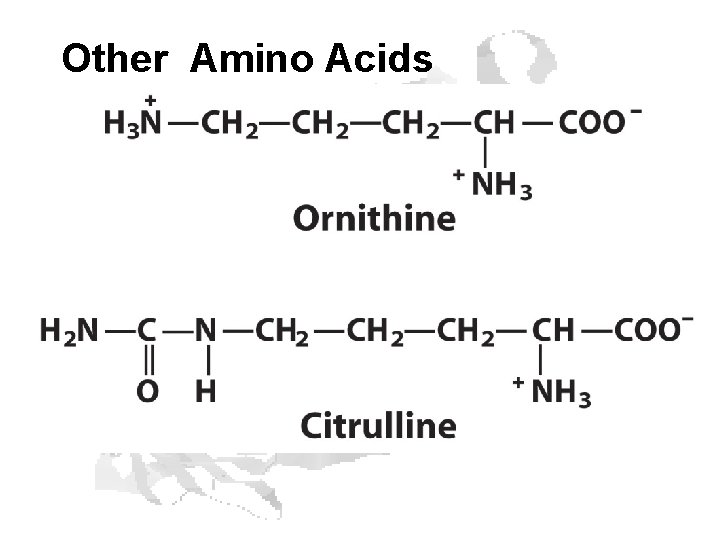
Other Amino Acids


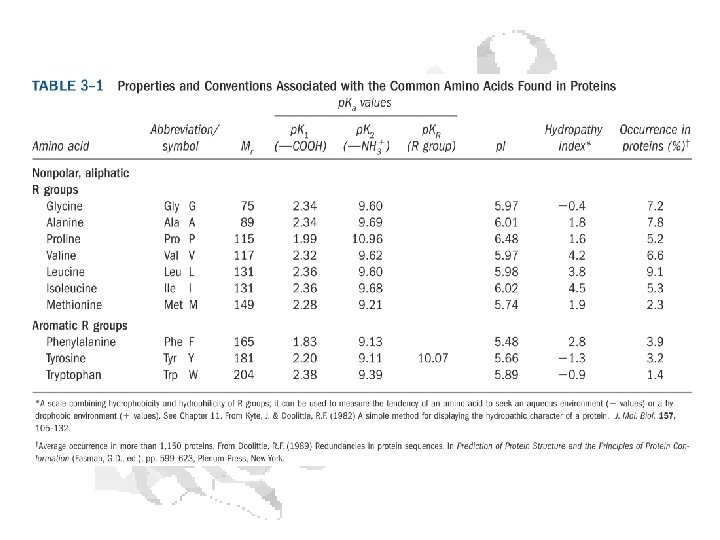
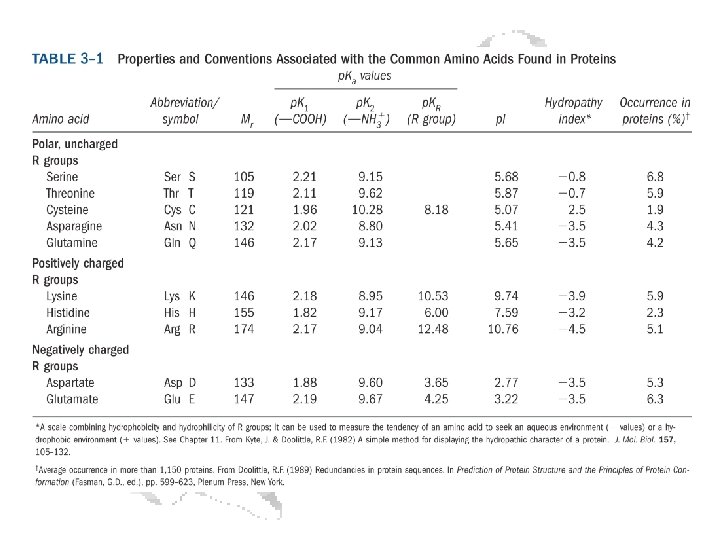
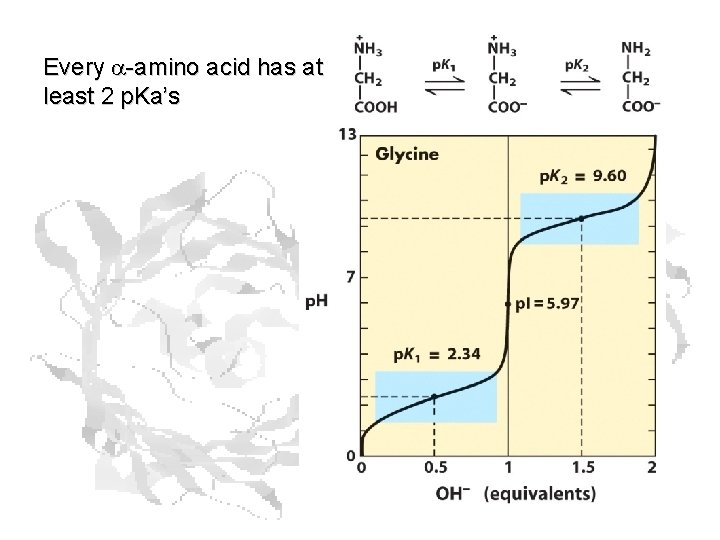
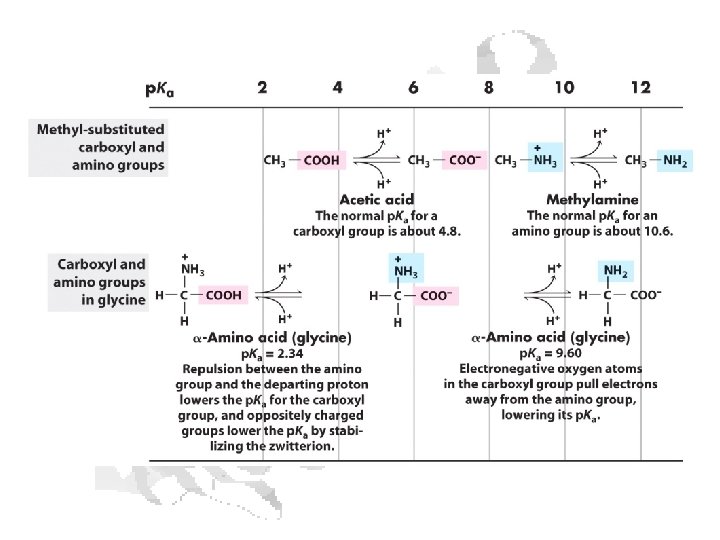
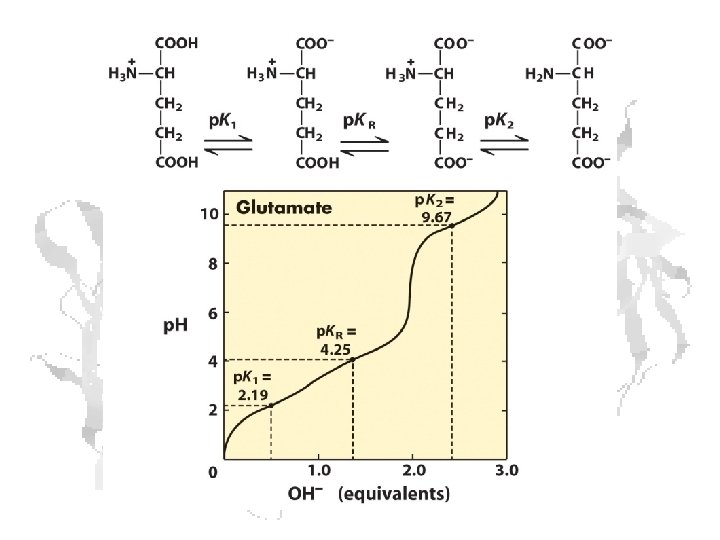
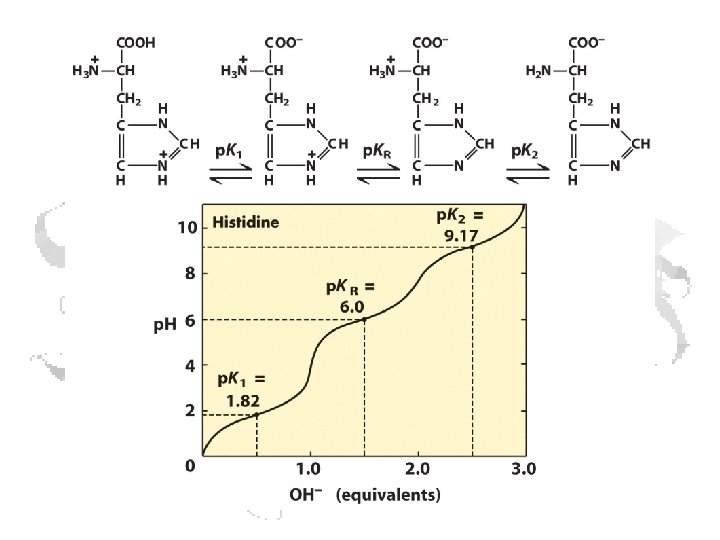
Every a-amino acid has at least 2 p. Ka’s




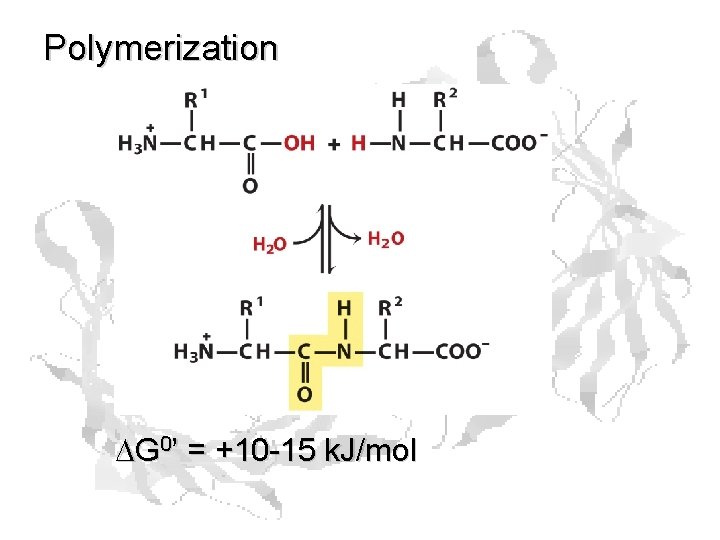
Polymerization DG 0’ = +10 -15 k. J/mol

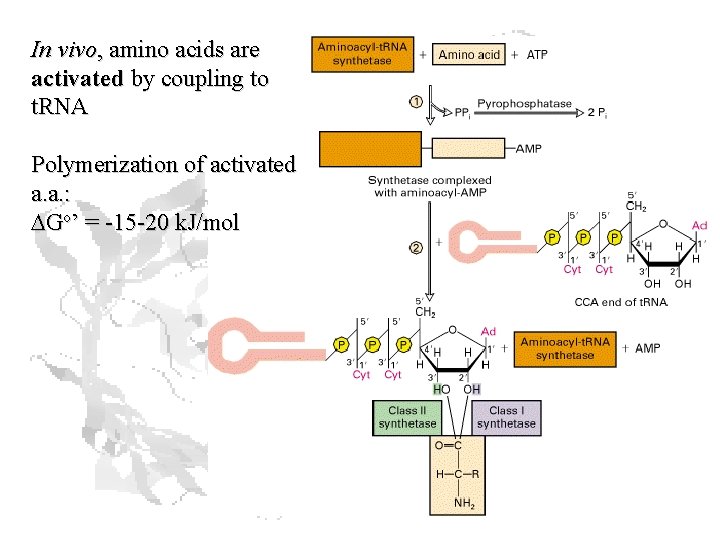
In vivo, amino acids are activated by coupling to t. RNA Polymerization of activated a. a. : DGo’ = -15 -20 k. J/mol

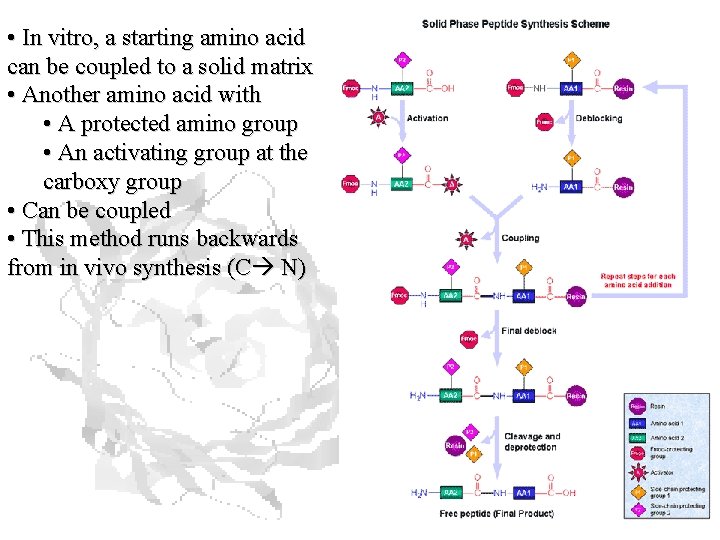
• In vitro, a starting amino acid can be coupled to a solid matrix • Another amino acid with • A protected amino group • An activating group at the carboxy group • Can be coupled • This method runs backwards from in vivo synthesis (C N)

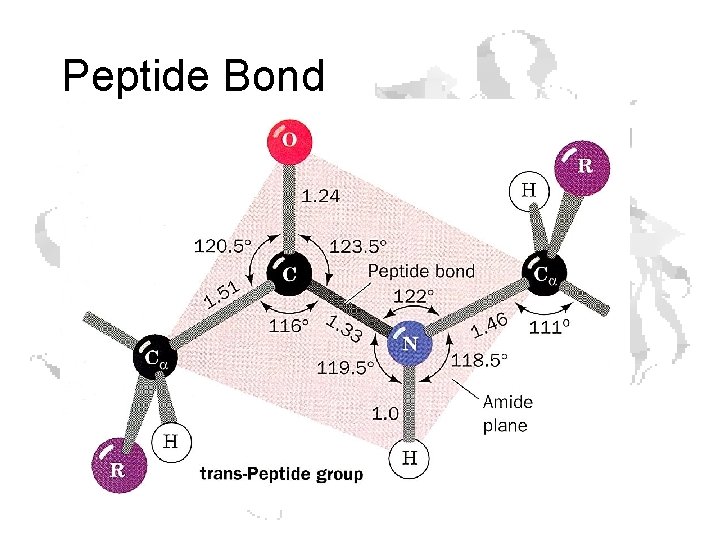
Peptide Bond

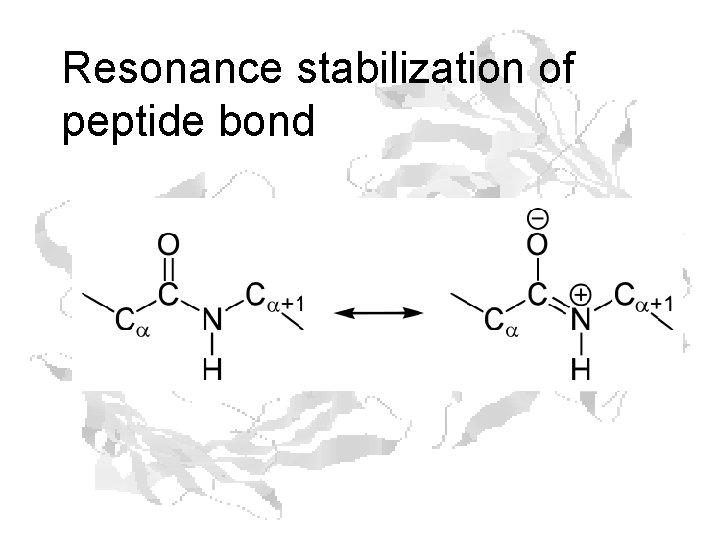
Resonance stabilization of peptide bond

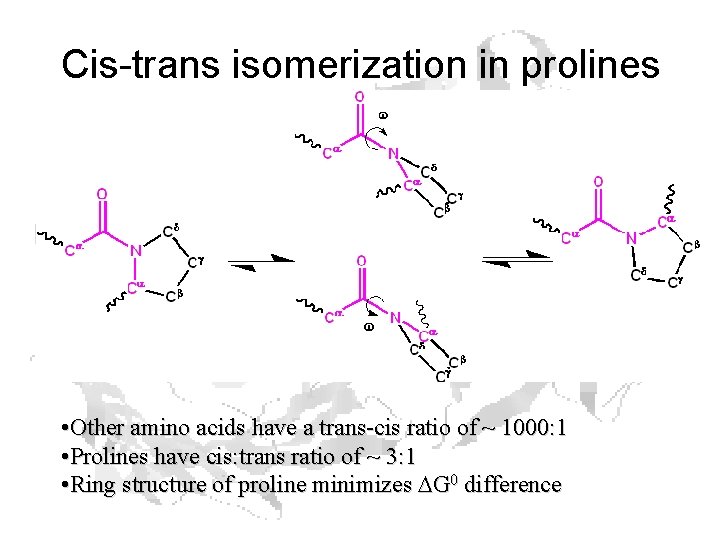
Cis-trans isomerization in prolines • Other amino acids have a trans-cis ratio of ~ 1000: 1 • Prolines have cis: trans ratio of ~ 3: 1 • Ring structure of proline minimizes DG 0 difference







MOLECULAR EVOLUTION


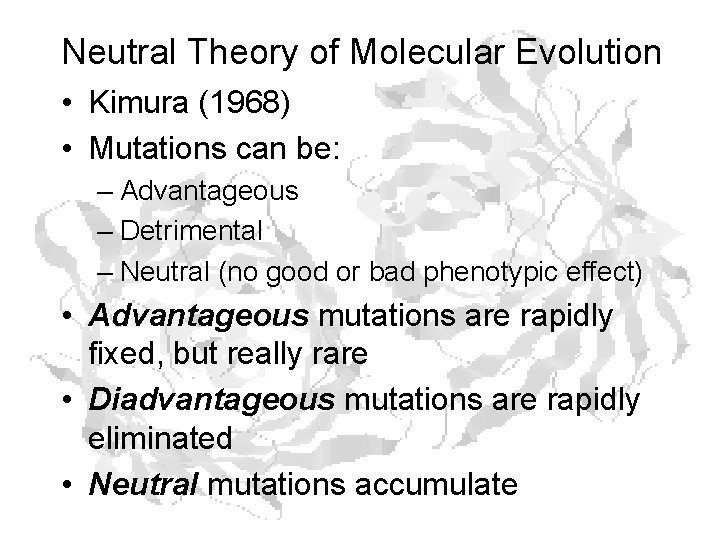
Neutral Theory of Molecular Evolution • Kimura (1968) • Mutations can be: – Advantageous – Detrimental – Neutral (no good or bad phenotypic effect) • Advantageous mutations are rapidly fixed, but really rare • Diadvantageous mutations are rapidly eliminated • Neutral mutations accumulate

What Happens to a Neutral Mutation? • Frequency subject to random chance • Will carrier of gene reproduce? • Many born but few survive – Partly selection – Mostly dumb luck • Gene can have two fates – Elimination (frequent – Fixation (rare)

Genetic Drift in Action Our green genes are evolutionarily superior! Ow! Never mind…

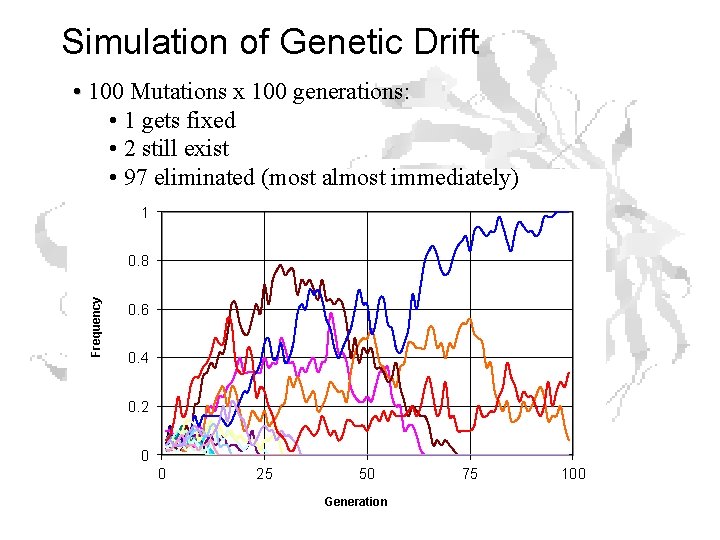
Simulation of Genetic Drift • 100 Mutations x 100 generations: • 1 gets fixed • 2 still exist • 97 eliminated (most almost immediately) 1 Frequency 0. 8 0. 6 0. 4 0. 2 0 0 25 50 Generation 75 100

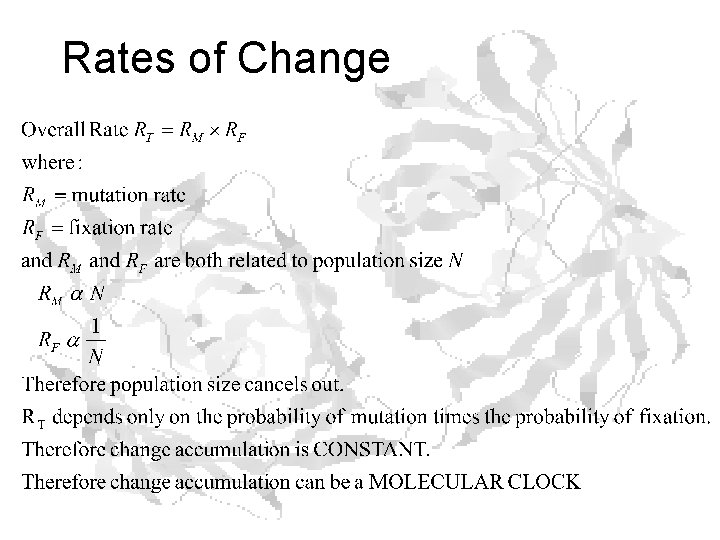
Rates of Change

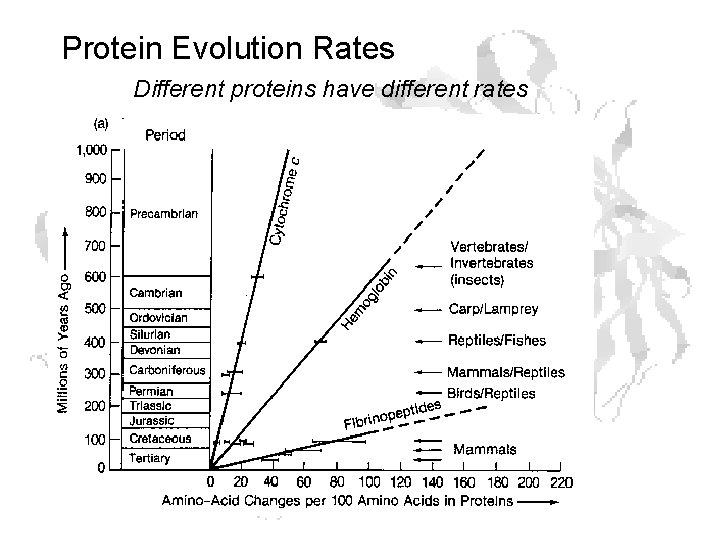
Protein Evolution Rates Different proteins have different rates

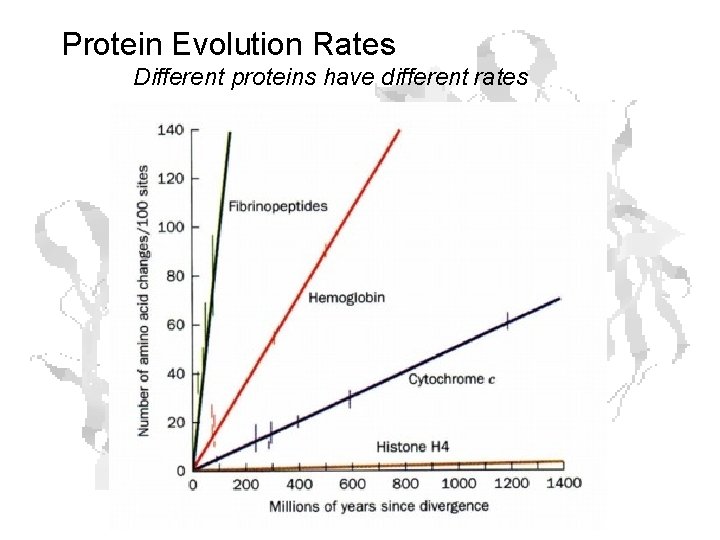
Protein Evolution Rates Different proteins have different rates

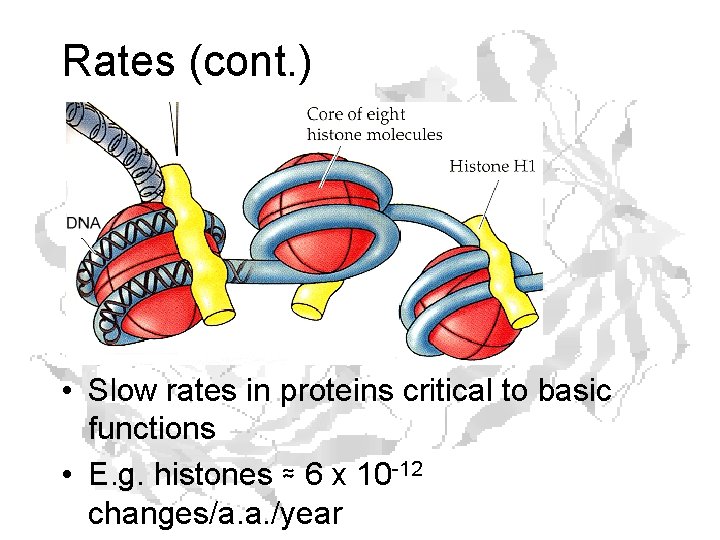
Rates (cont. ) • Slow rates in proteins critical to basic functions • E. g. histones ≈ 6 x 10 -12 changes/a. a. /year

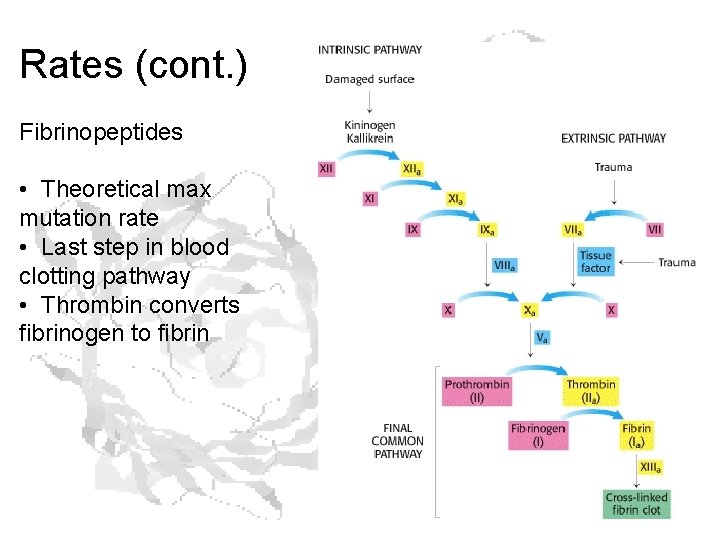
Rates (cont. ) Fibrinopeptides • Theoretical max mutation rate • Last step in blood clotting pathway • Thrombin converts fibrinogen to fibrin

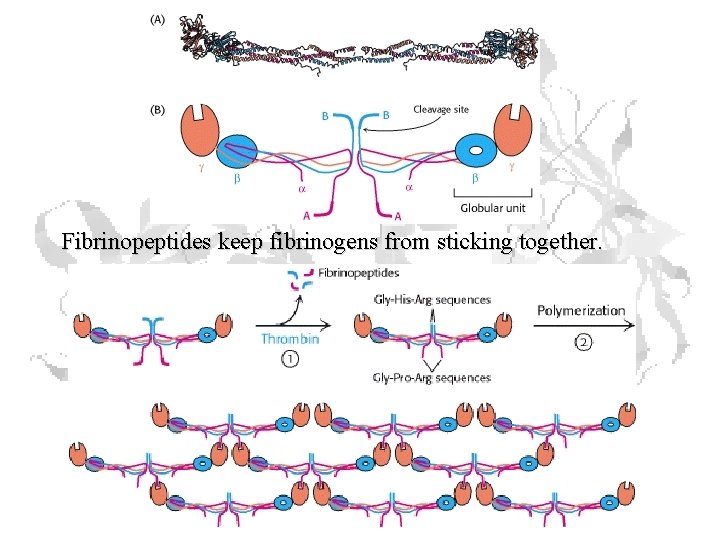
Fibrinopeptides keep fibrinogens from sticking together.

Rates (cont. ) • Only constraint on sequence is that it has to physically be there • Fibrinopeptide limit ≈ 9 x 10 -9 changes/a. a. /year

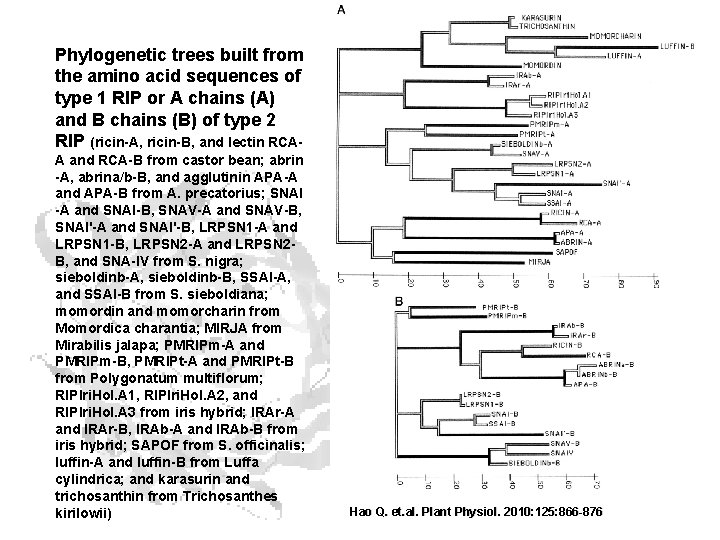
Amino acid sequences of several ribosome-inhibiting proteins

Phylogenetic trees built from the amino acid sequences of type 1 RIP or A chains (A) and B chains (B) of type 2 RIP (ricin-A, ricin-B, and lectin RCAA and RCA-B from castor bean; abrin -A, abrina/b-B, and agglutinin APA-A and APA-B from A. precatorius; SNAI -A and SNAI-B, SNAV-A and SNAV-B, SNAI'-A and SNAI'-B, LRPSN 1 -A and LRPSN 1 -B, LRPSN 2 -A and LRPSN 2 B, and SNA-IV from S. nigra; sieboldinb-A, sieboldinb-B, SSAI-A, and SSAI-B from S. sieboldiana; momordin and momorcharin from Momordica charantia; MIRJA from Mirabilis jalapa; PMRIPm-A and PMRIPm-B, PMRIPt-A and PMRIPt-B from Polygonatum multiflorum; RIPIri. Hol. A 1, RIPIri. Hol. A 2, and RIPIri. Hol. A 3 from iris hybrid; IRAr-A and IRAr-B, IRAb-A and IRAb-B from iris hybrid; SAPOF from S. officinalis; luffin-A and luffin-B from Luffa cylindrica; and karasurin and trichosanthin from Trichosanthes kirilowii) Hao Q. et. al. Plant Physiol. 2010: 125: 866 -876

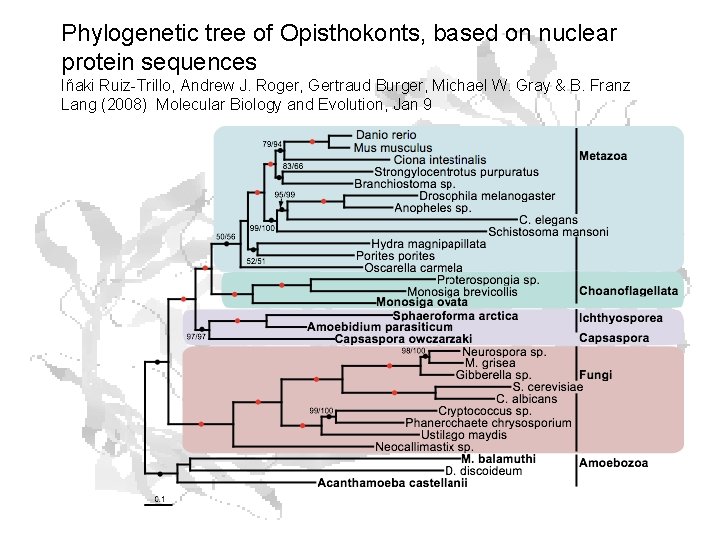
Phylogenetic tree of Opisthokonts, based on nuclear protein sequences Iñaki Ruiz-Trillo, Andrew J. Roger, Gertraud Burger, Michael W. Gray & B. Franz Lang (2008) Molecular Biology and Evolution, Jan 9