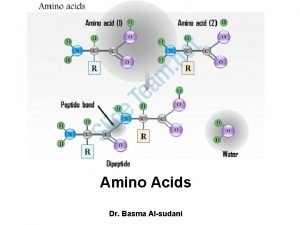
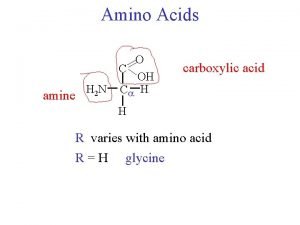
CLASSIFICATION OF AMINO ACIDS R group at neutral




























































- Slides: 92




CLASSIFICATION OF AMINO ACIDS * R group at neutral p. H * Based on nutritional/physiological roles

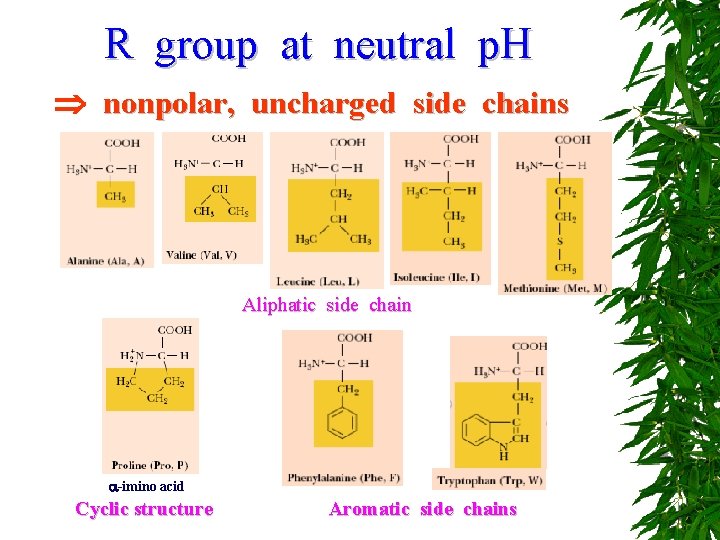
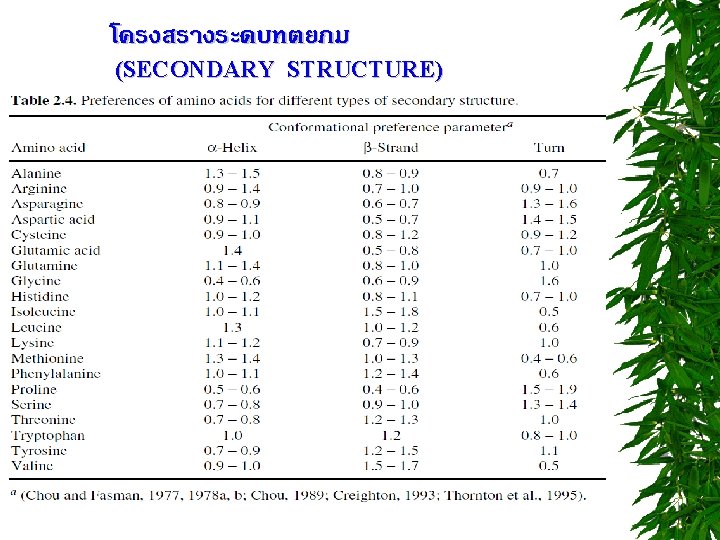
R group at neutral p. H nonpolar, uncharged side chains Aliphatic side chain -imino acid Cyclic structure Aromatic side chains

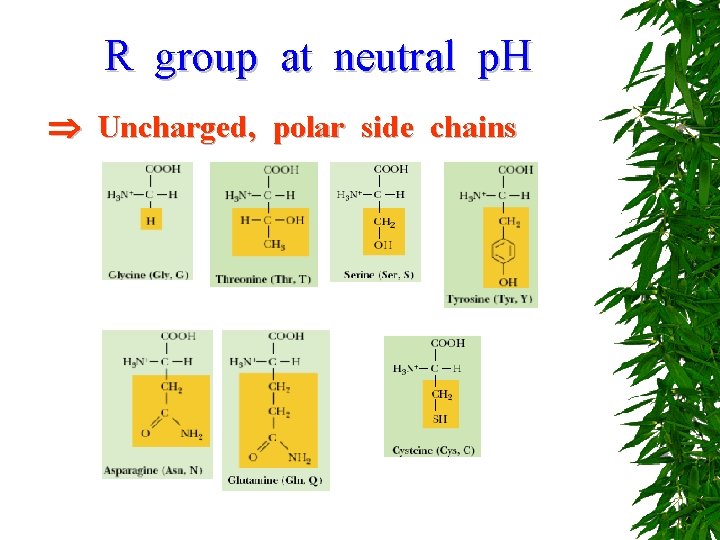
R group at neutral p. H Uncharged, polar side chains

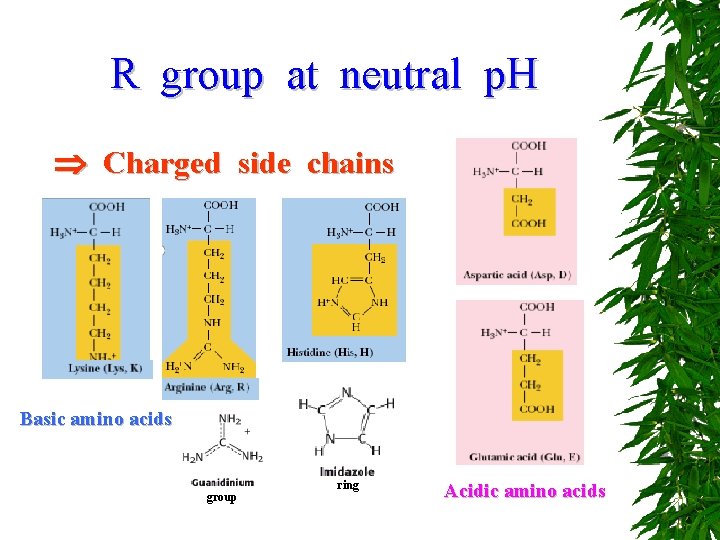
R group at neutral p. H Charged side chains Basic amino acids group ring Acidic amino acids




Based on nutritional/physiological roles Essential amino acids valine, leucine, isoleucine, phenylalanine, tryptophan, methionine, threonine, histidine (infant), lysine, arginine (semi-essential) Non-essential amino acids glycine, alanine, proline, serine, cysteine, tyrosine, asparagine, glutamine, aspartic acid, glutamic acid


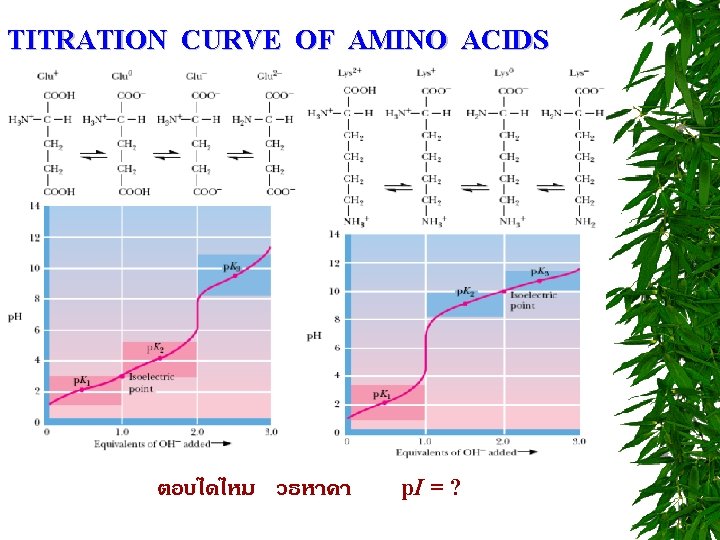
TITRATION CURVE OF AMINO ACIDS ตอบไดไหม วธหาคา p. I = ?


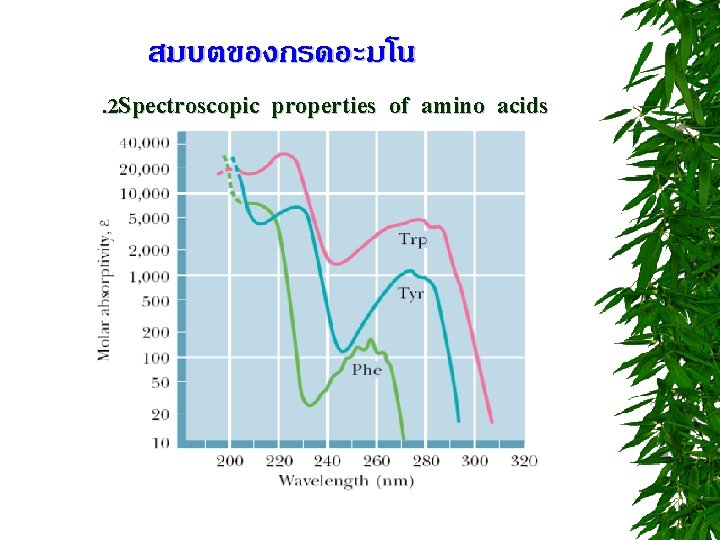
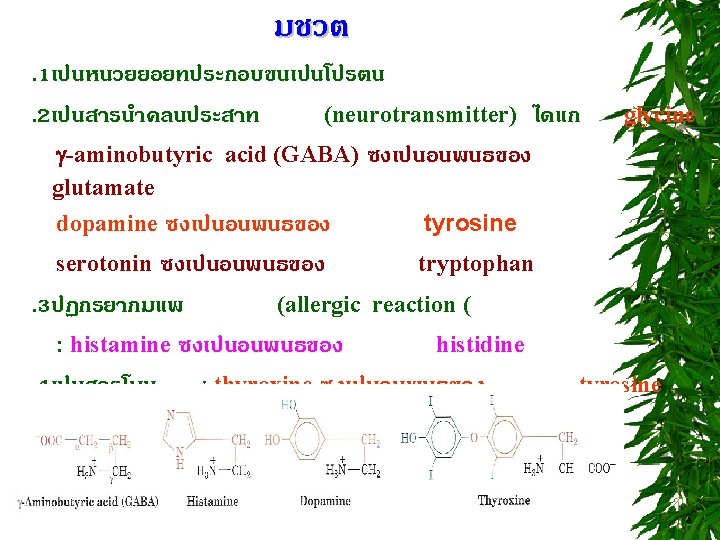
สมบตของกรดอะมโน. 2 Spectroscopic properties of amino acids





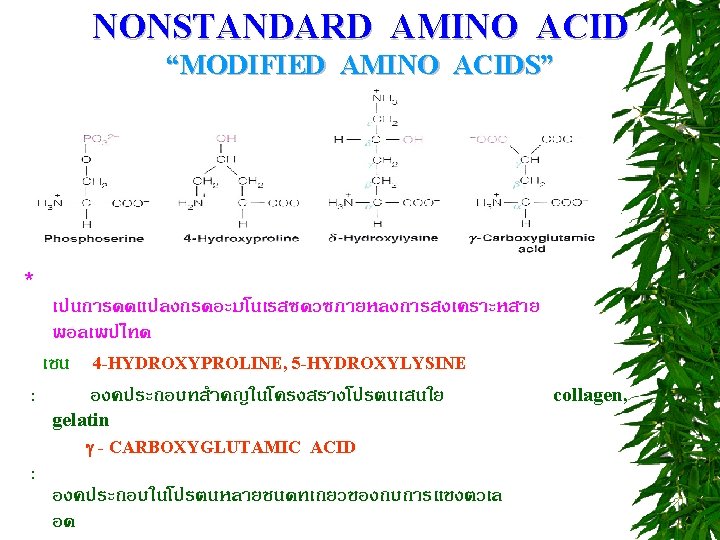
NONSTANDARD AMINO ACID “MODIFIED AMINO ACIDS” กรดอะมโนเรสซดวซบางชนดในโปรตน เชน histone อาจถกเตมหม methyl, หม acetyl, หม phosphate ตวอยาง o–phoserine, N–acetylserine, 3 -methylhistidine







PROTEINS






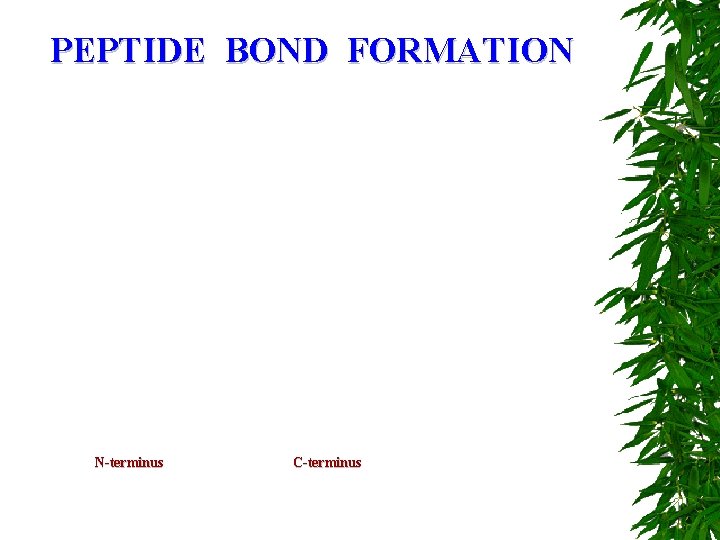
PEPTIDE BOND FORMATION N-terminus C-terminus









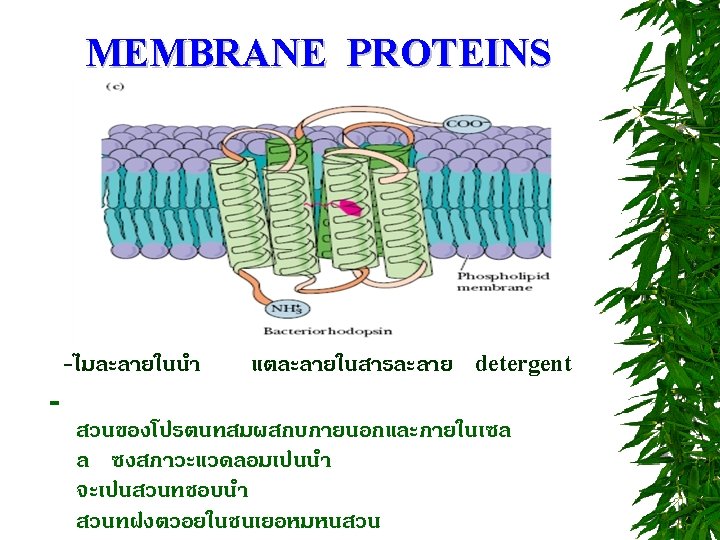
CLASSIFICATION OF PROTEINS BASED ON SHAPE FIBROUS PROTEINS (SCLEROPROTEINS) GLOBULAR PROTEINS (SPHEROPROTEINS) MEMBRANE PROTEINS




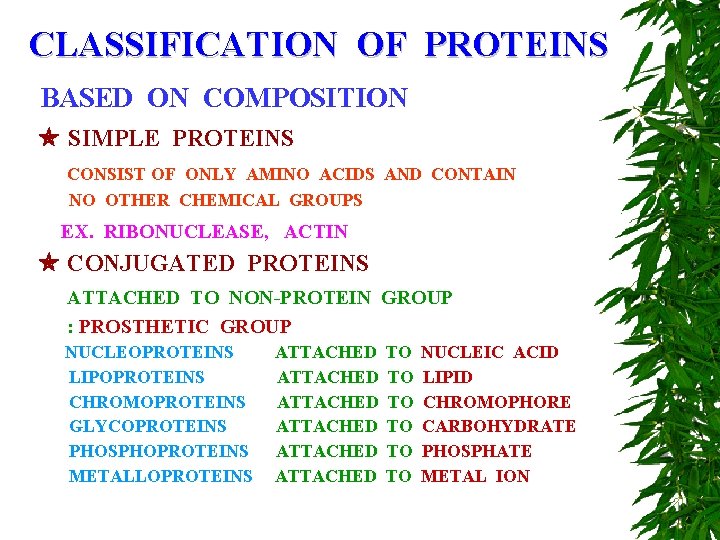
CLASSIFICATION OF PROTEINS BASED ON COMPOSITION SIMPLE PROTEINS CONSIST OF ONLY AMINO ACIDS AND CONTAIN NO OTHER CHEMICAL GROUPS EX. RIBONUCLEASE, ACTIN CONJUGATED PROTEINS ATTACHED TO NON-PROTEIN GROUP : PROSTHETIC GROUP NUCLEOPROTEINS LIPOPROTEINS CHROMOPROTEINS GLYCOPROTEINS PHOSPHOPROTEINS METALLOPROTEINS ATTACHED ATTACHED TO TO TO NUCLEIC ACID LIPID CHROMOPHORE CARBOHYDRATE PHOSPHATE METAL ION



BIOLOGICAL ROLES OF PROTEINS 1. ENERGY SOURCE 2. STRUCTURE EX : COLLAGEN, FIBOIN, ELASTIN 3. MOVEMENT EX : ACTIN, TUBULIN, CYTOSKELETON PROTEINS 4. PROTECTIVE EX : KERATIN, FIBRINOGEN AND THROMBIN, IMMUNOGLOBULIN 5. REGULATION HORMONE, TRANSCRIPTIONAL FACTOR 6. TRANSPORT EX : GLUCOSE TRANSPORTER, Na+-K+ ATPase 7. CATALYSIS ENZYME










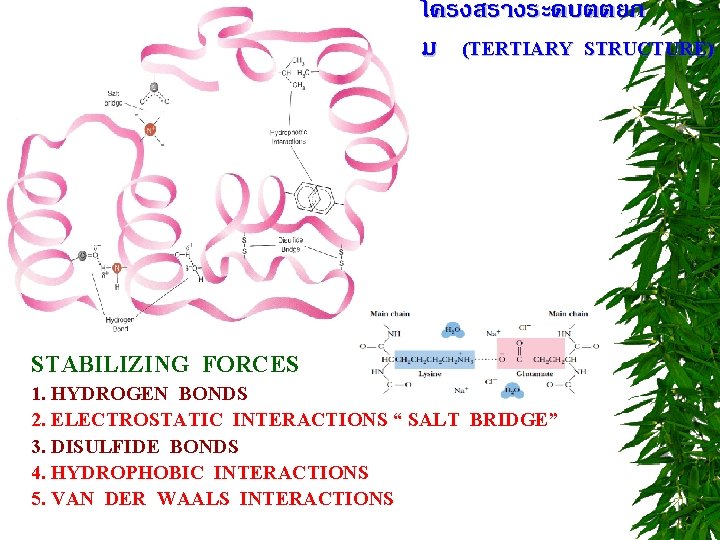
โครงสรางระดบตตยภ ม (TERTIARY STRUCTURE) STABILIZING FORCES 1. HYDROGEN BONDS 2. ELECTROSTATIC INTERACTIONS “ SALT BRIDGE” 3. DISULFIDE BONDS 4. HYDROPHOBIC INTERACTIONS 5. VAN DER WAALS INTERACTIONS



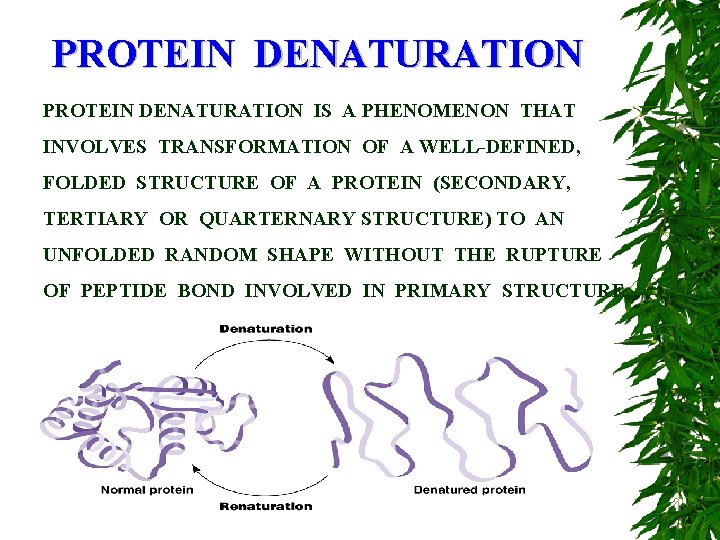
PROTEIN DENATURATION IS A PHENOMENON THAT INVOLVES TRANSFORMATION OF A WELL-DEFINED, FOLDED STRUCTURE OF A PROTEIN (SECONDARY, TERTIARY OR QUARTERNARY STRUCTURE) TO AN UNFOLDED RANDOM SHAPE WITHOUT THE RUPTURE OF PEPTIDE BOND INVOLVED IN PRIMARY STRUCTURE.

PROTEIN DENATURATION EFFECTS : - DECREASED SOLUBILITY : UNMASKING OF HYDROPHOBIC PORTION - LOST BIOLOGICAL ACTIVITY : CATALYTIC PROPERTY : IMMUNOLOGICAL PROPERTY - INCREASE SUSCEPTIBILITY TO BREAKDOWN BY PROTEASE - INABILITY TO CRYSTALIZE

DENATURATING CONDITIONS 1. 2. 3. 4. 5. 6. 7. 8. STRONG ACID AND BASE ORGANIC SOLVENT DETERGENT REDUCING AGENTS SALT CONCENTRATION HEAVY METAL IONS TEMPERATURE CHANGE MECHANICAL STRESS

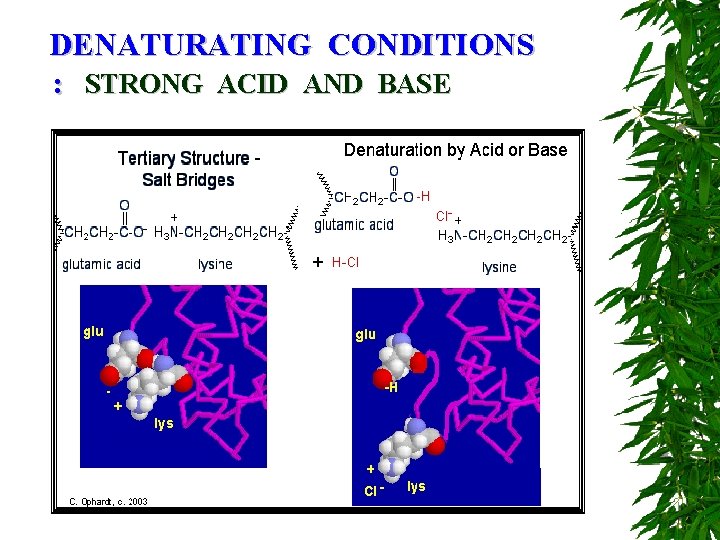
DENATURATING CONDITIONS : STRONG ACID AND BASE

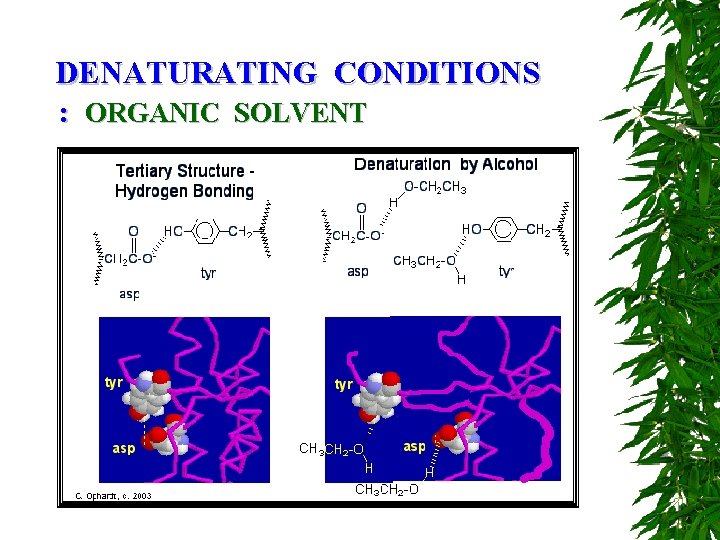
DENATURATING CONDITIONS : ORGANIC SOLVENT

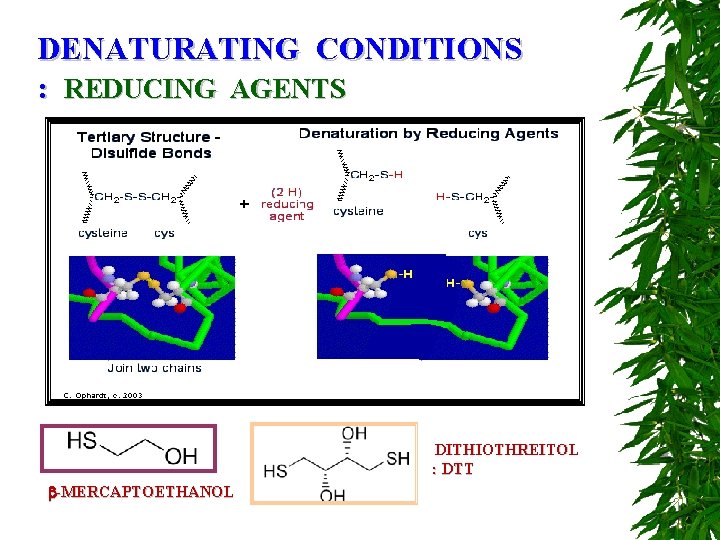
DENATURATING CONDITIONS : REDUCING AGENTS DITHIOTHREITOL : DTT -MERCAPTOETHANOL

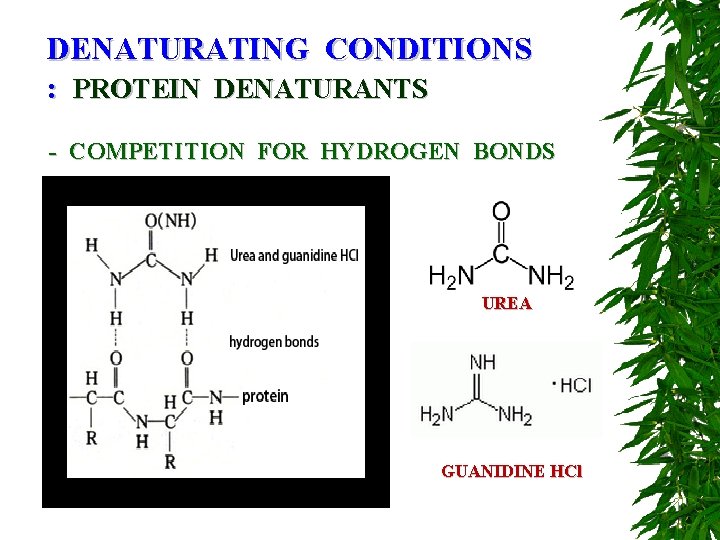
DENATURATING CONDITIONS : PROTEIN DENATURANTS - COMPETITION FOR HYDROGEN BONDS UREA GUANIDINE HCl

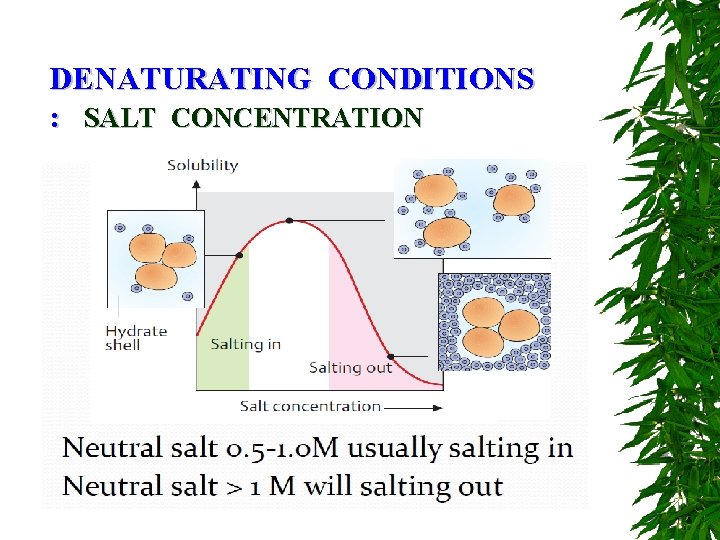
DENATURATING CONDITIONS : SALT CONCENTRATION

DENATURATING CONDITIONS : TEMPERATURE CHANGE HEAT : DISRUPTS HYDROGEN BONDS AND WEAK INTERACTION IN PROTEIN STRUCTURE DUE TO INCREASING OF TRANSLATIONAL AND VIBRATIONAL ENERGY COLD : FREEZING TEMPERATURE CAN DENATURE SOME PROTEINS

DENATURATING CONDITIONS : MECHANICAL STRESS AGITATION : SHEARING OF HYDROGEN BONDS

DENATURATING CONDITIONS : IRRADIATION - THE EFFECT DEPENDS ON THE WAVELENGHT AND ENERGY INVOLVED - RADIATION CAUSE : OXIDATION OF AMINO ACID RESIDUES : RUPTURE OF COVALENT BOND : IONIZATION : FORMATION OF PROTEIN FREE RADICAL




TECHNIQUES IN PROTEIN STUDY

PROTEIN PURIFICATION : PROTEIN EXTRACTION

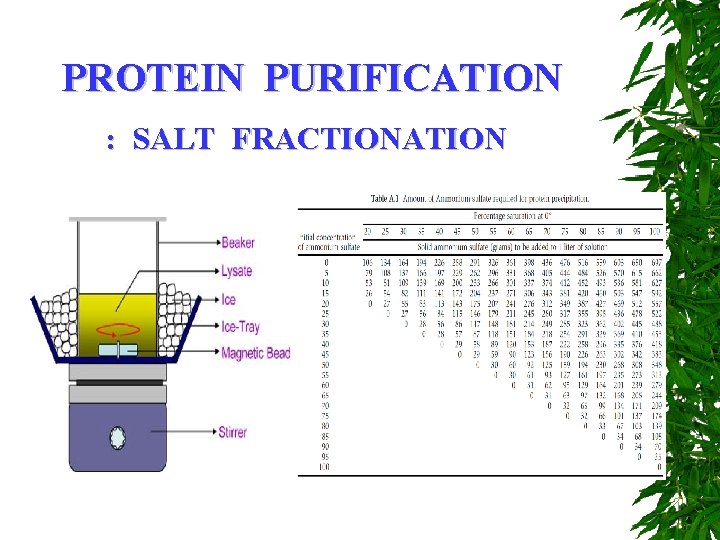
PROTEIN PURIFICATION : SALT FRACTIONATION

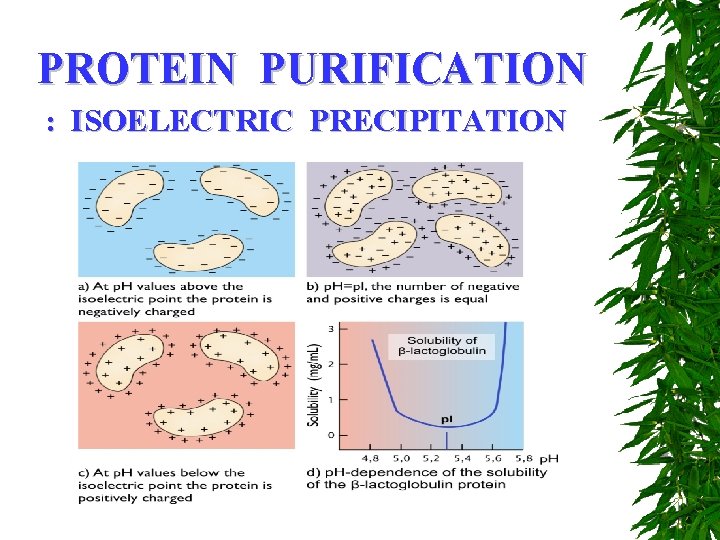
PROTEIN PURIFICATION : ISOELECTRIC PRECIPITATION

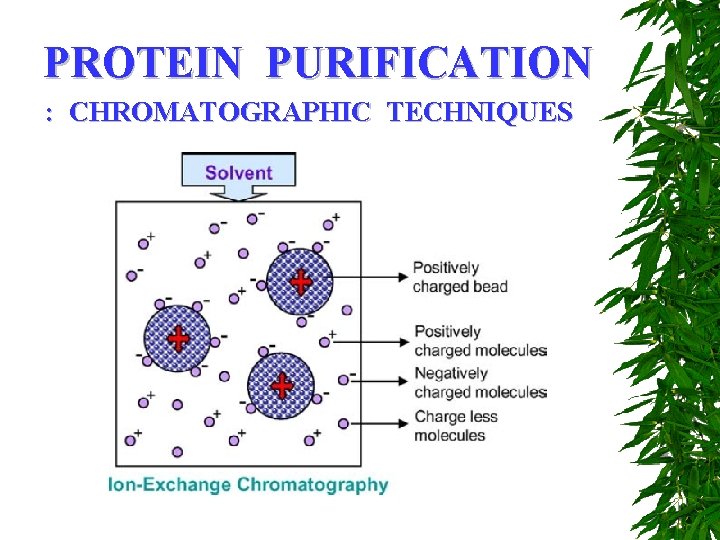
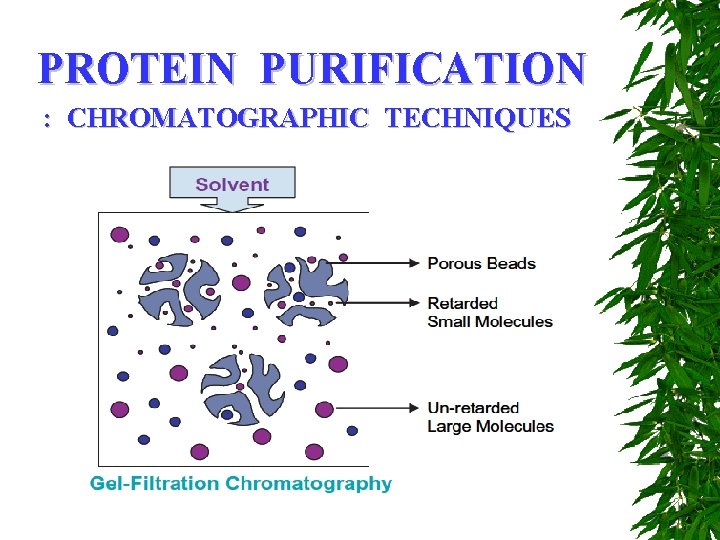
PROTEIN PURIFICATION : CHROMATOGRAPHIC TECHNIQUES

PROTEIN PURIFICATION : CHROMATOGRAPHIC TECHNIQUES

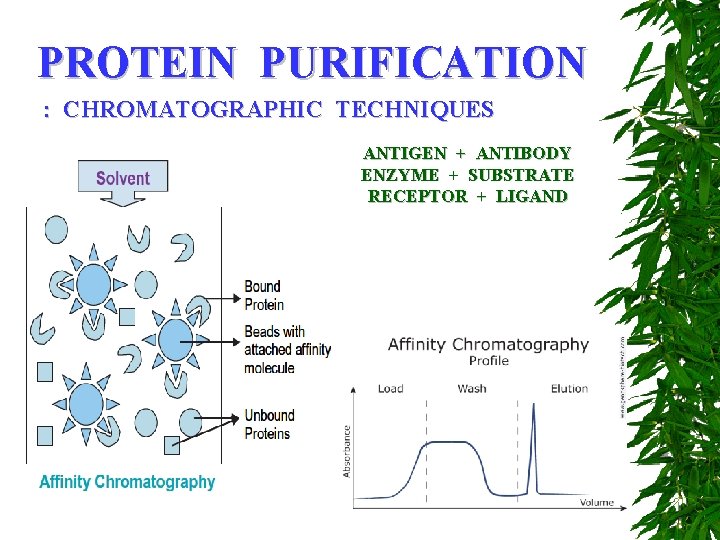
PROTEIN PURIFICATION : CHROMATOGRAPHIC TECHNIQUES ANTIGEN + ANTIBODY ENZYME + SUBSTRATE RECEPTOR + LIGAND

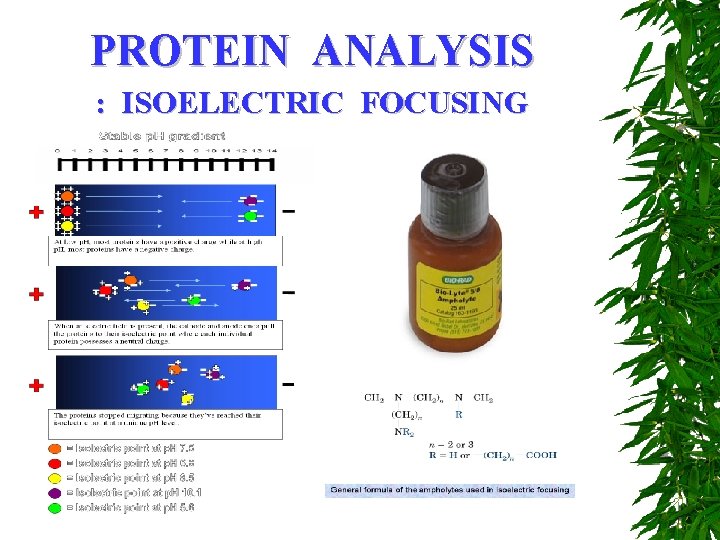
PROTEIN ANALYSIS : ISOELECTRIC FOCUSING

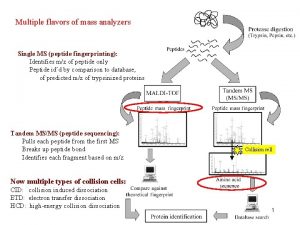
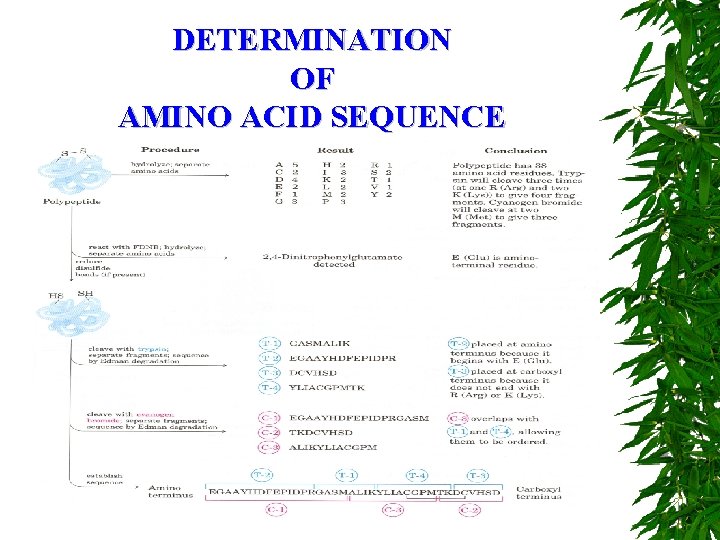
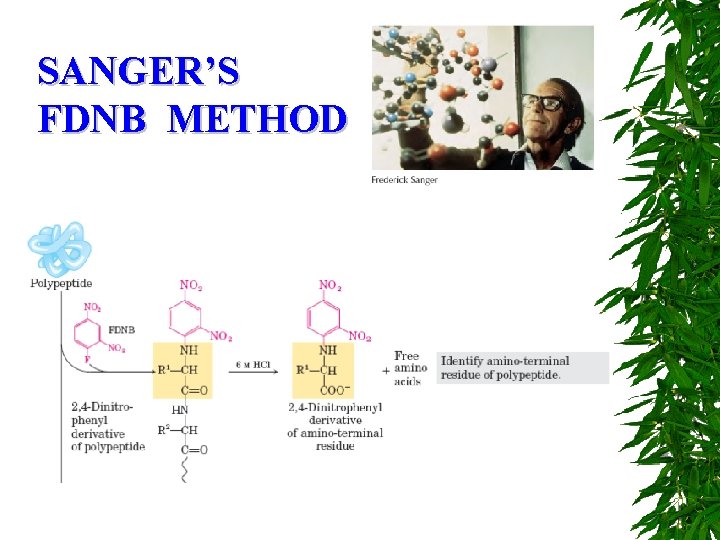
DETERMINATION OF AMINO ACID SEQUENCE SANGER’S FDNB METHOD DANSYL CHLORIDE EDMAN DEGRADATION

SPECIFICITY OF SOME POLYPEPTIDE CLEAVING REAGENTS

DETERMINATION OF AMINO ACID SEQUENCE

SANGER’S FDNB METHOD

DANSYL CHLORIDE Yellow fluorescence

EDMAN DEGRADATION พฒนาวธขนโดย เปนวธทใชในเครอง Pehr Edman amino acid sequencer Phenylisothiocyanate TFA : Trifluoroacetic acid PTH : Phenylthiohydantoin

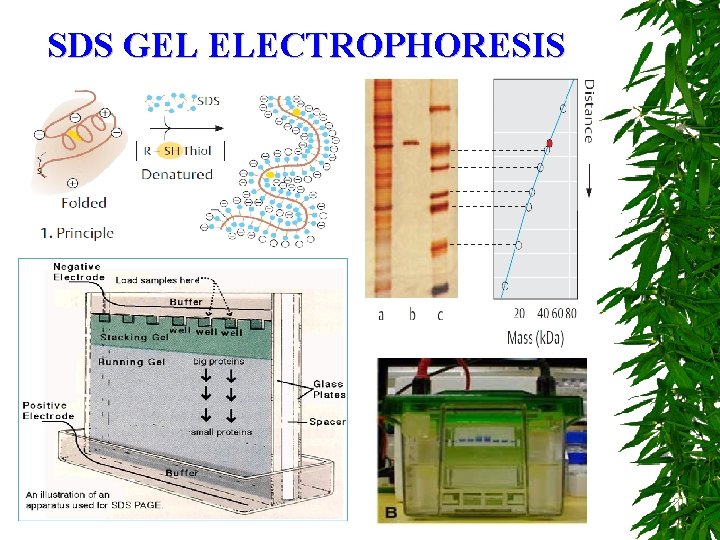
SDS GEL ELECTROPHORESIS


 What is the r group in amino acids
What is the r group in amino acids Non essential amino acids mnemonics
Non essential amino acids mnemonics Classification of amino acids
Classification of amino acids Amino acids classification
Amino acids classification Amino acids classification
Amino acids classification Quaternary structure of protein
Quaternary structure of protein Amino group and carboxyl group
Amino group and carboxyl group Amino group and carboxyl group
Amino group and carboxyl group Translation
Translation Titration curves of amino acids
Titration curves of amino acids Titration curve of amino acids
Titration curve of amino acids Deamination of amino acids
Deamination of amino acids Properties of amino acids slideshare
Properties of amino acids slideshare Deamination of glutamine
Deamination of glutamine 20 amino acid structure
20 amino acid structure Right handed amino acids
Right handed amino acids Non essential amino acids in food
Non essential amino acids in food Titration curve of amino acids
Titration curve of amino acids Gluconeogenes
Gluconeogenes Purely glucogenic amino acids
Purely glucogenic amino acids Glutamate oxidative deamination
Glutamate oxidative deamination Chemsheets
Chemsheets Properties of amino acids
Properties of amino acids Chymotrypsin cleaves which amino acids
Chymotrypsin cleaves which amino acids Serylglycyltyrosylalanylleucine
Serylglycyltyrosylalanylleucine Diphthamide
Diphthamide Non essential amino acids mnemonics
Non essential amino acids mnemonics Dehydration synthesis of amino acids
Dehydration synthesis of amino acids Aromatic amino acids
Aromatic amino acids Phenol containing amino acids
Phenol containing amino acids Dextro amino acid
Dextro amino acid Peptide bond dehydration synthesis
Peptide bond dehydration synthesis Properties of amino acids
Properties of amino acids Biomedical importance of amino acids
Biomedical importance of amino acids Are amino acids negatively charged
Are amino acids negatively charged Conditionally essential amino acids
Conditionally essential amino acids Polar and non polar amino acids
Polar and non polar amino acids Amino acids are building blocks of
Amino acids are building blocks of Oxidative deamination of amino acids
Oxidative deamination of amino acids Glucogenic amino acid
Glucogenic amino acid Difference between hydrophobic and hydrophilic amino acids
Difference between hydrophobic and hydrophilic amino acids Non essential amino acids mnemonics
Non essential amino acids mnemonics What is protien
What is protien Quantitative qualitative estimation
Quantitative qualitative estimation Protein structure
Protein structure Protein metabolism notes
Protein metabolism notes Transdeamination of amino acids
Transdeamination of amino acids 17/35
17/35 Sp hybridization of nitrogen
Sp hybridization of nitrogen Salt bridge amino acids
Salt bridge amino acids Ketogenic amino acids
Ketogenic amino acids Upon hydrolysis of fibron which amino acids are produced
Upon hydrolysis of fibron which amino acids are produced Alpha carbon
Alpha carbon Acid base properties of amino acids
Acid base properties of amino acids Transdeamination of amino acids
Transdeamination of amino acids Ketogenic amino acids
Ketogenic amino acids Titration curve of glycine
Titration curve of glycine What is made of amino acids
What is made of amino acids Wikipedia amino acids
Wikipedia amino acids Meister cycle
Meister cycle What is made of amino acids
What is made of amino acids Importance of sulphur containing amino acids
Importance of sulphur containing amino acids Charged amino acids
Charged amino acids Milady chemical texture services test
Milady chemical texture services test Protein amino acids
Protein amino acids Protein amino acids
Protein amino acids Acid base chemistry of amino acids
Acid base chemistry of amino acids Net charges of amino acids
Net charges of amino acids 191 amino acid
191 amino acid Nitrogen removal from amino acids
Nitrogen removal from amino acids Urea cycle definition
Urea cycle definition Examples of physical function of art
Examples of physical function of art Amino acids table
Amino acids table Structure of amino acid
Structure of amino acid Mixed amino acids
Mixed amino acids Properties of amino acids slideshare
Properties of amino acids slideshare Carbamoyl phosphate synthetase reaction
Carbamoyl phosphate synthetase reaction Importance of amino acids
Importance of amino acids 20 amino acids structures
20 amino acids structures Aromatic amino acids
Aromatic amino acids How many codons are needed to specify three amino acids?
How many codons are needed to specify three amino acids? Amino acids are joined together in proteins by
Amino acids are joined together in proteins by Qualitative tests for amino acids
Qualitative tests for amino acids Chapter grabber
Chapter grabber A _________bond joins amino acids together.
A _________bond joins amino acids together. Acetylated amino group
Acetylated amino group Carbonyl group in carboxylic acids
Carbonyl group in carboxylic acids Where did wwi begin
Where did wwi begin Is kno3 acidic basic or neutral
Is kno3 acidic basic or neutral Saponifiable and non saponifiable lipids
Saponifiable and non saponifiable lipids Thermoneutral environment for neonates
Thermoneutral environment for neonates There is no neutral ground in the universe
There is no neutral ground in the universe Vega greek
Vega greek