PROTEIN STRUCTURE Amino Acid Monomers Amino acids are













- Slides: 96

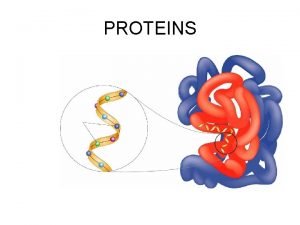
PROTEIN STRUCTURE

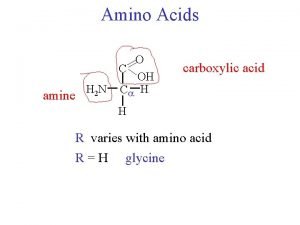
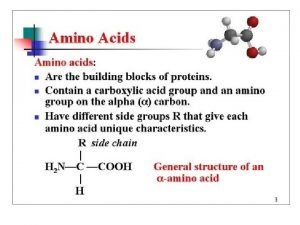
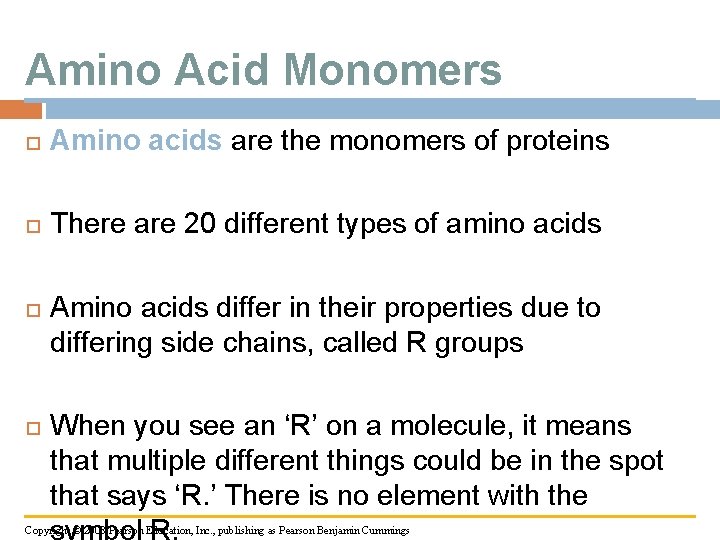
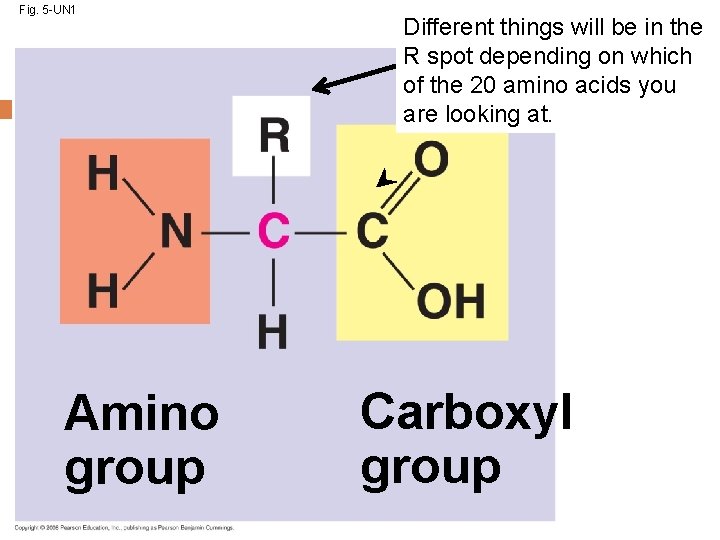
Amino Acid Monomers Amino acids are the monomers of proteins There are 20 different types of amino acids Amino acids differ in their properties due to differing side chains, called R groups When you see an ‘R’ on a molecule, it means that multiple different things could be in the spot that says ‘R. ’ There is no element with the symbol R. Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings

Fig. 5 -UN 1 Amino group Different things will be in the R spot depending on which of the 20 amino acids you are looking at. Carboxyl group

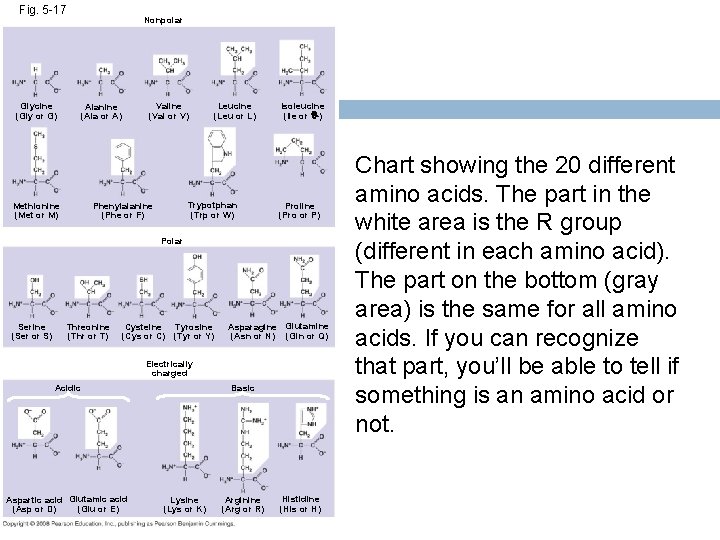
Fig. 5 -17 Glycine (Gly or G) Nonpolar Valine (Val or V) Alanine (Ala or A) Methionine (Met or M) Leucine (Leu or L) Trypotphan (Trp or W) Phenylalanine (Phe or F) Isoleucine (Ile or I) Proline (Pro or P) Polar Serine (Ser or S) Threonine (Thr or T) Cysteine (Cys or C) Tyrosine (Tyr or Y) Asparagine Glutamine (Asn or N) (Gln or Q) Electrically charged Acidic Aspartic acid Glutamic acid (Glu or E) (Asp or D) Basic Lysine (Lys or K) Arginine (Arg or R) Histidine (His or H) Chart showing the 20 different amino acids. The part in the white area is the R group (different in each amino acid). The part on the bottom (gray area) is the same for all amino acids. If you can recognize that part, you’ll be able to tell if something is an amino acid or not.

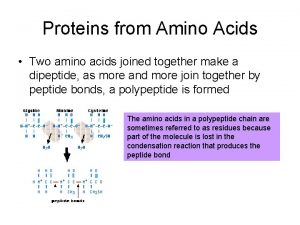
Polypeptides are polymers built from amino acids A protein consists of one or more polypeptides Polypeptides range in length from a few to more than a thousand monomers Each polypeptide has its amino acids in a different order. Some polypeptides have more of some kinds of amino acids, others include different kinds of amino acids. Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings

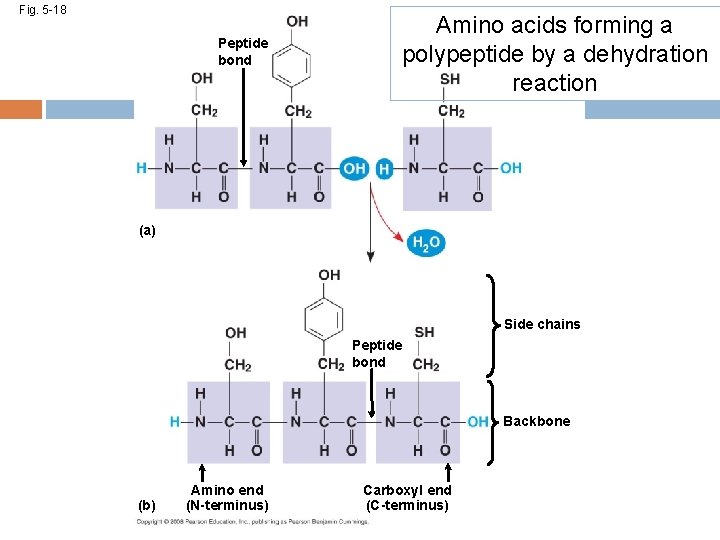
Fig. 5 -18 Peptide bond Amino acids forming a polypeptide by a dehydration reaction (a) Side chains Peptide bond Backbone (b) Amino end (N-terminus) Carboxyl end (C-terminus)

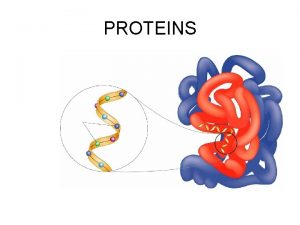
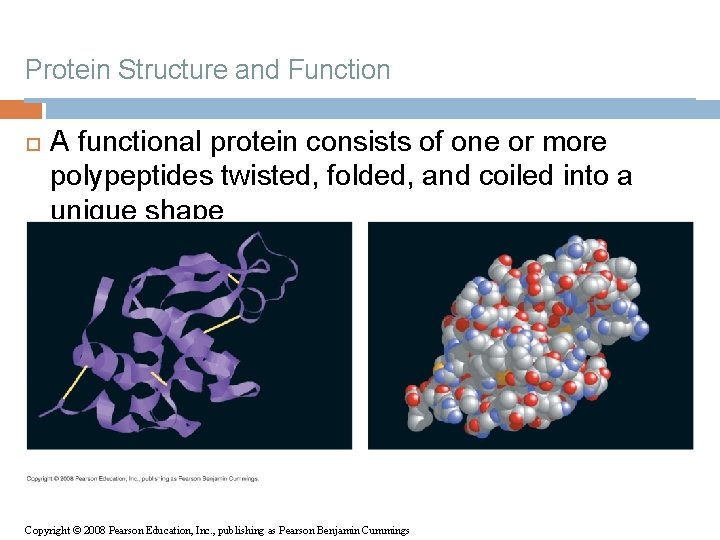
Protein Structure and Function A functional protein consists of one or more polypeptides twisted, folded, and coiled into a unique shape Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings

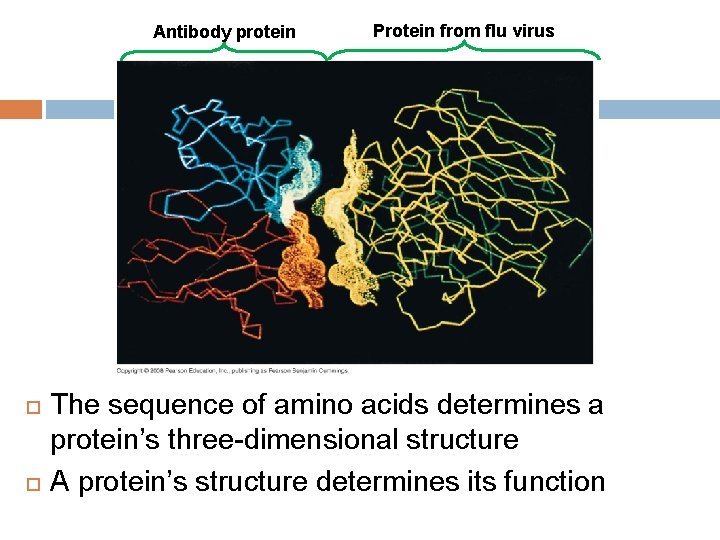
Antibody protein Protein from flu virus The sequence of amino acids determines a protein’s three-dimensional structure A protein’s structure determines its function

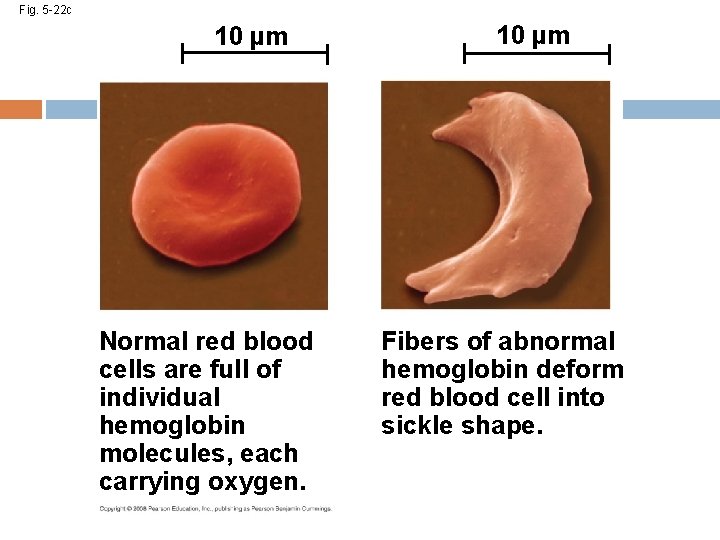
Sickle-Cell Disease: A Change in Protein Structure A slight change in protein structure can affect a protein’s structure and ability to function Sickle-cell disease, an inherited blood disorder, results from a single wrong amino acid in the protein hemoglobin The DNA gives instructions about which amino acids to put together to make a protein. If there’s a mistake in the DNA (called a mutation), you can get a protein with a wrong amino acid; the mutated protein may not function well Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings

Fig. 5 -22 c 10 µm Normal red blood cells are full of individual hemoglobin molecules, each carrying oxygen. 10 µm Fibers of abnormal hemoglobin deform red blood cell into sickle shape.

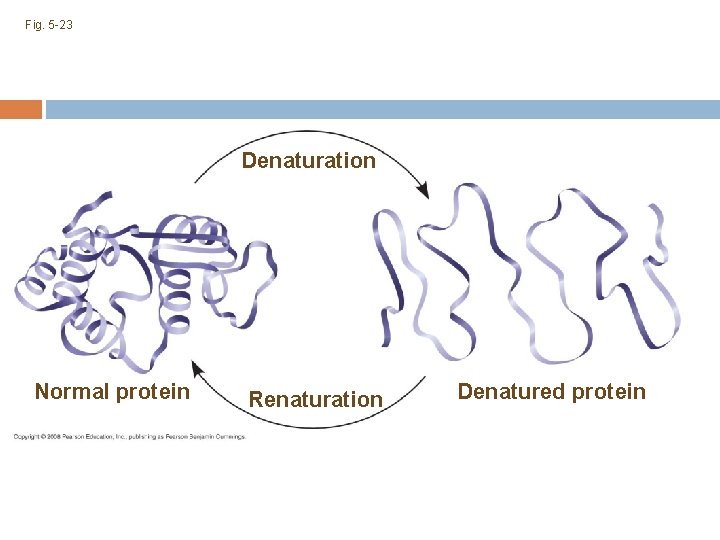
What Determines Protein Structure? In addition to the sequence of amino acids, physical and chemical conditions can affect protein shape Alterations in p. H, salt concentration, temperature, or other environmental factors can cause a protein to unravel This loss of a protein’s original shape is called denaturation A denatured protein is biologically inactive (one reason doctors worry about very high fevers) Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings

Fig. 5 -23 Denaturation Normal protein Renaturation Denatured protein

Functions of Proteins: Lots! Structure: -Proteins are embedded in cell membranes with phospholipids -Proteins direct DNA to fold into chromosomes before cell division Image from: http: //employees. csbsju. edu/hjakubowski/classes/ch 331/dna/chromosome. gif

Proteins control gene expression by turning genes on and off http: //www. cat. cc. md. us/courses/bio 141/lecguide/unit 4/genetics/protsyn/regulation/ionoind. html

Images from: http: //www. imac. auckland. ac. nz/vaccines/vacc_graph. htm Proteins fight germs. Antibodies are proteins. ANTIBODIES ATTACK & KILL THEM

Proteins help with transport. Proteins in cell membranes move molecules in and out of cells. Image from: http: //bio. winona. msus. edu/berg/ANIMTNS/Fac. Diff. htm

Proteins help with transport. Hemoglobin in blood helps transport oxygen all over the body. Image from: http: //www. cellsalive. com/pics/cover 4. gif

Some proteins act as hormones. Eating carbohydrates puts glucose in your bloodstream. Insulin is a protein hormone that controls blood glucose. Image from: http: //www. cibike. org/Cartoon. Eating. gif

Insulin function image by Riedell using Glycogen image modified from: http: //www. msu. edu/course/lbs/145/smith/s 02/graphics/campbell_5. 6. gif

People with diabetes can’t make enough insulin, so glucose stays in their blood instead of being stored by cells. Shots can replace the insulin and help to reduce blood sugar. Image modified from: http: //sonya. lanecurrent. net/Health/Images/meds. gif

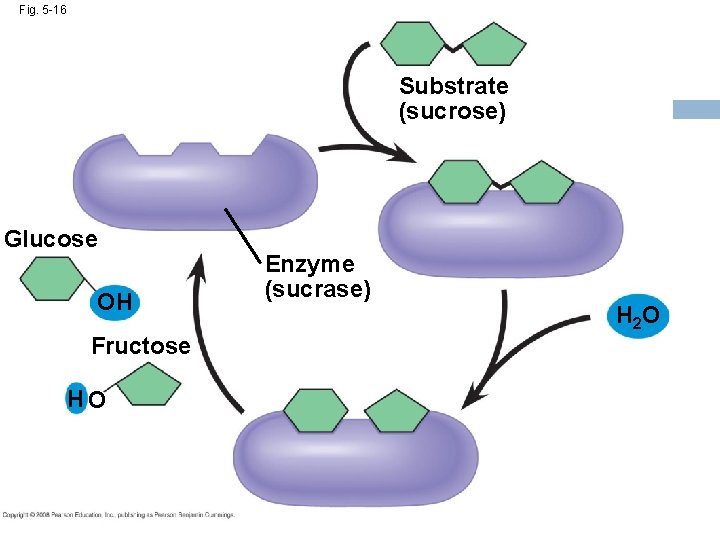
Fig. 5 -16 Substrate (sucrose) Glucose OH Fructose HO Enzyme (sucrase) H 2 O

PROTEIN SYNTHESIS

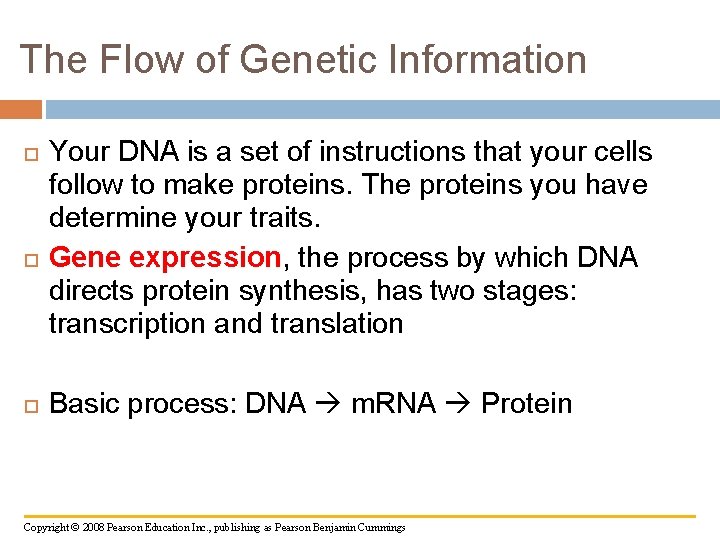
The Flow of Genetic Information Your DNA is a set of instructions that your cells follow to make proteins. The proteins you have determine your traits. Gene expression, the process by which DNA directs protein synthesis, has two stages: transcription and translation Basic process: DNA m. RNA Protein Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

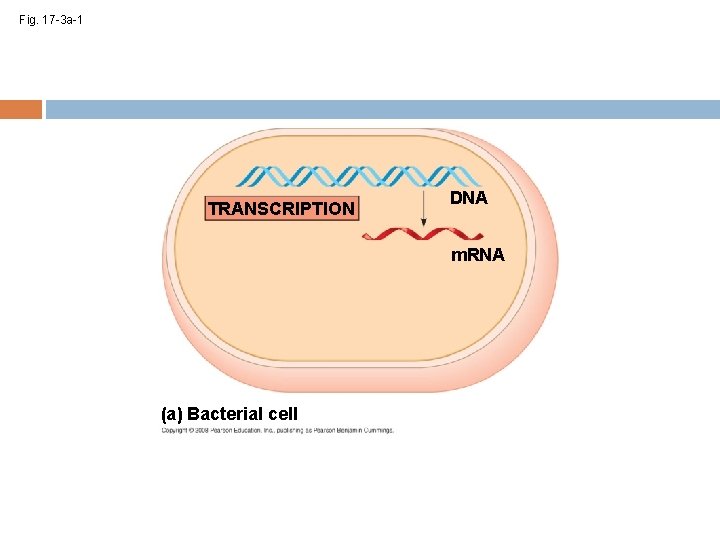
Fig. 17 -3 a-1 TRANSCRIPTION DNA m. RNA (a) Bacterial cell

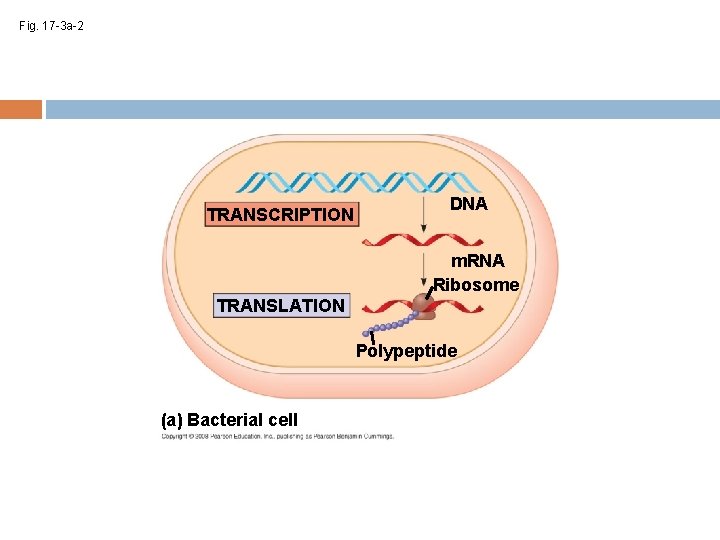
Fig. 17 -3 a-2 TRANSCRIPTION DNA m. RNA Ribosome TRANSLATION Polypeptide (a) Bacterial cell

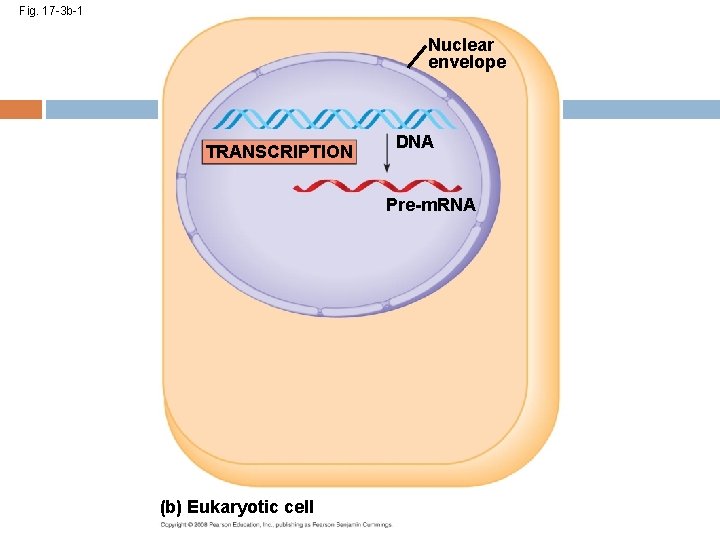
Fig. 17 -3 b-1 Nuclear envelope TRANSCRIPTION DNA Pre-m. RNA (b) Eukaryotic cell

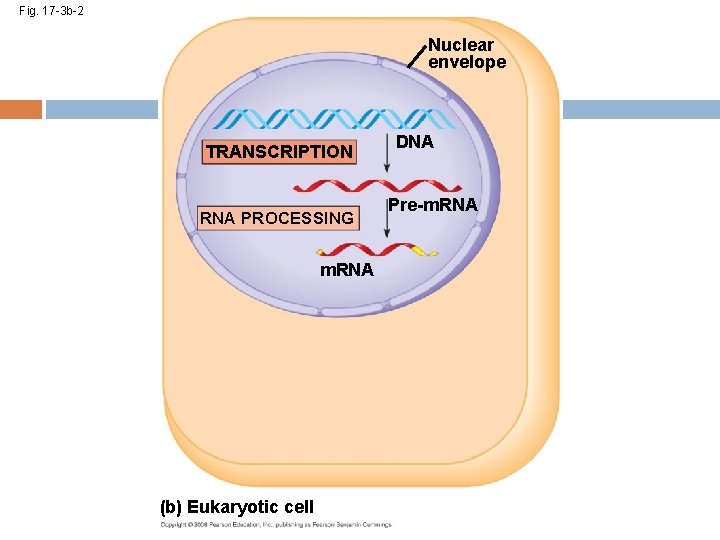
Fig. 17 -3 b-2 Nuclear envelope TRANSCRIPTION RNA PROCESSING m. RNA (b) Eukaryotic cell DNA Pre-m. RNA

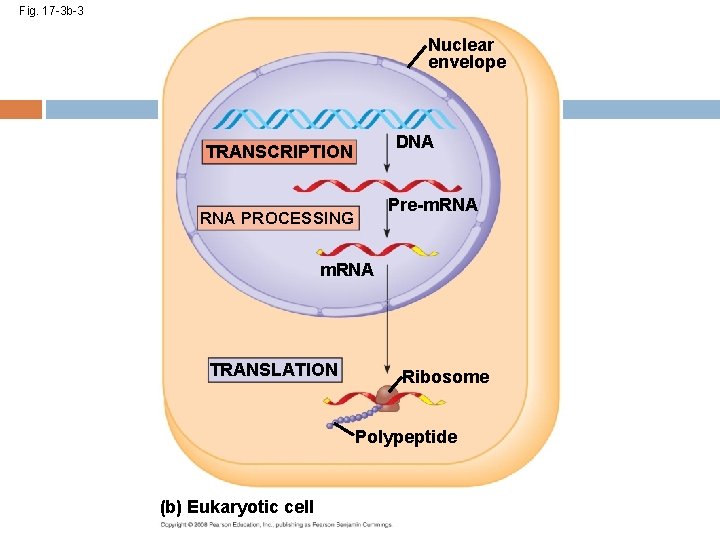
Fig. 17 -3 b-3 Nuclear envelope DNA TRANSCRIPTION Pre-m. RNA PROCESSING m. RNA TRANSLATION Ribosome Polypeptide (b) Eukaryotic cell

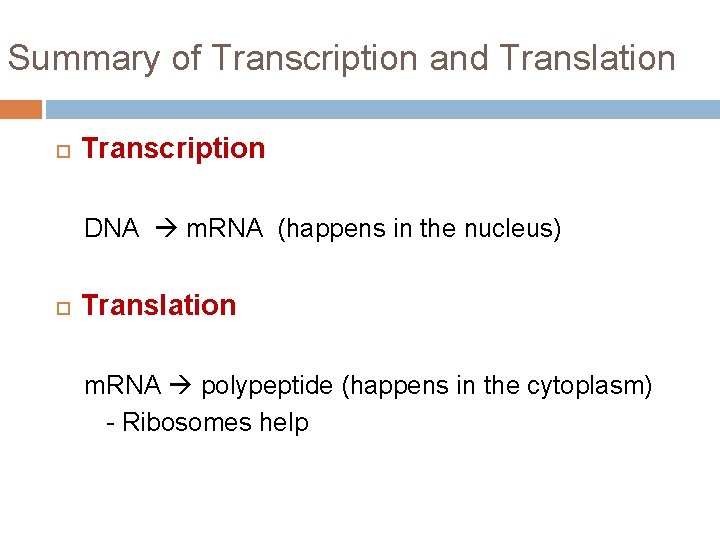
Summary of Transcription and Translation Transcription DNA m. RNA (happens in the nucleus) Translation m. RNA polypeptide (happens in the cytoplasm) - Ribosomes help

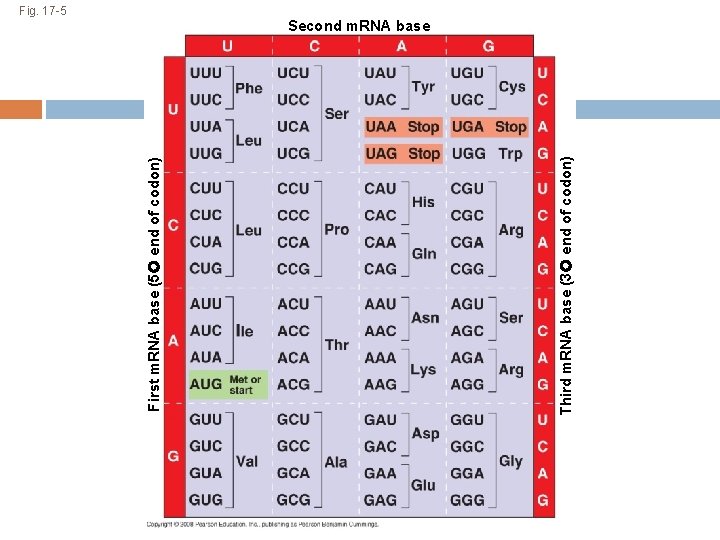
The Genetic Code How does DNA tell us which kinds of amino acids to put together for each protein? There are 20 amino acids, but there are only four nitrogenous bases in DNA How many bases correspond to an amino acid? Remember – Proteins are strings of amino acid monomers put together Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Codons: Triplets of Bases Every 3 DNA nucleotides represent a triplet, which your ribosomes read like a word. Each 3 -letter “word” on the DNA codes for a specific amino acid Example: AGT on the DNA tells the ribosome to add the amino acid Serine Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

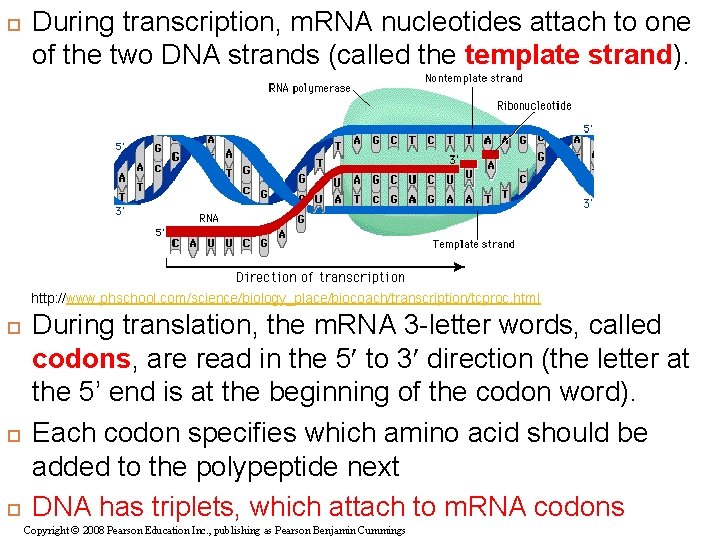
During transcription, m. RNA nucleotides attach to one of the two DNA strands (called the template strand). http: //www. phschool. com/science/biology_place/biocoach/transcription/tcproc. html During translation, the m. RNA 3 -letter words, called codons, are read in the 5 to 3 direction (the letter at the 5’ end is at the beginning of the codon word). Each codon specifies which amino acid should be added to the polypeptide next DNA has triplets, which attach to m. RNA codons Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

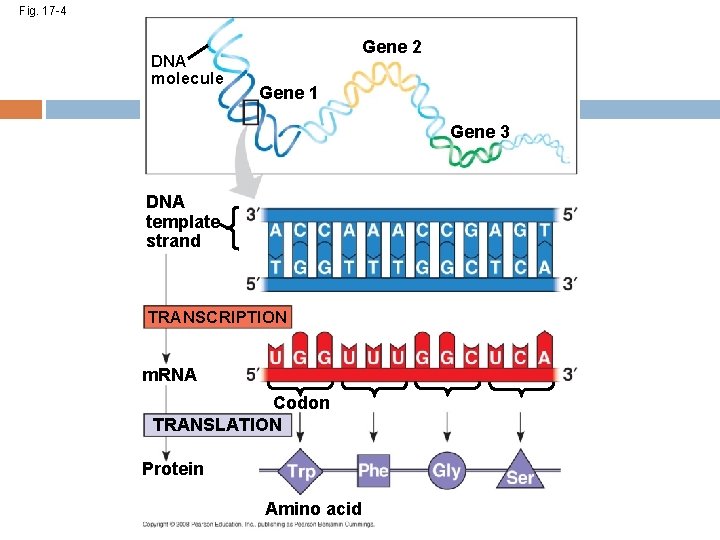
Fig. 17 -4 DNA molecule Gene 2 Gene 1 Gene 3 DNA template strand TRANSCRIPTION m. RNA Codon TRANSLATION Protein Amino acid

Cracking the Code All 64 codons were deciphered by the mid-1960 s Of the 64 triplets, 61 code for amino acids; 3 triplets are “stop” signals to end translation Just as some words mean the same thing, multiple codons can give the same amino acid Codons must be read in the correct reading frame (correct groupings) in order for the specified polypeptide to be produced Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Reading frames For the sequence ‘AUGCCGAC’, you could start at the beginning and read the codons “AUG, CCG” If you used a different reading frame, you could read “UGC, CGA” or “GCC, GAC” If you start in the wrong reading frame, you’ll combine the wrong amino acids to make the wrong protein. Since AUG is the start codon, ribosomes look for an AUG and start reading there.

Third m. RNA base (3 end of codon) First m. RNA base (5 end of codon) Fig. 17 -5 Second m. RNA base

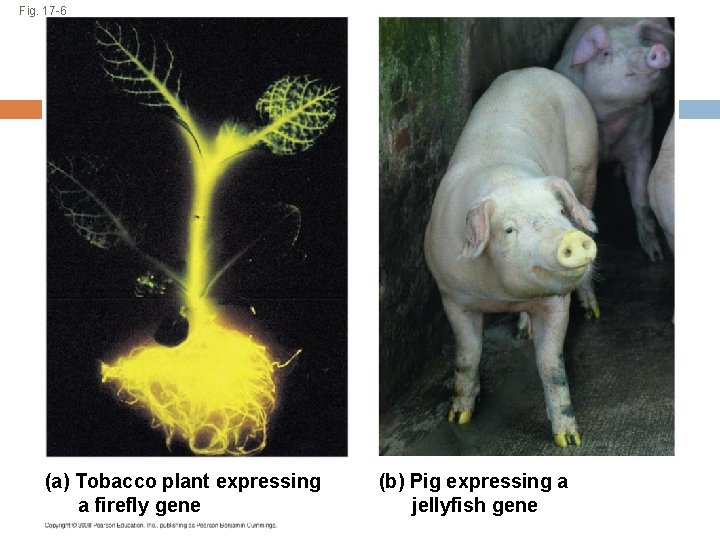
Evolution of the Genetic Code The genetic code is nearly universal, shared by the simplest bacteria to the most complex animals Genes can be transcribed and translated after being transplanted from one species to another The same sequence of nitrogenous bases in the DNA will code for the same amino acids and proteins in almost all species. Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -6 (a) Tobacco plant expressing a firefly gene (b) Pig expressing a jellyfish gene

http: //projecthdesign. com/wp-content/uploads/2007/11/landmineflowers 1. jpg http: //www. bbc. co. uk/news/scienceenvironment-14882008 http: //en. wikipedia. org/wiki/File: Glo. Fish. jpg http: //www. wired. com/science/plan etearth/news/2008/01/gm_insects#

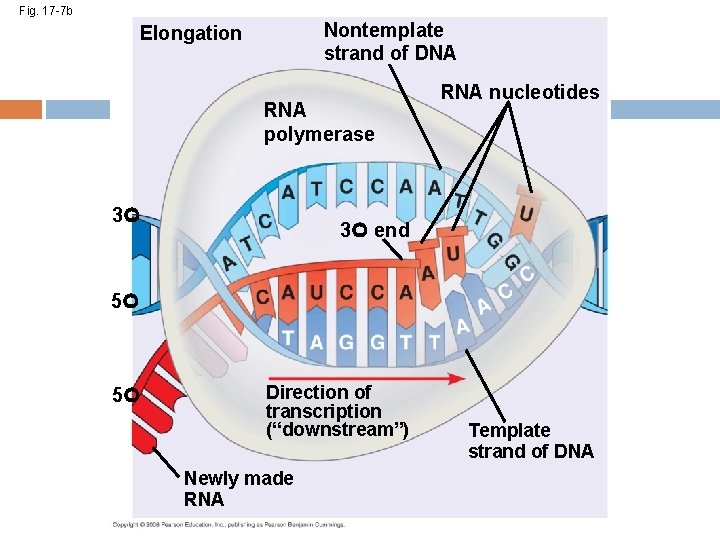
Details of Transcription RNA synthesis is catalyzed by RNA polymerase, which pries the DNA strands apart and hooks together the RNA nucleotides RNA synthesis follows the same base-pairing rules as DNA, except uracil substitutes for thymine RNA has U instead of T Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

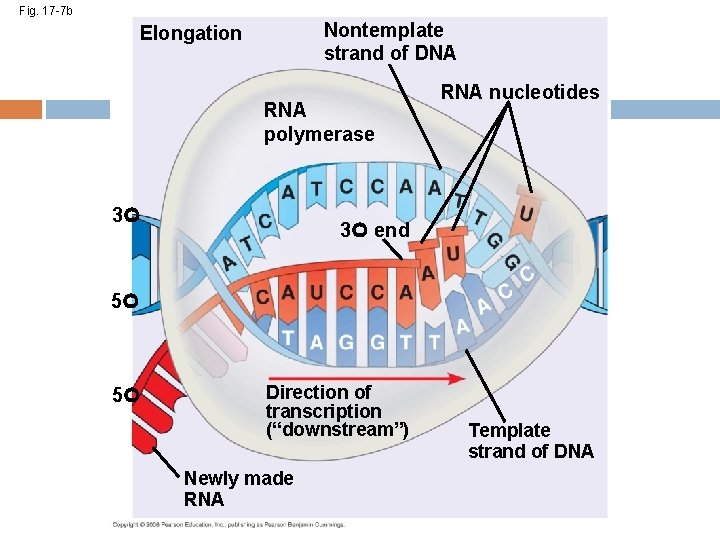
Fig. 17 -7 b Nontemplate strand of DNA Elongation RNA polymerase 3 RNA nucleotides 3 end 5 5 Direction of transcription (“downstream”) Newly made RNA Template strand of DNA

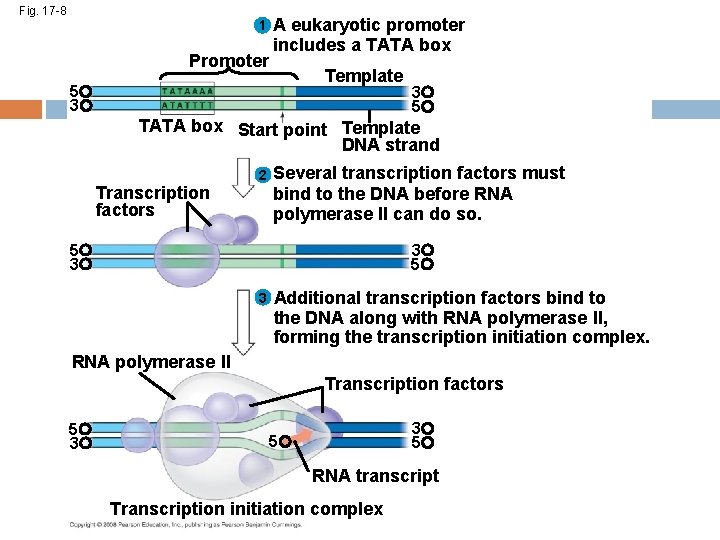
Fig. 17 -8 1 Promoter A eukaryotic promoter includes a TATA box Template 5 3 3 5 TATA box Start point Template DNA strand 2 Transcription factors Several transcription factors must bind to the DNA before RNA polymerase II can do so. 5 3 3 5 3 Additional transcription factors bind to the DNA along with RNA polymerase II, forming the transcription initiation complex. RNA polymerase II Transcription factors 5 3 3 5 5 RNA transcript Transcription initiation complex

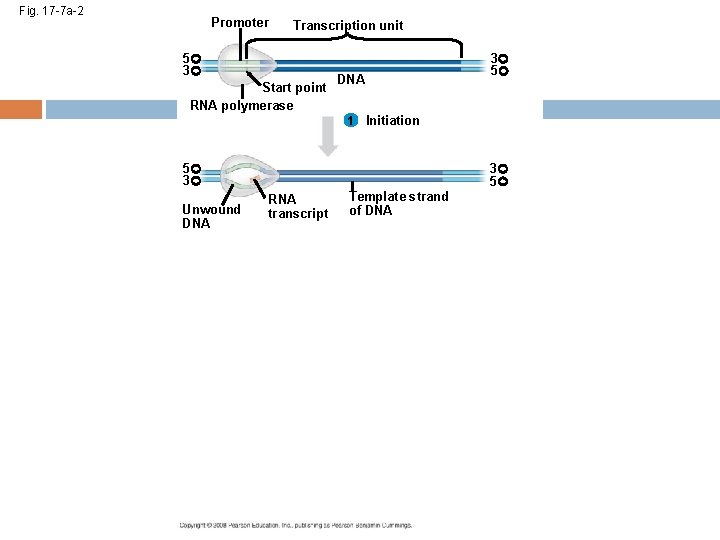
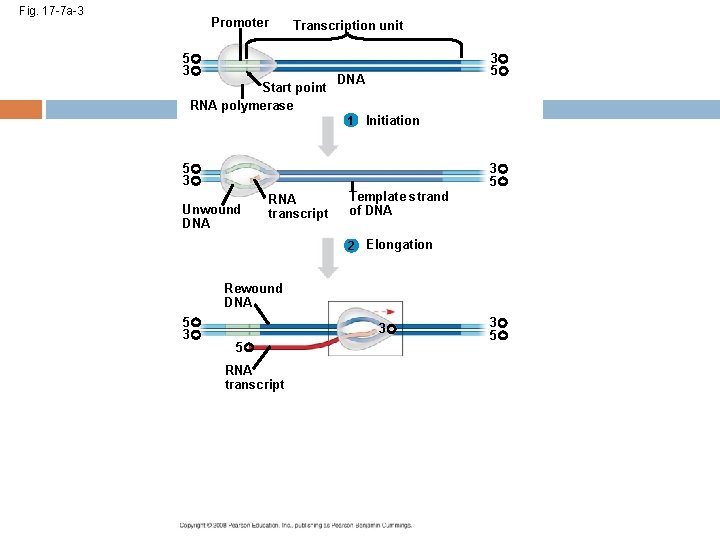
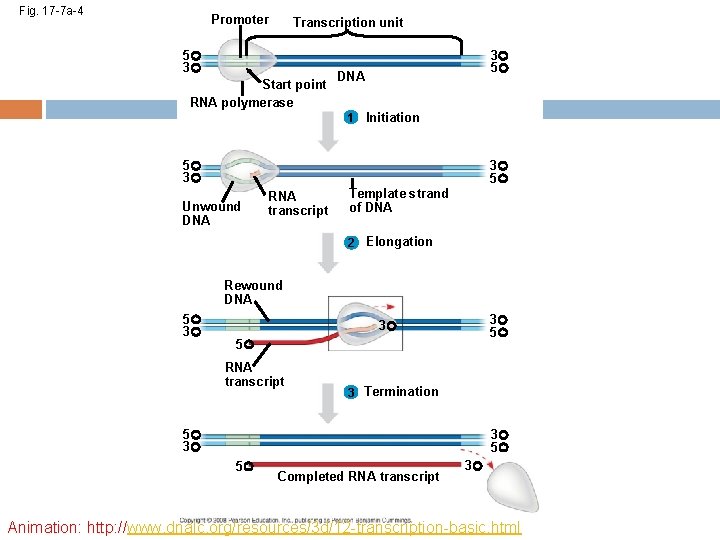
The DNA sequence where RNA polymerase first attaches is called the promoter; in bacteria, the sequence signaling the end of transcription is called the terminator The stretch of DNA that is transcribed is called a transcription unit Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

RNA Polymerase Binding and Initiation of Transcription Before transcription can start, Transcription factors attach to the promoter region and help the RNA polymerase bind onto the DNA in the right place A promoter called a TATA box is crucial in forming the initiation complex in eukaryotes Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

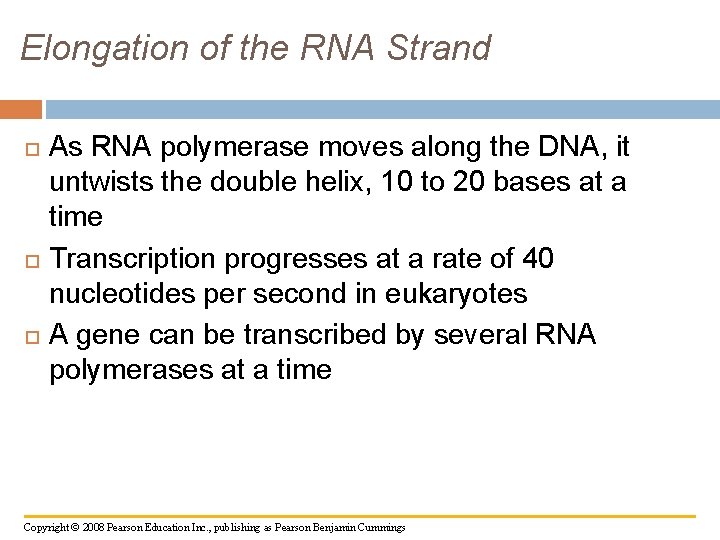
Elongation of the RNA Strand As RNA polymerase moves along the DNA, it untwists the double helix, 10 to 20 bases at a time Transcription progresses at a rate of 40 nucleotides per second in eukaryotes A gene can be transcribed by several RNA polymerases at a time Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -7 a-2 Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation 5 3 Unwound DNA 3 5 RNA transcript Template strand of DNA

Fig. 17 -7 b Nontemplate strand of DNA Elongation RNA polymerase 3 RNA nucleotides 3 end 5 5 Direction of transcription (“downstream”) Newly made RNA Template strand of DNA

Fig. 17 -7 a-3 Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation 5 3 3 5 Unwound DNA RNA transcript Template strand of DNA 2 Elongation Rewound DNA 5 3 3 5 RNA transcript 3 5

Fig. 17 -7 a-4 Promoter Transcription unit 5 3 Start point RNA polymerase 3 5 DNA 1 Initiation 5 3 3 5 Unwound DNA RNA transcript Template strand of DNA 2 Elongation Rewound DNA 5 3 3 5 RNA transcript 3 Termination 5 3 3 5 5 Completed RNA transcript 3 Animation: http: //www. dnalc. org/resources/3 d/12 -transcription-basic. html

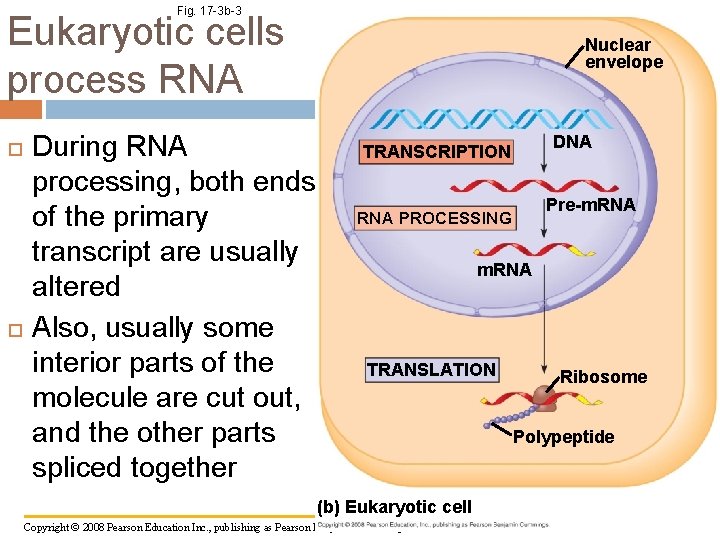
Fig. 17 -3 b-3 Eukaryotic cells process RNA Nuclear envelope During RNA processing, both ends of the primary transcript are usually altered Also, usually some interior parts of the molecule are cut out, and the other parts spliced together DNA TRANSCRIPTION Pre-m. RNA PROCESSING m. RNA TRANSLATION (b) Eukaryotic cell Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings Ribosome Polypeptide

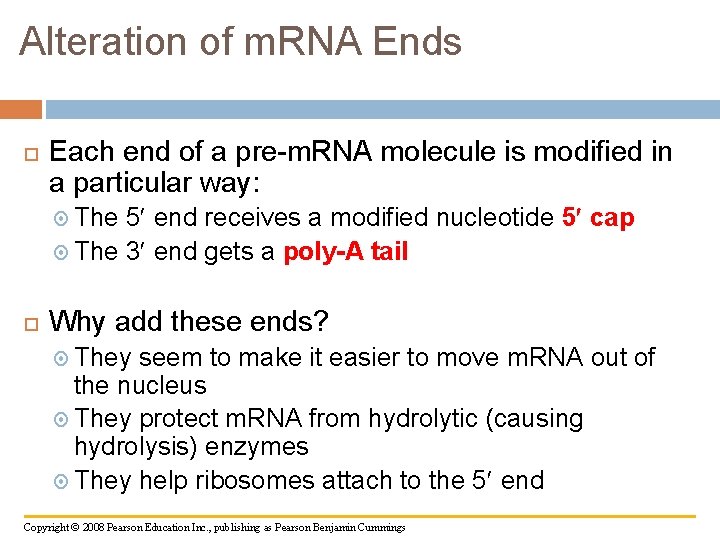
Alteration of m. RNA Ends Each end of a pre-m. RNA molecule is modified in a particular way: The 5 end receives a modified nucleotide 5 The 3 end gets a poly-A cap tail Why add these ends? They seem to make it easier to move m. RNA out of the nucleus They protect m. RNA from hydrolytic (causing hydrolysis) enzymes They help ribosomes attach to the 5 end Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

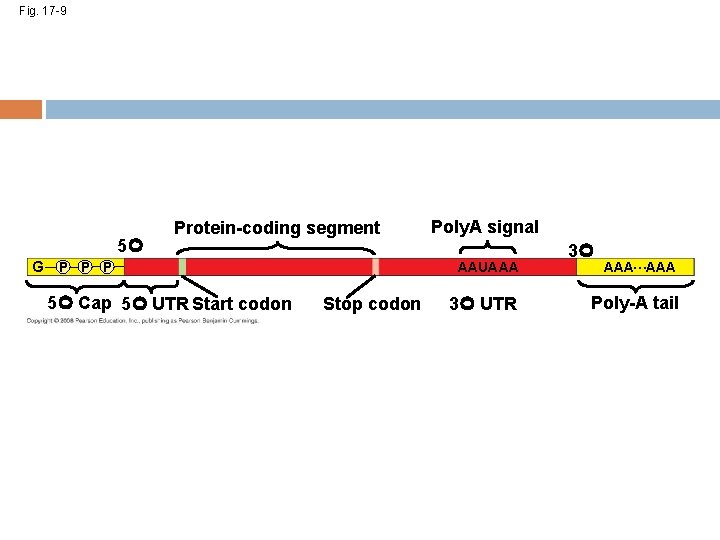
Fig. 17 -9 5 G Protein-coding segment P P P 5 Cap 5 UTR Start codon Poly. A signal AAUAAA Stop codon 3 UTR 3 AAA…AAA Poly-A tail

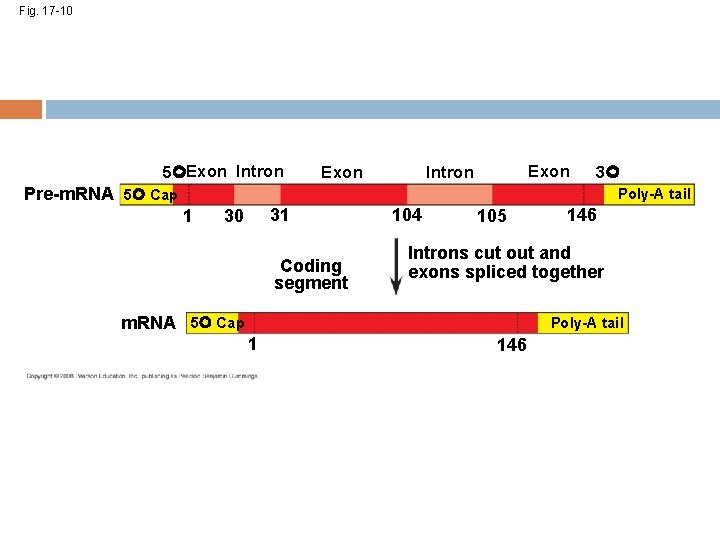
Exons, Introns, and RNA Splicing • • Most eukaryotic genes and their RNA transcripts have pieces that don’t code for useful proteins. These noncoding regions are called introns The other parts are called exons because they are eventually expressed (translated into amino acid sequences) RNA splicing removes the introns and joins the exons together, creating m. RNA with a continuous coding sequence Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -10 5 Exon Intron 3 Pre-m. RNA 5 Cap Poly-A tail 1 31 30 Coding segment 104 105 146 Introns cut out and exons spliced together m. RNA 5 Cap Poly-A tail 1 146

The Functional and Evolutionary Importance of Introns Some genes can code for more than one kind of protein, depending on which segments are treated as exons during RNA splicing These variations are called alternative RNA splicing Because of alternative RNA splicing, the number of different proteins an organism can produce is much greater than its number of genes http: //www. dnalc. org/view/16941 -2 D-Animation-of-Alternative-RNA-Splicing. html http: //www. dnalc. org/view/16940 -Alternative-RNA-Splicing. html Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

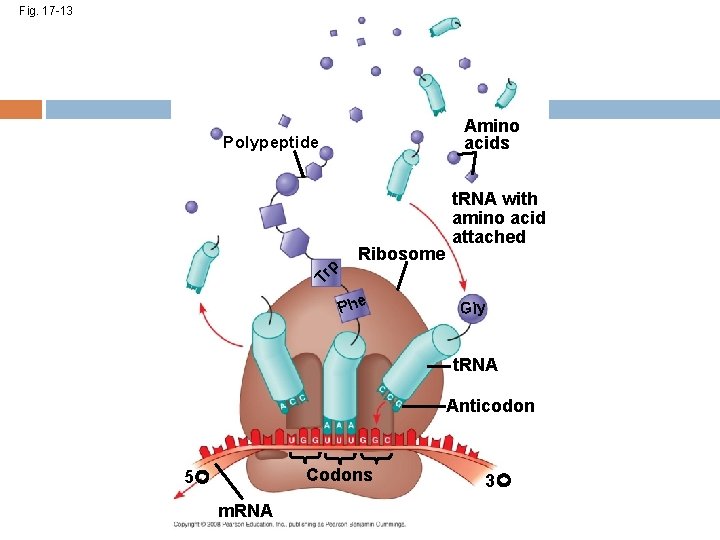
After RNA processing, the m. RNA is translated. A cell translates an m. RNA message into protein with the help of transfer RNA (t. RNA) Molecules of t. RNA are not identical: Each carries a specific amino acid on one end Each has an anticodon on the other end; the anticodon base-pairs with a complementary codon on m. RNA Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -13 Amino acids Polypeptide Tr p Ribosome t. RNA with amino acid attached Phe Gly t. RNA Anticodon Codons 5 m. RNA 3

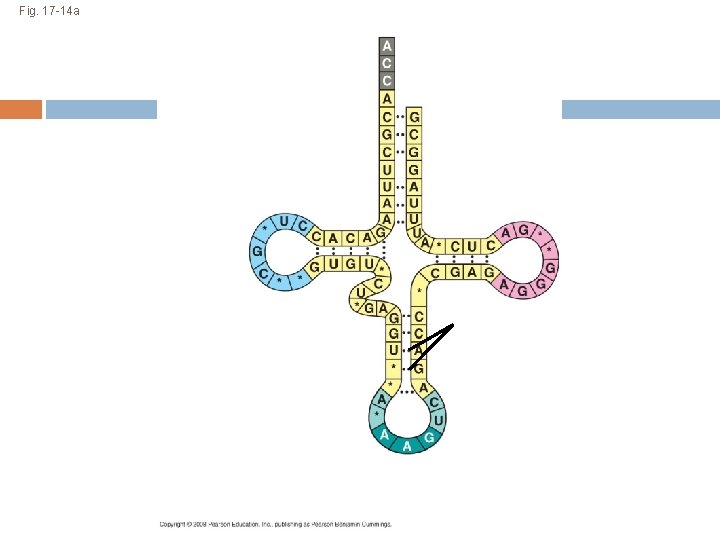
The Structure and Function of Transfer RNA A t. RNA molecule consists of a single RNA strand that is only about 80 nucleotides long Flattened into one plane to reveal its base pairing, a t. RNA molecule looks like a cloverleaf Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -14 a 3 Amino acid attachment site 5 Hydrogen bonds Anticodon (a) Two-dimensional structure

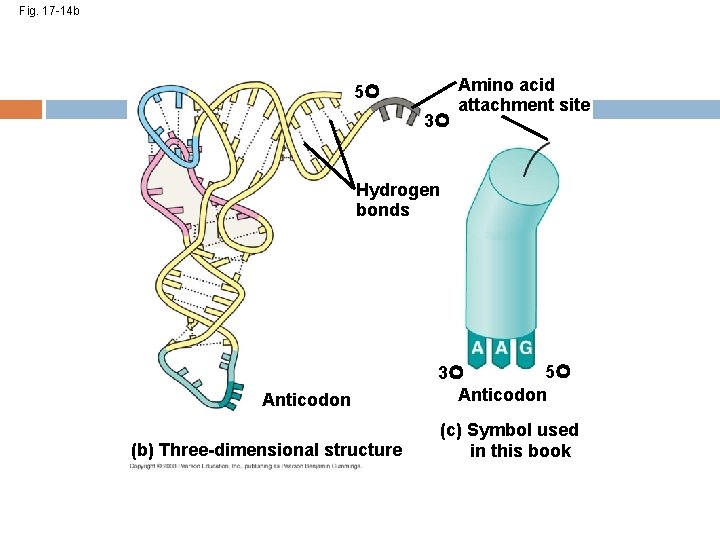
Because of hydrogen bonds, t. RNA actually twists and folds into a three-dimensional molecule t. RNA is roughly L-shaped Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -14 b 5 3 Amino acid attachment site Hydrogen bonds Anticodon (b) Three-dimensional structure 5 3 Anticodon (c) Symbol used in this book

Accurate translation requires two steps: First: a correct match between a t. RNA and an amino acid (t. RNA with an amino acid attached is “charged” t. RNA) Second: a correct match between the t. RNA anticodon and an m. RNA codon Flexible pairing at the third base of a codon is called wobble and allows some t. RNAs to bind to more than one codon Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Ribosomes help t. RNA anticodons to pair up with specific m. RNA codons in protein synthesis The two ribosomal subunits (large and small) are made of proteins and ribosomal RNA (r. RNA) Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

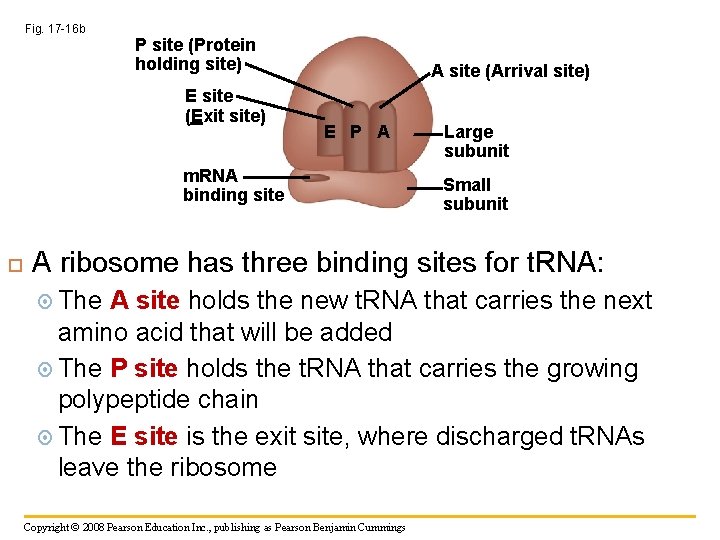
Fig. 17 -16 b P site (Protein holding site) E site (Exit site) A site (Arrival site) E P A m. RNA binding site Large subunit Small subunit A ribosome has three binding sites for t. RNA: The A site holds the new t. RNA that carries the next amino acid that will be added The P site holds the t. RNA that carries the growing polypeptide chain The E site is the exit site, where discharged t. RNAs leave the ribosome Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

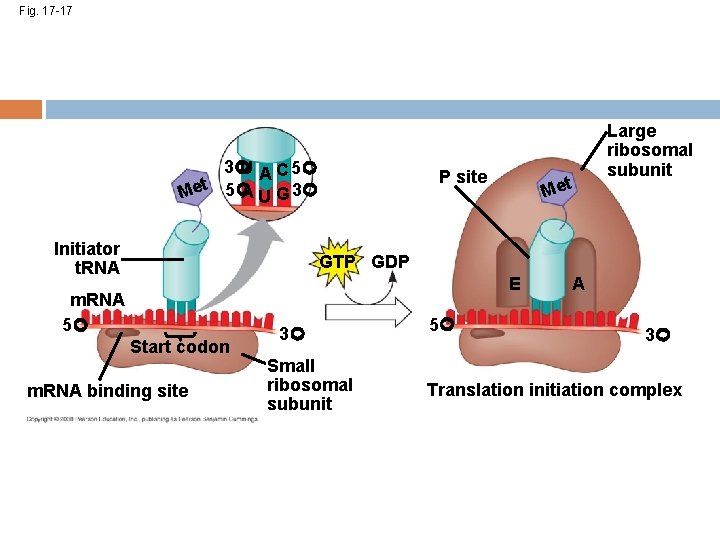
Initiation of Translation First, a small ribosomal subunit binds with m. RNA and a special initiator t. RNA Then the small subunit moves along the m. RNA until it reaches the start codon (AUG) Proteins called initiation factors bring in the large subunit The initiator t. RNA sticks to the AUG codon. What is the anticodon on this t. RNA? Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -17 3 U A C 5 A U G 3 Met 5 Initiator t. RNA P site Met Large ribosomal subunit GTP GDP E m. RNA 5 Start codon m. RNA binding site 3 Small ribosomal subunit 5 A 3 Translation initiation complex

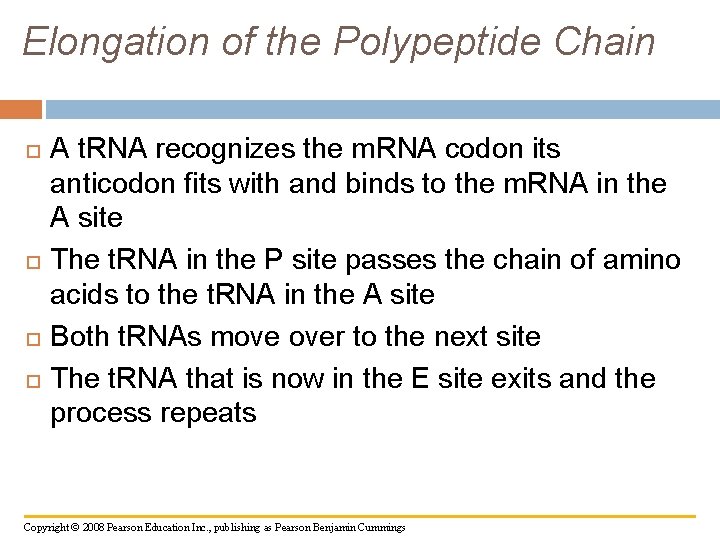
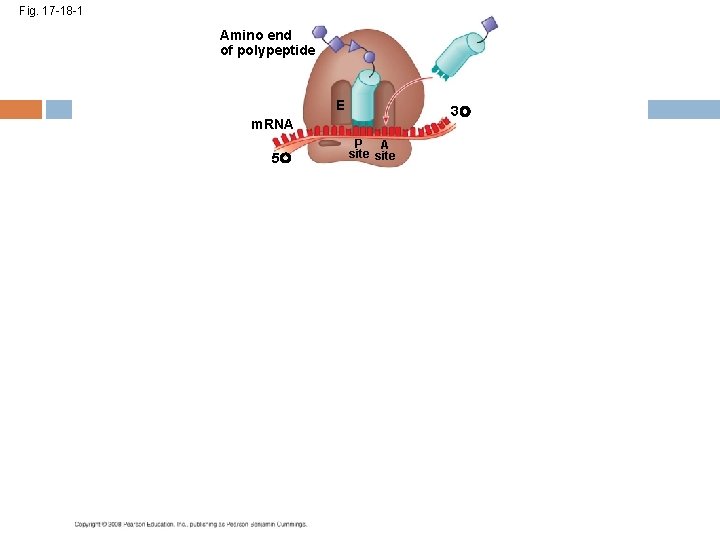
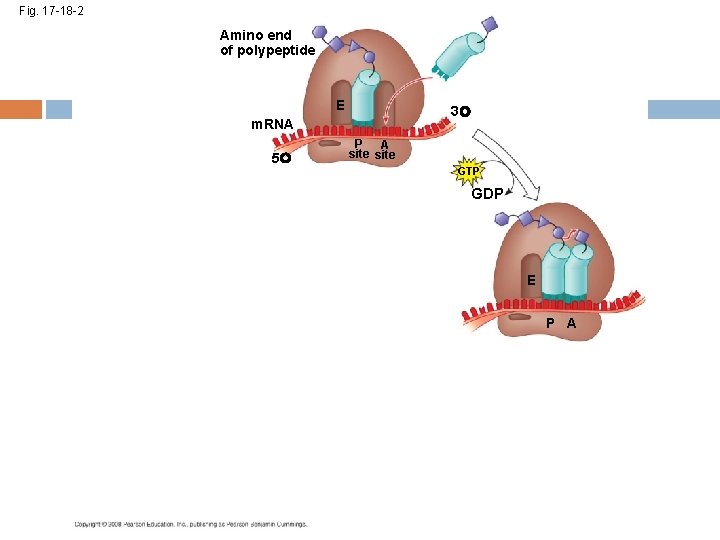
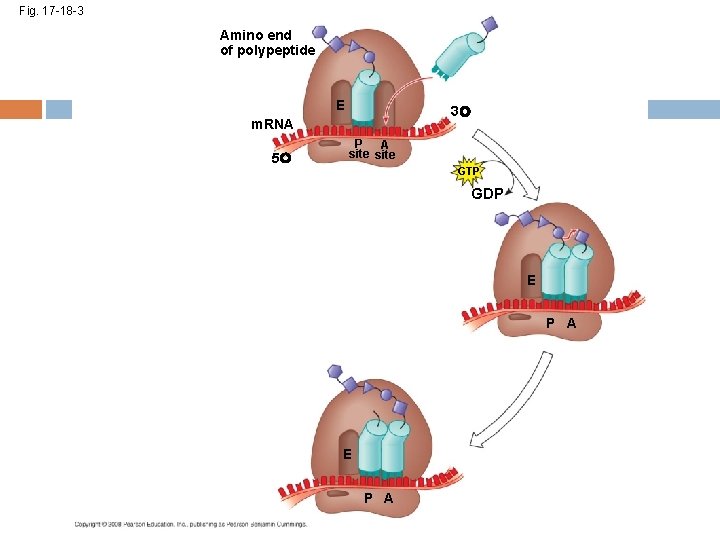
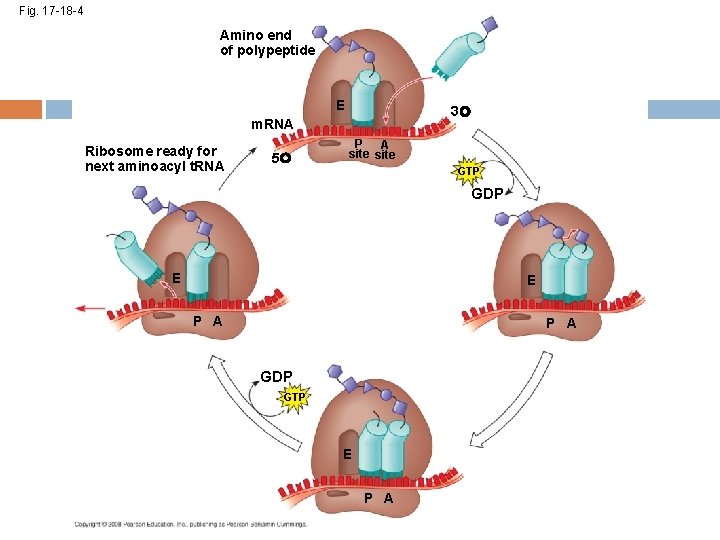
Elongation of the Polypeptide Chain A t. RNA recognizes the m. RNA codon its anticodon fits with and binds to the m. RNA in the A site The t. RNA in the P site passes the chain of amino acids to the t. RNA in the A site Both t. RNAs move over to the next site The t. RNA that is now in the E site exits and the process repeats Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -18 -1 Amino end of polypeptide E 3 m. RNA 5 P A site

Fig. 17 -18 -2 Amino end of polypeptide E 3 m. RNA 5 P A site GTP GDP E P A

Fig. 17 -18 -3 Amino end of polypeptide E 3 m. RNA 5 P A site GTP GDP E P A

Fig. 17 -18 -4 Amino end of polypeptide E 3 m. RNA Ribosome ready for next aminoacyl t. RNA 5 P A site GTP GDP E E P A GDP GTP E P A

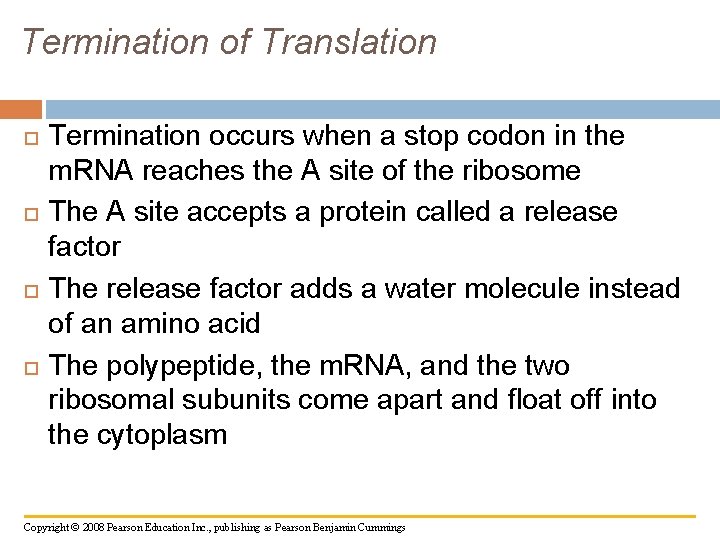
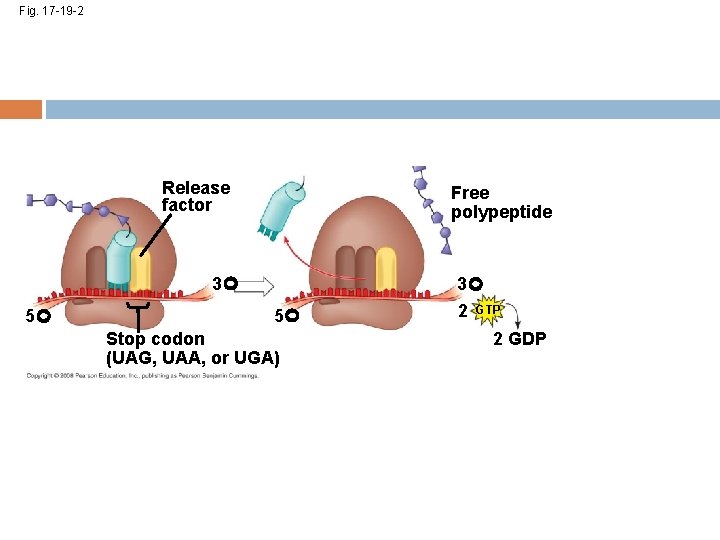
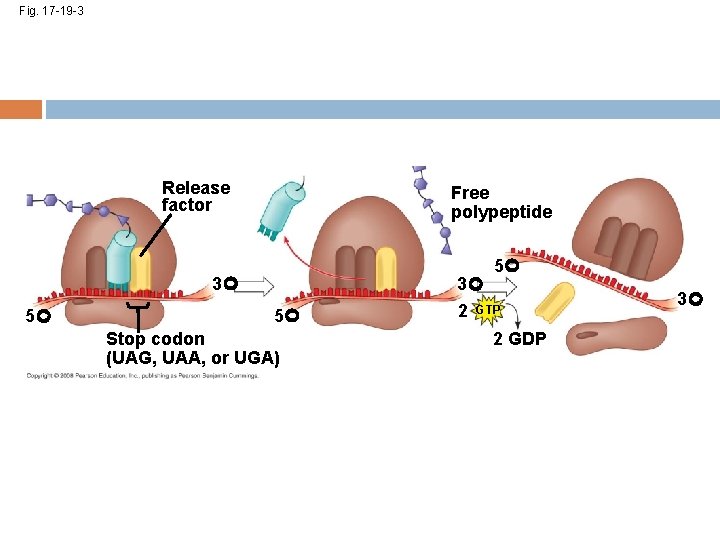
Termination of Translation Termination occurs when a stop codon in the m. RNA reaches the A site of the ribosome The A site accepts a protein called a release factor The release factor adds a water molecule instead of an amino acid The polypeptide, the m. RNA, and the two ribosomal subunits come apart and float off into the cytoplasm Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -19 -1 Release factor 3 5 Stop codon (UAG, UAA, or UGA)

Fig. 17 -19 -2 Release factor Free polypeptide 3 5 5 Stop codon (UAG, UAA, or UGA) 3 2 GTP 2 GDP

Fig. 17 -19 -3 Release factor Free polypeptide 5 3 5 5 Stop codon (UAG, UAA, or UGA) 3 2 GTP 2 GDP 3

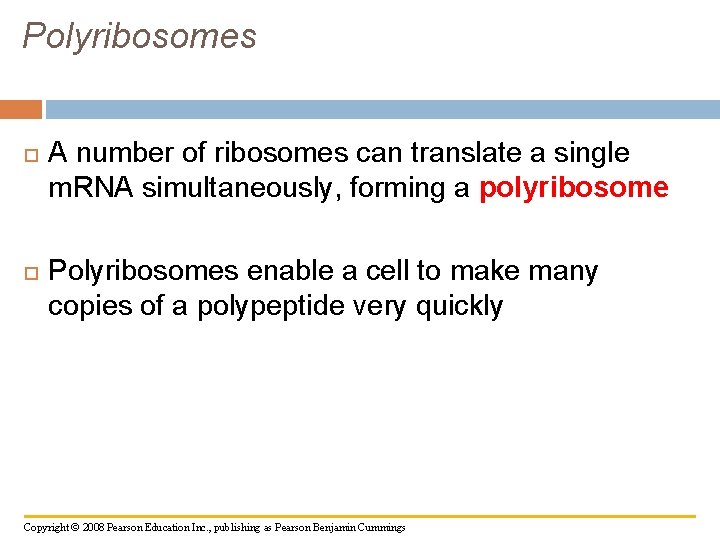
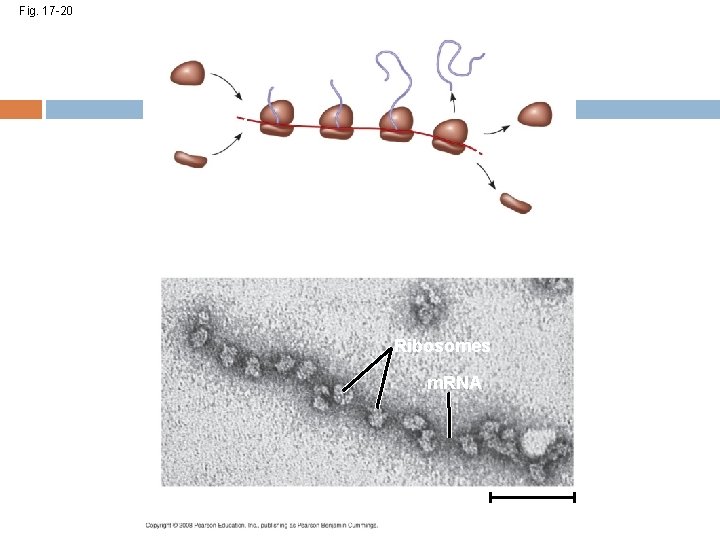
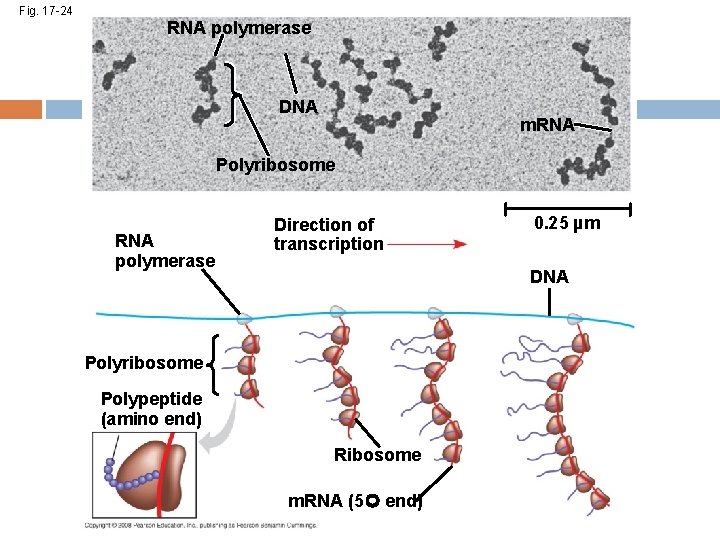
Polyribosomes A number of ribosomes can translate a single m. RNA simultaneously, forming a polyribosome Polyribosomes enable a cell to make many copies of a polypeptide very quickly Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -20 Completed polypeptide Growing polypeptides Incoming ribosomal subunits Start of m. RNA (5 end) (a) Polyribosome End of m. RNA (3 end) Ribosomes m. RNA (b) 0. 1 µm

Targeting Polypeptides to Specific Locations Cells have 2 kinds of ribosomes: free ribosomes and bound ribosomes Free ribosomes mostly synthesize proteins that are used in the cytoplasm Bound ribosomes make proteins of the endomembrane system and proteins that are secreted from the cell Ribosomes are identical and can switch from free to bound Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

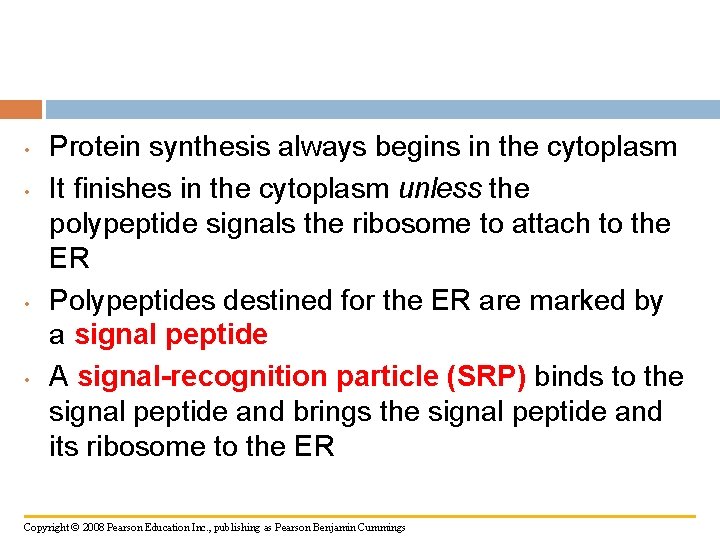
• • Protein synthesis always begins in the cytoplasm It finishes in the cytoplasm unless the polypeptide signals the ribosome to attach to the ER Polypeptides destined for the ER are marked by a signal peptide A signal-recognition particle (SRP) binds to the signal peptide and brings the signal peptide and its ribosome to the ER Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

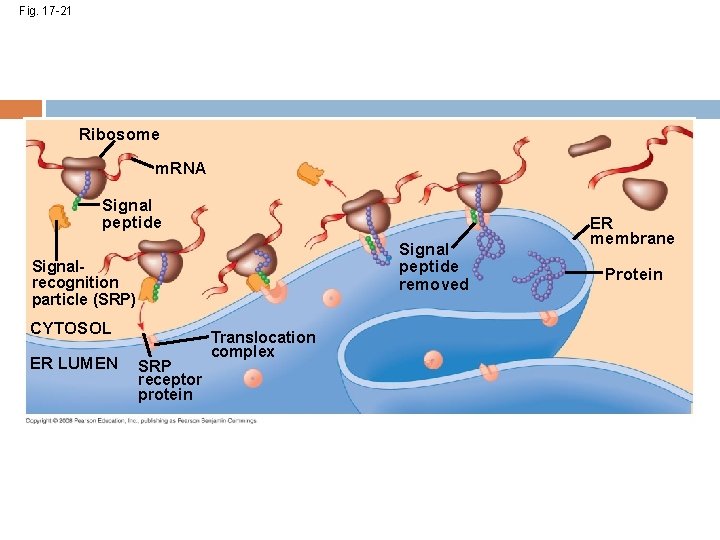
Fig. 17 -21 Ribosome m. RNA Signal peptide removed Signalrecognition particle (SRP) CYTOSOL ER LUMEN SRP receptor protein Translocation complex ER membrane Protein

Types of Mutations are changes in the genetic material of a cell or virus Point mutations are changes in just one base pair of a gene The change of a single nucleotide in a DNA template strand can lead to the production of an abnormal protein Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

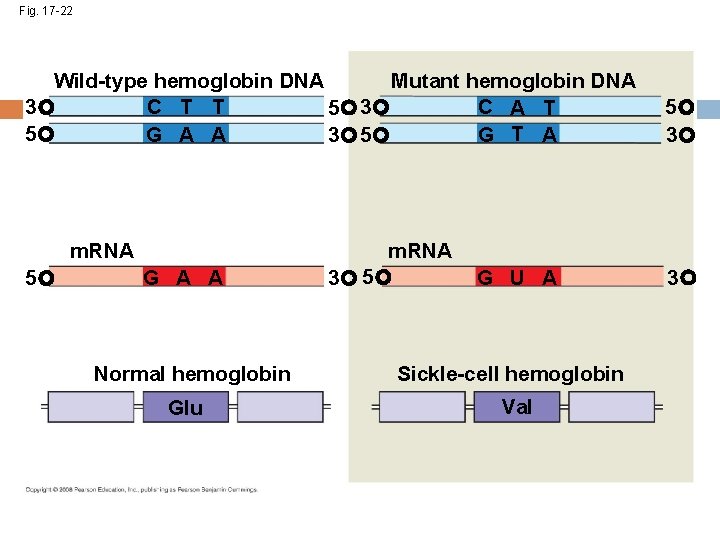
Fig. 17 -22 Wild-type hemoglobin DNA Mutant hemoglobin DNA C T T C A T 3 5 G T A G A A 3 5 m. RNA 5 G A A Normal hemoglobin Glu m. RNA 3 5 G U A Sickle-cell hemoglobin Val 5 3 3

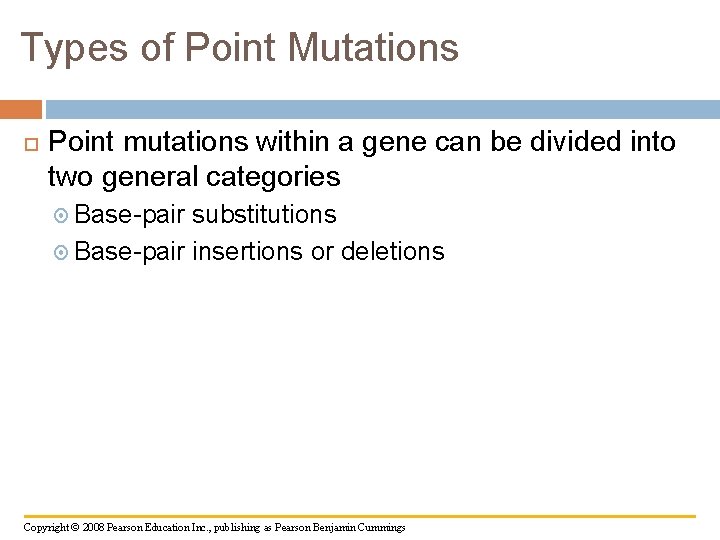
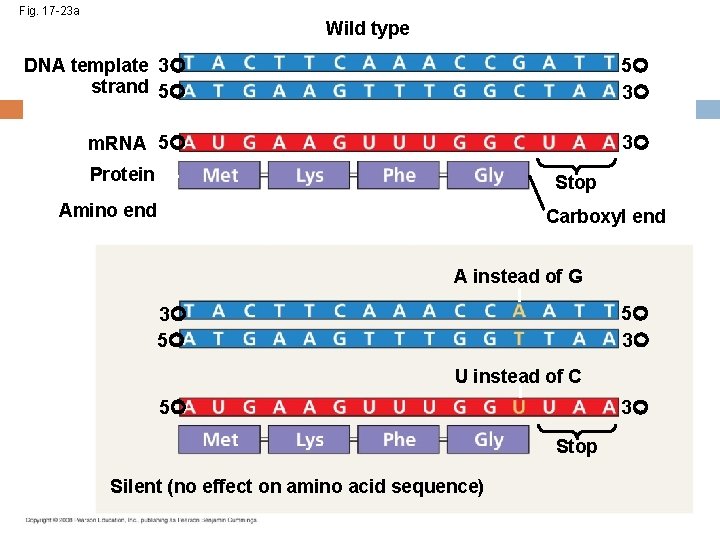
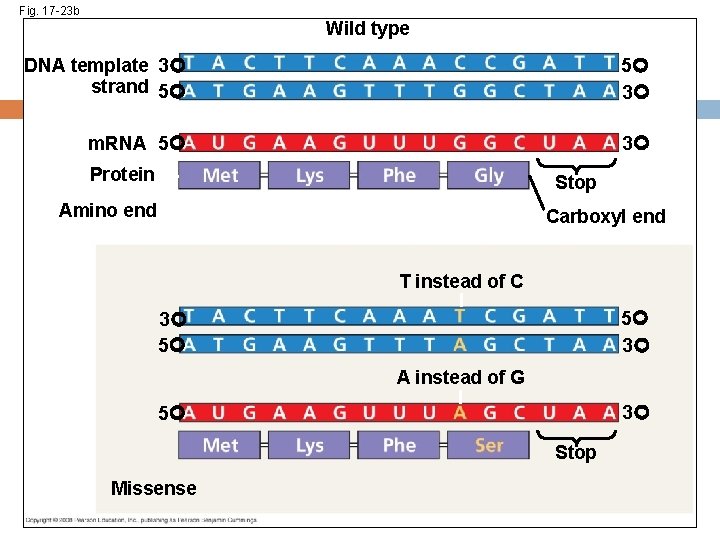
Types of Point Mutations Point mutations within a gene can be divided into two general categories Base-pair substitutions Base-pair insertions or deletions Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -23 a Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end A instead of G 5 3 3 5 U instead of C 5 3 Stop Silent (no effect on amino acid sequence)

Fig. 17 -23 b Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end T instead of C 5 3 3 5 A instead of G 3 5 Stop Missense

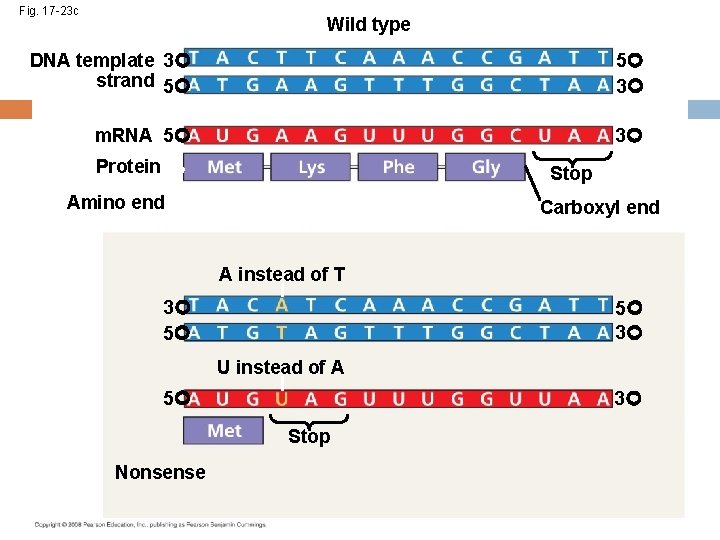
Fig. 17 -23 c Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end A instead of T 3 5 5 3 U instead of A 5 3 Stop Nonsense

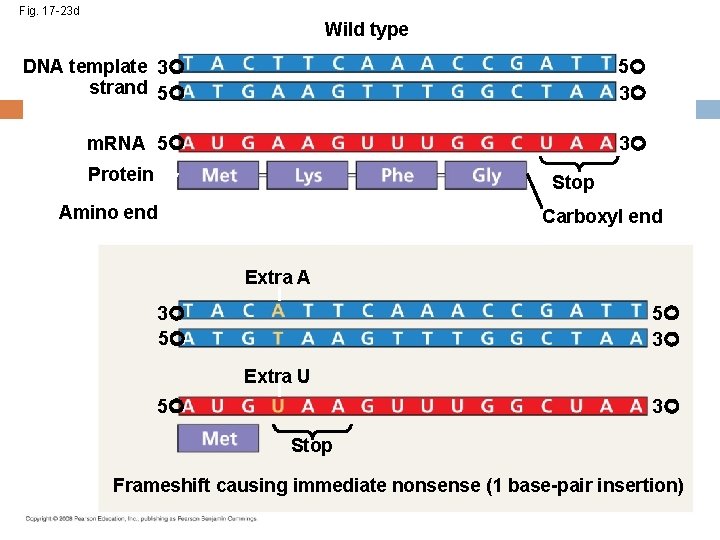
Fig. 17 -23 d Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end Extra A 5 3 3 5 Extra U 5 3 Stop Frameshift causing immediate nonsense (1 base-pair insertion)

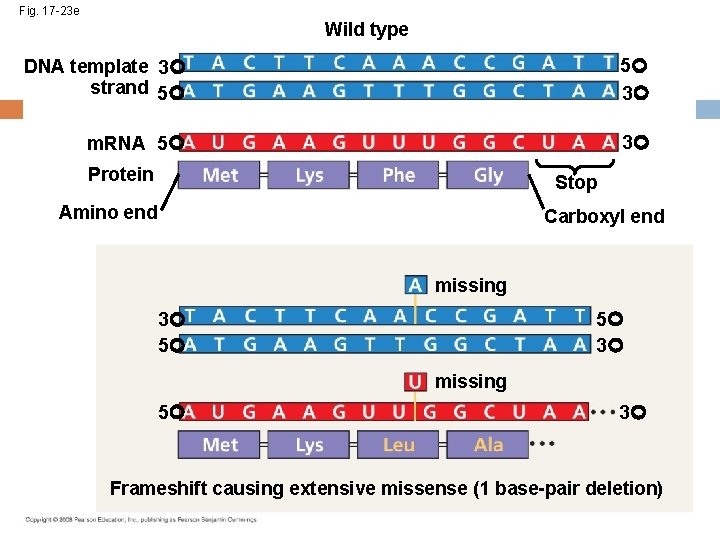
Fig. 17 -23 e Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end missing 5 3 3 5 missing 5 3 Frameshift causing extensive missense (1 base-pair deletion)

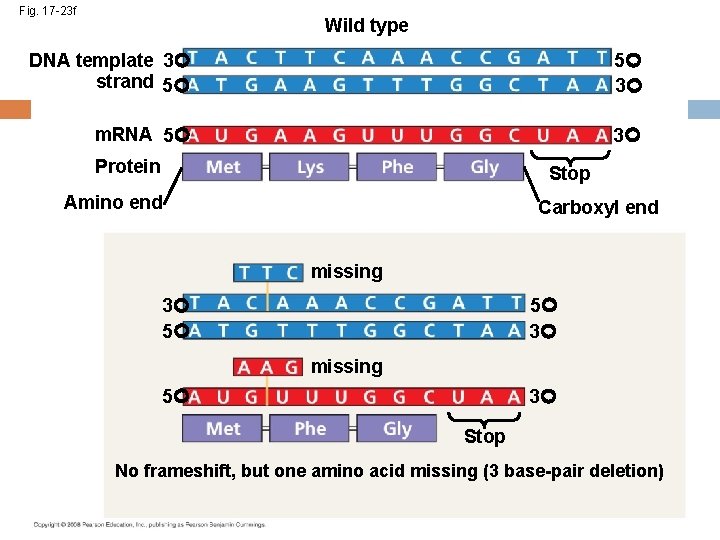
Fig. 17 -23 f Wild type DNA template 3 strand 5 5 3 m. RNA 5 3 Protein Stop Amino end Carboxyl end missing 5 3 3 5 missing 5 3 Stop No frameshift, but one amino acid missing (3 base-pair deletion)

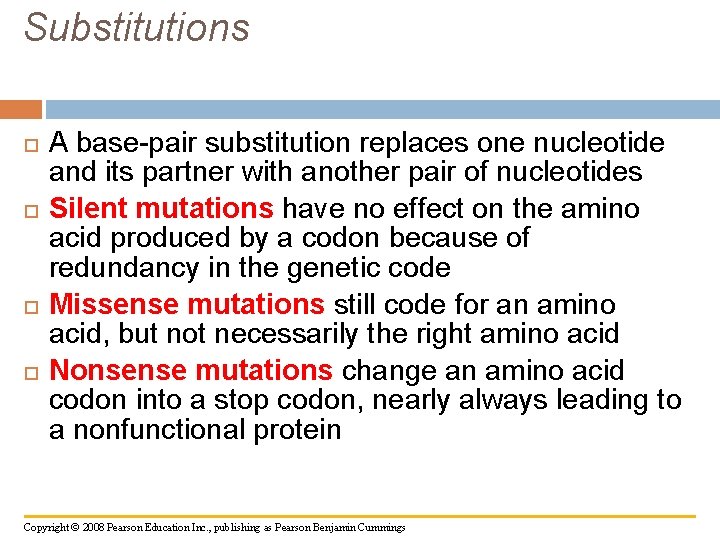
Substitutions A base-pair substitution replaces one nucleotide and its partner with another pair of nucleotides Silent mutations have no effect on the amino acid produced by a codon because of redundancy in the genetic code Missense mutations still code for an amino acid, but not necessarily the right amino acid Nonsense mutations change an amino acid codon into a stop codon, nearly always leading to a nonfunctional protein Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Insertions and Deletions Insertions and deletions are additions or losses of nucleotide pairs in a gene Insertion or deletion of nucleotides may alter the reading frame, producing a frameshift mutation These mutations can change the protein much more than a substitution does, and can be disastrous Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Comparing Gene Expression in Bacteria, Archaea, and Eukarya Bacteria have a terminator sequence, archaea and eukarya do not Bacteria (and probably archaea) can transcribe and translate the same gene at the same time Eukaryotes must do these steps separately, because transcription happens in the nucleus and translation happens in the cytoplasm Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

Fig. 17 -24 RNA polymerase DNA m. RNA Polyribosome RNA polymerase Direction of transcription 0. 25 µm DNA Polyribosome Polypeptide (amino end) Ribosome m. RNA (5 end)

What Is a Gene? Revisiting the Question The idea of the gene itself is a unifying concept of life We have considered a gene as: A discrete unit of inheritance A region of specific nucleotide sequence in a chromosome A DNA sequence that codes for a specific polypeptide chain Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings

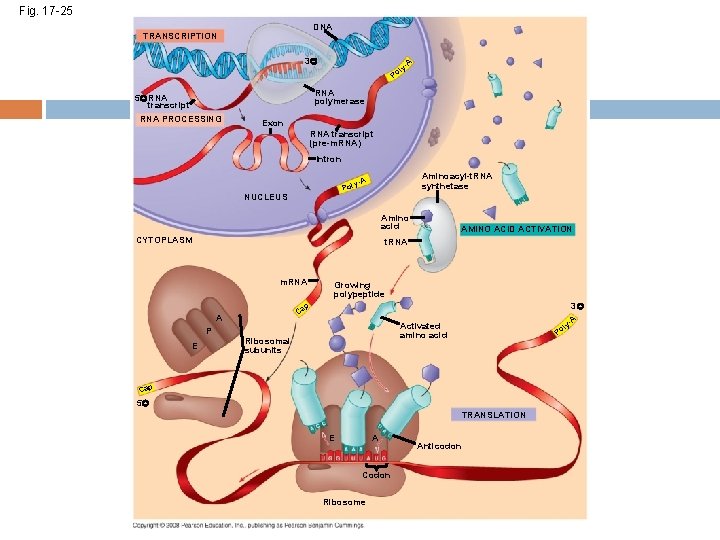
Fig. 17 -25 DNA TRANSCRIPTION 3 l Po A y- RNA polymerase 5 RNA transcript RNA PROCESSING Exon RNA transcript (pre-m. RNA) Intron Aminoacyl-t. RNA synthetase y-A Pol NUCLEUS Amino acid CYTOPLASM AMINO ACID ACTIVATION t. RNA m. RNA Growing polypeptide 3 p Ca A P E A y- Activated amino acid Ribosomal subunits l Po Cap 5 TRANSLATION E A Codon Ribosome Anticodon

In summary, a gene can be defined as a piece of DNA that can be expressed to produce a functional protein and is inherited by organisms from their parents Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
 Mikael ferm
Mikael ferm Protein amino acids
Protein amino acids B-pleated sheet
B-pleated sheet Nucleic acid building block
Nucleic acid building block Monomers protein
Monomers protein Dogma of biology
Dogma of biology Oxidative deamination of glutamate
Oxidative deamination of glutamate Acid base properties of amino acids
Acid base properties of amino acids Amino acid name
Amino acid name Acid base chemistry of amino acids
Acid base chemistry of amino acids Amino acid structure
Amino acid structure Amino acid structure
Amino acid structure Properties of amino acids
Properties of amino acids 20 amino acid structure
20 amino acid structure Aspartate titration curve
Aspartate titration curve Titration curve of amino acids
Titration curve of amino acids Deamination of amino acids
Deamination of amino acids Amino acids
Amino acids Oxidative deamination of amino acids
Oxidative deamination of amino acids Chirality definition
Chirality definition Non essential amino acids in food
Non essential amino acids in food Titration plot
Titration plot Link reaction
Link reaction Isoleucine ketogenic glucogenic
Isoleucine ketogenic glucogenic Glutamate oxidative deamination
Glutamate oxidative deamination Amino acids groups
Amino acids groups Amino acids classification
Amino acids classification Www.chemsheets.co.uk
Www.chemsheets.co.uk Properties of amino acids
Properties of amino acids Epsilon amino
Epsilon amino Serylglycyltyrosylalanylleucine
Serylglycyltyrosylalanylleucine Which amino acids have ionizable side chains
Which amino acids have ionizable side chains Non essential amino acids mnemonics
Non essential amino acids mnemonics Dehydration synthesis of amino acids
Dehydration synthesis of amino acids Aromatic amino acids
Aromatic amino acids Phenol containing amino acids
Phenol containing amino acids Millon's test positive result
Millon's test positive result Dehydration synthesis of amino acids
Dehydration synthesis of amino acids Properties of amino acids
Properties of amino acids Biomedical importance of amino acids
Biomedical importance of amino acids Are amino acids negatively charged
Are amino acids negatively charged Conditionally essential amino acids
Conditionally essential amino acids Essential aa mnemonic
Essential aa mnemonic Is glycine polar or nonpolar
Is glycine polar or nonpolar Amino acids classification
Amino acids classification Amino acids characteristics
Amino acids characteristics Glucogenic amino acid
Glucogenic amino acid Difference between hydrophobic and hydrophilic amino acids
Difference between hydrophobic and hydrophilic amino acids Non essential amino acids mnemonics
Non essential amino acids mnemonics Peptide stereochemistry
Peptide stereochemistry Quantitative estimation of amino acids by ninhydrin method
Quantitative estimation of amino acids by ninhydrin method Protein structure
Protein structure Biliverdin color
Biliverdin color Transdeamination of amino acids
Transdeamination of amino acids Glucose alanine cycle
Glucose alanine cycle Salt bridges in proteins
Salt bridges in proteins Protein ionization
Protein ionization Upon hydrolysis of fibron which amino acids are produced
Upon hydrolysis of fibron which amino acids are produced Alpha carbon amino acid
Alpha carbon amino acid Transdeamination of amino acids
Transdeamination of amino acids Nitrogen balance
Nitrogen balance Glutamate titration curve
Glutamate titration curve What is made of amino acids
What is made of amino acids Wikipedia amino acids
Wikipedia amino acids Meister cycle
Meister cycle What are enzymes made of
What are enzymes made of Importance of sulphur containing amino acids
Importance of sulphur containing amino acids Charged amino acids
Charged amino acids Chemical texture
Chemical texture Net charges of amino acids
Net charges of amino acids 191 amino acid
191 amino acid Amino acids classification
Amino acids classification Nitrogen removal from amino acids
Nitrogen removal from amino acids Amino acid urea
Amino acid urea Difference between glycolysis and pentose phosphate pathway
Difference between glycolysis and pentose phosphate pathway Amino acids table
Amino acids table Francis crick
Francis crick Mixed amino acids
Mixed amino acids Properties of amino acids
Properties of amino acids Glucogenic amino acids
Glucogenic amino acids Importance of amino acids
Importance of amino acids 20 amino acids structures
20 amino acids structures Kr
Kr Abcdrna
Abcdrna Amino acids are joined together in proteins by
Amino acids are joined together in proteins by Qualitative tests for amino acids
Qualitative tests for amino acids Chapter 12 dna and rna
Chapter 12 dna and rna A _________bond joins amino acids together.
A _________bond joins amino acids together. Ornitin protein
Ornitin protein Fawaz aldabbagh
Fawaz aldabbagh Monomers building blocks
Monomers building blocks Monomers of lipids
Monomers of lipids Cellulose starch glycogen
Cellulose starch glycogen Alpha glucose vs beta glucose
Alpha glucose vs beta glucose Specialty monomers
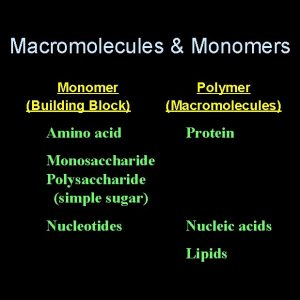
Specialty monomers Macromolecules and their monomers
Macromolecules and their monomers Monomers that make up proteins
Monomers that make up proteins