Chapter 4 Amino acids 1 Structures and general

























![n We obtain the Henderson-Hasselbalch equation: [A-] p. H = p. Ka + log n We obtain the Henderson-Hasselbalch equation: [A-] p. H = p. Ka + log](https://slidetodoc.com/presentation_image_h/e38034594a262f9a858822578fe4402b/image-46.jpg)

- Slides: 51

Chapter 4 Amino acids 1. Structures and general properties 2. Peptide bonds 3. Classifications and characteristics 4. Acidic –basic properties

I. Overview n In this chapter we study the structures and properties of the monomeric units of proteins, the amino acids. n n It is from these substances that proteins are synthesized. Although more than 300 different amino acids have been described in nature: n Only twenty amino acids are commonly found as n Constituents of mammalian proteins, n Is the only amino acids that are coded for by DNA, the genetic material in the cell.

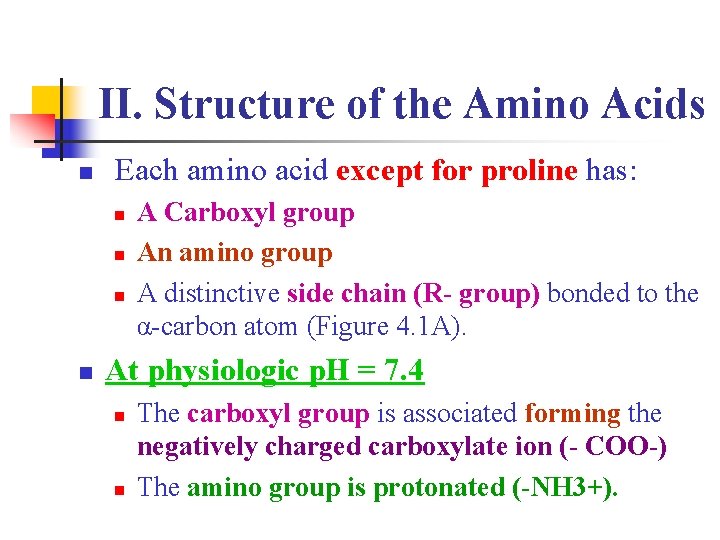
II. Structure of the Amino Acids n Each amino acid except for proline has: n n A Carboxyl group An amino group A distinctive side chain (R- group) bonded to the α-carbon atom (Figure 4. 1 A). At physiologic p. H = 7. 4 n n The carboxyl group is associated forming the negatively charged carboxylate ion (- COO-) The amino group is protonated (-NH 3+).

Figure 4. 1: Structural features of amino acids (shown in their fully protonated form)

n In proteins: n Almost all of these carboxyl and amino groups are combined in peptide linkage n These groups are not available for chemical reaction except for hydrogen bond formation.

n n Thus, it is the nature of the side chains that ultimately dictates the role of an amino acid plays in a protein. It is, useful to classify the amino acids according to the properties of their side chains, that is, whether they are: n Nonpolar (have an even distribution of n electrons) Polar (have an uneven distribution of electrons) such as acids and bases (Figure 4. 2)

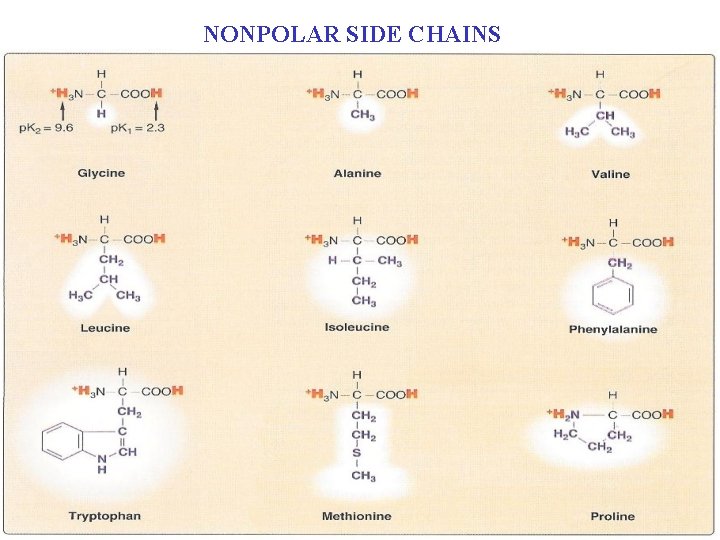
A. Amino acids with nonpolar side chains n Nine of the 20 amino acids have a nonpolar side chain that: n n n Does not bind or give off protons Do not participate in hydrogen or ionic bonds (see Figure 4. 2). The side chains of these amino acids can be thought of as “oily” or lipid-like, n A property that promotes hydrophobic interactions.

n n These amino acids are: Glycine: n First amino acid identified n Found to be component of gelatin n Has the smallest side chain an H atom n The most abundant amino acid in most proteins

Alanine, valine, leucine, and isoleucine: n n Have aliphatic hydrocarbon side chains ranging in size from n A methyl group for Alanine n n n Isomeric butyl groups for leucine and isoleucine These four amino acids are chemically uncreative They play an important role in n Establishing and maintaining the threedimensional structure of proteins because n Of their tendency to gather away from water.

Phenylalanine and tryptophan: n n Contain aromatic side chain, which are characterized by: n Bulk n Nonpolarity, which can buried in the hydrophobic interior of globular proteins. Phenylalanine has a phenyl moiety Tryptophan has an indol group, which can be regarded as indolealanine.

n Methionine: n Has a thiol ether side chain that n Resembles an n-butyl group in many of its physical properties

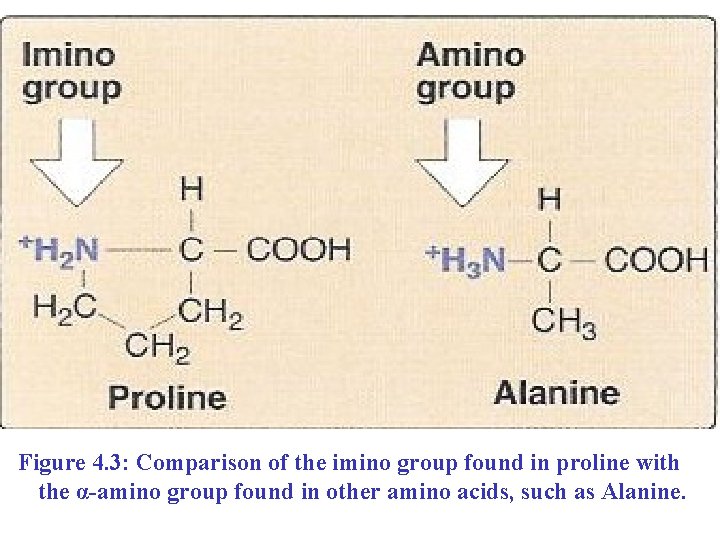
n Proline: n n n Its side chain is bonded to the nitrogen as well as to the α-carbon of the central compound. Because of the bound to nitrogen, proline is technically an imino (-NH-) rather than an amino (-NH 2) acid (Figure 4. 3). It is chemically Not reactive: n Its five-membered side chain, a pyrrolidine ring, restricts the geometry of the backbone chain of the protein that contains it.

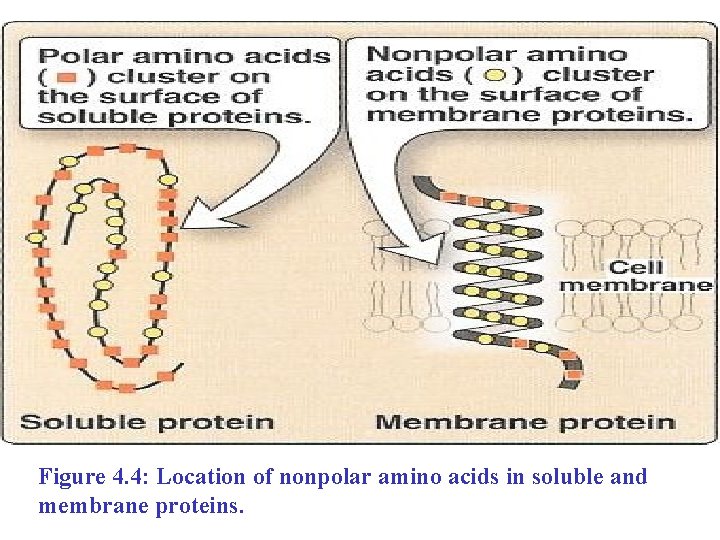
1. Location of nonpolar amino acids in proteins A) In proteins found in aqueous solutions, the side chains of the nonpolar amino acid n Tend to cluster together in the interior of the protein (Figure 4. 4). n n This phenomenon is the result of the hydrophobicity of the nonpolar R-groups, which act much like droplets of oil that coalesce in an aqueous environment. The nonpolar R-groups thus fill up the interior of the folded protein and help give it its three- dimensional shape.

B) In proteins that are located in a hydrophobic n environment, such as membrane: n The nonpolar R-groups are found on the outside surface of the protein, n Interacting with the lipid environment. These hydrophobic interactions are important in stabilizing protein structure.

NONPOLAR SIDE CHAINS

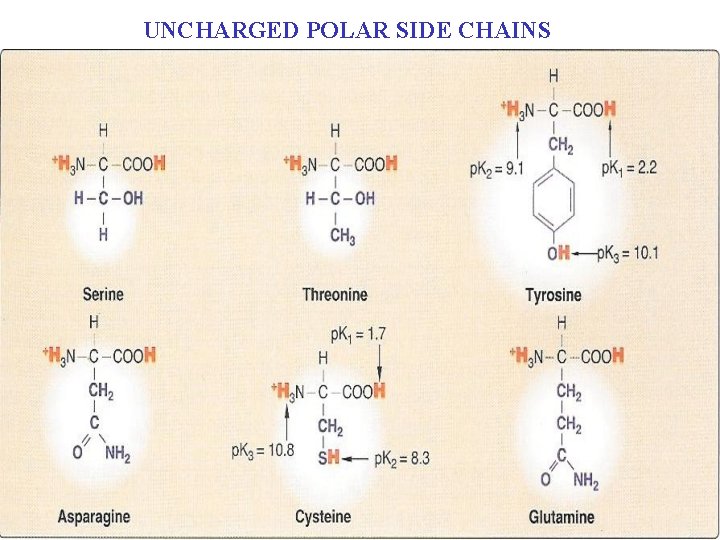
UNCHARGED POLAR SIDE CHAINS

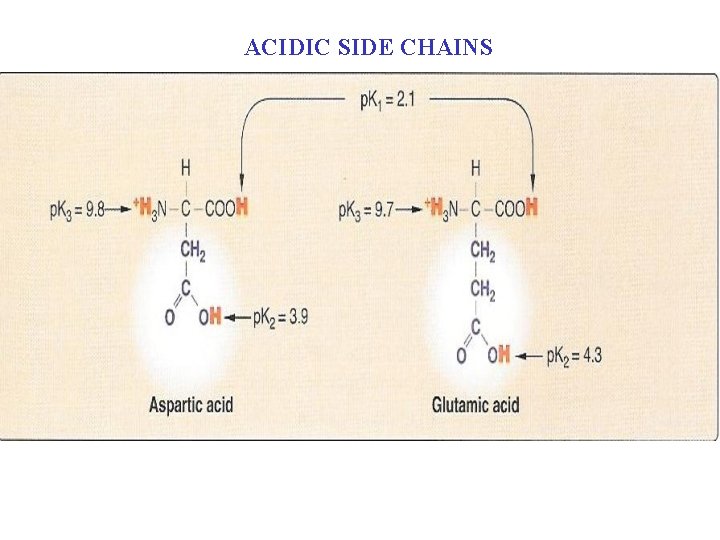
ACIDIC SIDE CHAINS

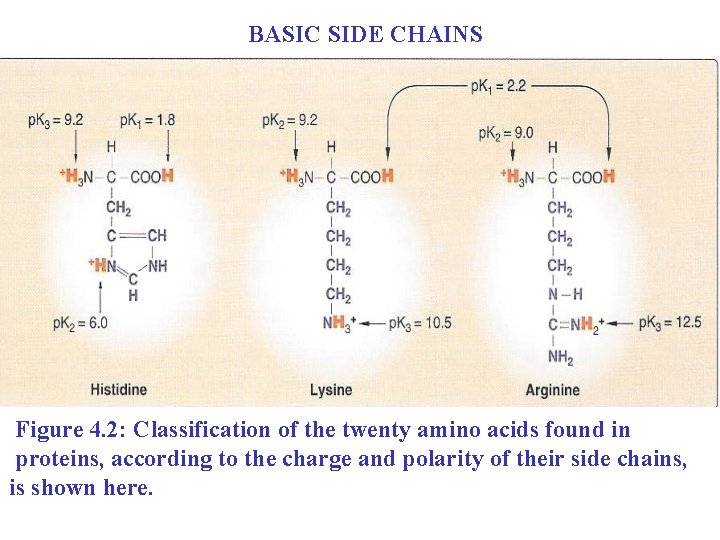
BASIC SIDE CHAINS Figure 4. 2: Classification of the twenty amino acids found in proteins, according to the charge and polarity of their side chains, is shown here.

Figure 4. 3: Comparison of the imino group found in proline with the α-amino group found in other amino acids, such as Alanine.

Figure 4. 4: Location of nonpolar amino acids in soluble and membrane proteins.

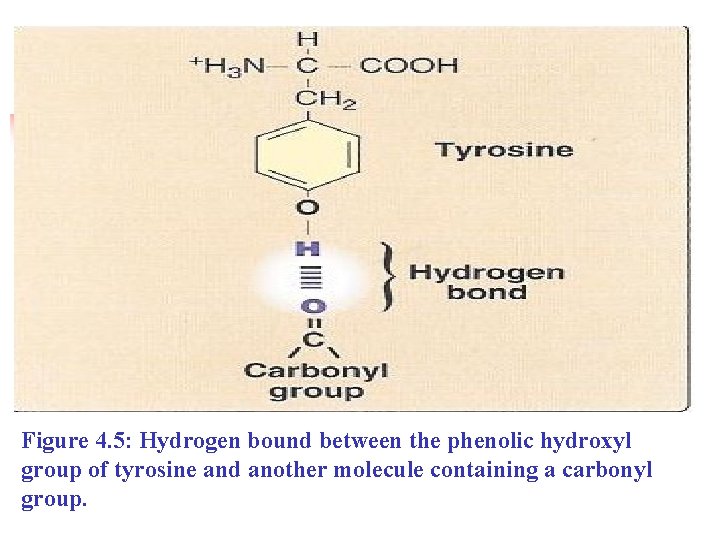
B. Amino acids with uncharged polar side chains n n They are six amino acids that have uncharged (zero net charge at neutral p. H) polar side chain. These amino acids are: Serine, threonine, and tyrosine: n Each contains a polar hydroxyl group of different sizes that can n Participate in hydrogen bond formation (Figure 4. 5).

Figure 4. 5: Hydrogen bound between the phenolic hydroxyl group of tyrosine and another molecule containing a carbonyl group.

n Serine threonine, and, rarely, tyrosine n n Contain a polar hydroxyl group that can n Serve as a site of attachment for structures such as a phosphate group. n The side chain of serine is an important component of the active site of many enzymes. The hydroxyl group of serine or threonine, can serve as a site of attachment for oligosaccharide chains in glycoproteins

Asparagines and glutamine: n n n Each contains a carbonyl group and an amide group, both of which can also participate in hydrogen bonds. The side chain of asparagines and glutamine can cross-link folded proteins by hydrogen bonds. They are the amides derived from two other amino acids, aspartate and glutamate. The amide group of asparagines, can serve as a site of attachment for oligosaccharide chains in glycoproteins

Cysteine: n n n The side chain of cysteine contains a sulfhydryl group (-SH), which is n An important component of the active site of many enzymes. In proteins, the –SH group of two cysteines can become oxidized to form a dimer, cystine n Which contains a covalent cross-link called a disulfide bond (-S-S-).

C. Amino acids with acidic side chains They are two amino acids with a polar and negatively charged side chain n n Aspartic acid and glutamic acid: n At neutral p. H they are proton donors: The side chains of these amino acids are fully ionized, containing a negatively charged carboxylate group (-COO-). They are, therefore, called aspartate or glutamate emphasize that these amino acids are negatively charged at physiologic p. H. n n

D. Amino acids with basic side chains n n They are three amino acids with a polar and positively charged side chain. The side chains of the basic amino acids accept protons (see Figure 4. 2). 1) Lysine: n The side chain contains a primary amine (NH 2) attached to the terminal carbon

It is a strong base; at p. H 7. 0, the predominant ionic species is the ammonium ion, -NH 3+. (The ammonium ion, of course, is a weak acid. ) n At physiologic p. H the side chains of lysine is: n Fully ionized n Positively charged n

2) Arginine: n n n Is the most basic amino acid Bears a guanidine group At physiologic p. H the side chains of arginine is n Fully ionized and n Positively charged to guanidinium ion

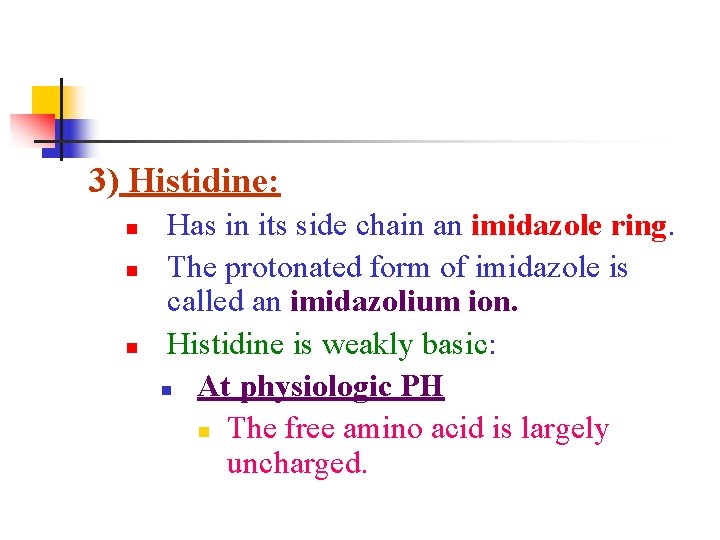
3) Histidine: n n n Has in its side chain an imidazole ring. The protonated form of imidazole is called an imidazolium ion. Histidine is weakly basic: n At physiologic PH n The free amino acid is largely uncharged.

When histidine is incorporated into a protein, its side chain can be either: n Positively charged n Neutral depending on the ionic environment provided by the polypeptide chains of the protein. n This is an important property of histidine that contributes to the role it plays in the functioning of proteins such as n n Hemoglobin

E. Abbreviations and symbols for the commonly occurring amino n n Each amino acid name has an associated n Three-letter abbreviation and n One-letter symbol (Figure 3. 6). These abbreviations are, in most cases, taken from the first three letters of the corresponding amino acid’s name; they are conversationally pronounced as read.

n The one-letter codes are determined by the following rules: n Unique first letter: If only one amino acid begins with a particular letter, then that letter is used as symbol. For example, I = isoleucine. n Most commonly occurring amino acids have priority: If more than one amino acid begins with a particular letter, the most common of these amino acids receives this letter as its symbol. n For example, Glycine is more common than glutamate, so G = glycine.

n n n Similar sounding names: Some one-letter symbols sound like the amino acid they represent. For example, F = Phenylalanine, or W = tryptophan Letter close to initial letter: For the remaining amino acids, as possible to the initial of the amino acid. Further B is assigned to Asx, signifying either aspartic acid or asparagines, Z is assigned to Glx, signifying either aspartic acid or glutamine, and X is assigned to an unidentified amino acid.

Figure 4. 6

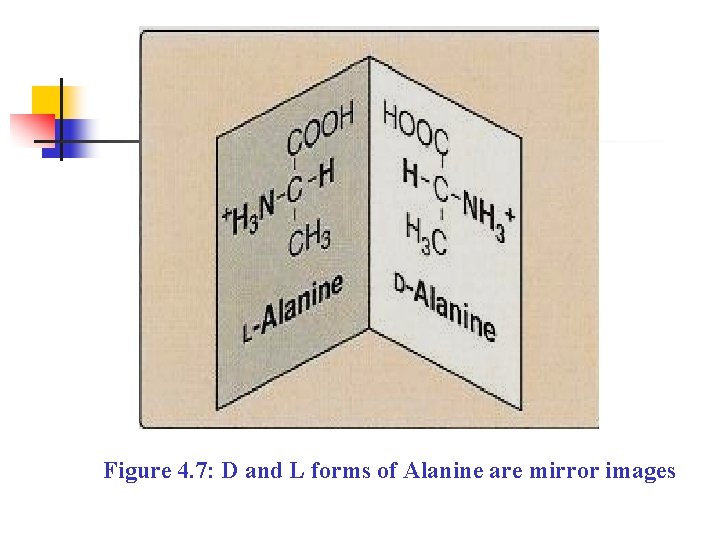
Figure 4. 7: D and L forms of Alanine are mirror images

F. Optical properties of amino acids n A chiral or optically active carbon atom: n n Is the α -carbon of each amino acid is attached to four different chemical groups and is, therefore: Glycine is the exception n Because its α -carbon has two hydrogen substituents and, therefore, is optically inactive.

n n Amino acids that have an a symmetric center at the α -carbon can exist in two forms, n Designated D and L, which are mirror images of each other (Figure 4. 7). The two forms in each pair are termed stereoisomers, optical isomers, or Enantiomers All amino acids found in proteins are of the Lconfiguration. However, D-amino acids are found in some antibiotics and bacterial cell walls.

III. General properties of amino acids n n n The p. K values of the 20 “standard” a-amino acids of proteins are tabulated in (Figure 4. 2). Here p. K 1 and p. K 2, respectively, refer to α-carboxylic acid and α-amino group, and p. KR refers to the side groups with acid-base properties. It indicates that the p. K values of the a α -carboxylic acid groups lie in a small range around 2. 2 so that above p. H 3. 5 these groups are almost entirely in their carboxylate forms.

n n The α -amino groups all have p. K values near 9. 4 and are therefore almost entirely in their ammonium ion forms below p. H 8. 0. This leads to a important structural point: n In the physiological p. H range, both the carboxylic acid and the amino groups of αamino acids are: n Completely ionized (Fig. 4. 2).

n n An amino acid can therefore act as either an acid or a base. Substances with this property are said to be amphoteric and are referred to as ampholytes (amphoteric electrolytes). Molecules that bear charged groups of opposite polarity are known as zwitterions.

n Because amino acids are zwitterions: Their physical properties are characteristic of ionic compounds. n Most α -amino acids have melting points near 300°C, whereas n Their nonionic derivatives usually melt around 100°C. n Amino acids, like other ionic compounds, are more soluble in polar solvent than in nonpolar solvents. Indeed, most α -amino acids are very soluble in water but: n Are Insoluble in most organic solvents. n n

1) Acid – base properties n Amino acids in aqueous solution contain: Weakly acidic α-carboxyl groups n Weakly basic α-amino groups n Ionizable group in its side chain. Both free amino acids and some amino acids combined in peptide linkages can act as buffers. The quantitative relationship between the concentration of a weak acid (HA) and its conjugate base (A-) is described by the Henderson-Hasselbalch equation. n n n

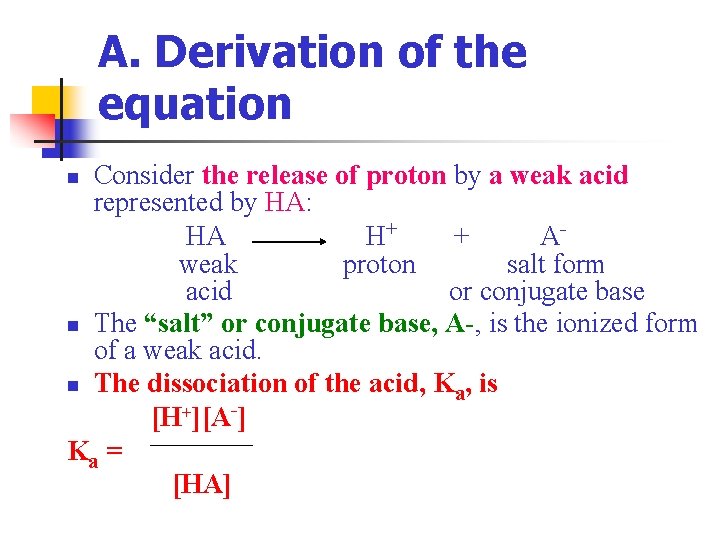
A. Derivation of the equation Consider the release of proton by a weak acid represented by HA: HA H+ + Aweak proton salt form acid or conjugate base n The “salt” or conjugate base, A-, is the ionized form of a weak acid. n The dissociation of the acid, Ka, is [H+][A-] Ka = [HA] n

n n n The larger the Ka, the stronger the acid n Because most of the HA has been converted into H+ and A-. The smaller the Ka, the weaker the acid n The less acid has dissociated By solving for the [H+] in the above equation, taking the logarithm of both sides of the equation, multiplying both sides of the equation by – 1, Substituting p. H = -log [H+] p. Ka = -log Ka
![n We obtain the HendersonHasselbalch equation A p H p Ka log n We obtain the Henderson-Hasselbalch equation: [A-] p. H = p. Ka + log](https://slidetodoc.com/presentation_image_h/e38034594a262f9a858822578fe4402b/image-46.jpg)
n We obtain the Henderson-Hasselbalch equation: [A-] p. H = p. Ka + log [HA] (1. 13)

B. Buffers n n A buffer is a solution that resists change in p. H following the addition of an acid or base. A buffer can be created by: Mixing a weak acid (HA) with its conjugate base (A-). If an acid such as HCI is added to such a solution, A- can neutralize it, in the process being converted to HA. If abase is added, HA can neutralize it, in the process being converted to A-. n n n

n Maximum buffering capacity occurs at p. H equal to the p. Ka: But a conjugate acid/base pair can still serve as an effective buffer when the p. H of solution is within approximately +or - 1 p. H unit of the p. Ka If the amounts of HA and A- are equal: n The p. H is equal to the p. K a n n

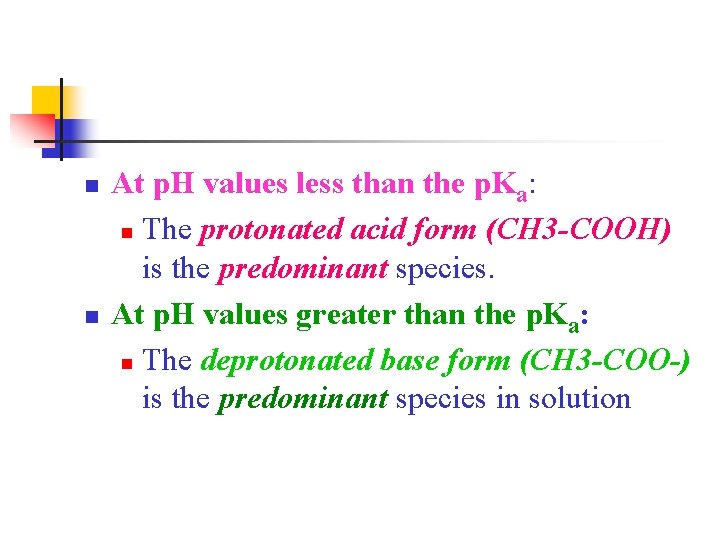
n n At p. H values less than the p. Ka: n The protonated acid form (CH 3 -COOH) is the predominant species. At p. H values greater than the p. Ka: n The deprotonated base form (CH 3 -COO-) is the predominant species in solution

2) Peptide bonds properties n n n The α - amino acids polymerize, through the elimination of a water molecule: n The resulting CO-NH linkage, which is known as a peptide bond. Polymers composed of two, three, a few (310) amino acid residues are called “peptides” Proteins are molecules that consist of one or more polypeptide chains.

n n These polypeptides range in length form: n ~40 to~33, 000 amino acid residues n Few have more than 1500 residues Polypeptides are linear polymers: n That is, each amino acid residue is linked to its neighbors in a head-to-tail fashion rather than forming branched chains.