Tecniche per lanalisi di mutazioni Vincenzo Nigro Dipartimento















- Slides: 51

Tecniche per l’analisi di mutazioni Vincenzo Nigro Dipartimento di Patologia Generale, Seconda Università degli Studi di Napoli Telethon Institute of Genetics and Medicine (TIGEM)

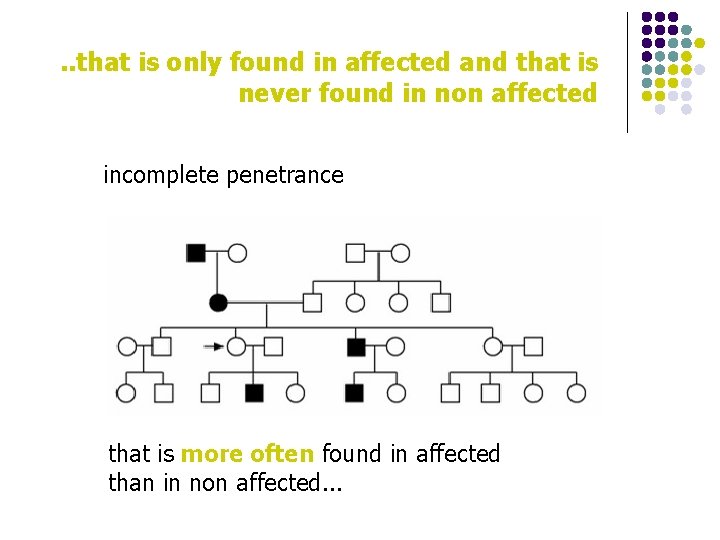
What is a mutation? a variation of the DNA sequence…. l . . that is only found in affected individuals l . . that is never found in non affected individuals l . . that accounts for the pathological process/status l . . that, when corrected in time, disease is rescued

. . that is only found in affected and that is never found in non affected incomplete penetrance that is more often found in affected than in non affected. . .

50. 000 private variants = innocuous differences belonging to one family CCCCAGCCTCCTTGCCAACGCCCCCTTTCCCTCTCCCCCTCCCGCTCGGCGCTGACC CCCCATCCCCACCCCCGTGGGAACACTGGGAGCCTGCACTCCACAGACCCTCTCCTT GCCTCTTCCCTCAGCCTCCGCTCCCCGCCCTCTTCCCGGCCCAGGGCGCCG GCCCACCCTTCCCTCCGCCGCCCCCCGGCCGCGGGGAGGACATGGCCGCGCACAG GCCGGTGGAATGGGTCCAGGCCGTGGTCAGCCGCTTCGACGAGCAGCTTCCAATAA AAACAGGACAGCAGAACACACATACCAAAGTCAGTACTGAGCACAACAAGGAATGTC TAATCAATATTTCCAAATACAAGTTTTCTTTGGTTATAAGCGGCCTCACTACTATTTTAA AGAATGTTAACAATATGAGAATATTTGGAGAAGCTGCTGAAAAAAATTTATATCTCTCT CAGTTGATTATATTGGATACACTGGAAAAATGTCTTGCTGGGCAACCAAAGGACACAA TGAGATTAGATGAAACGATGCTGGTCAAACAGTTGCTGCCAGAAATCTGCCATTTTCT TCACACCTGTCGTGAAGGAAACCAGCATGCAGCTGAACTTCGGAATTCTGCCTCTGG GGTTTTATTTTCTCTCAGCTGCAACAACTTCAATGCAGTCTTTAGTCGCATTTCTACCA GGTTACAGGAATTAACTGTTCAGAAGACAATGTTGATGTTCATGATATAGAATTG TTACAGTATATCAATGTGGATTGTGCAAAATTAAAACGACTCCTGAAGGAAACAGCAT TTAAATTTAAAGCCCTAAAGAAGGTTGCGCAGTTATAAATAGCCTGGAAAA GGCATTTTGGAACTGGGTAGAAAATTATCCAGATGAATTTACAAAACTGTACCAGATC CCACAGACTGATATGGCTGAATGTGCAGAAAAGCTATTTGACTTGGTGGATGGTTTTG CTGAAAGCACCAAACGTAAAGCAGCAGTTTGGCCACTACAAATCATTCTCCTTATCTT GTGTCCAGAAATAATCCAGGATATATCCAAAGACGTGGTTGATGAAAACAACATGAAT AAGAAGTTATTTCTGGACAGTCTACGAAAAGCTCTTGCTGGCCATGGAGGAAGTAGG CAGCTGACAGAAAGTGCTGCAATTGCCTGTGTCAAACTGTGTAAAGCAAGTACTTACA TCAATTGGGAAGATAACTCTGTCATTTTCCTACTTGTTCAGTCCATGGTGGTTGATCTT AAGAACCTGCTTTTTAATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATG TGGATCTAATGATTGACTGCCTTGTTTCTTGCTTTCGTATAAGCCCTCACAACAACCAA CACTTTAAGATCTGCCTGGCTCAGAATTCACCTTCTACATTTCACTATGTGCTGGTAAA TTCACTCCATCGAATCATCACCAATTCCGCATTGGTGGCCTAAGATTGATGCT GTGTATTGTCACTCGGTTGAACTTCGAAATATGTTTGGTGAAACACTTCATAAAGCAGT GCAAGGTTGTGGAGCACACCCAGCAATACGAATGGCACCGAGTCTTACATTTAAAGA AAAAGTAACAAGCCTTAAATTTAAAGAAAAACCTACAGACCTGGAGACAAGAAGCTAT AAGTATCTTCTCTTGTCCATGGTGAAACTAATTCATGCAGATCCAAAGCTCTTGCTTTG TAATCCAAGAAAACAGGGGCCCGAAACCCAAGGCAGTACAGCAGAATTACAGG GCTCGTCCAACTGGTCCCTCAGTCACACATGCCAGAGATTGCTCAGGAAGCAATGGA GGCTCTGCTGGTTCTTCATCAGTTAGATAGCATTGATTTGTGGAATCCTGATGCTCCT GTAGAAACATTTTGGGAGATTAGCTCACAAATGCTTTTTTACATCTGCAAGAAATTAAC TAGTCATCAAATGCTTAGTAGCACAGAAATTCTCAAGTGGTTGCGGGAAATATTGATC TGCAGGAATAAATTTCTTCTTAAAAATAAGCAGATAGAAGTTCCTGTCACTTTC CCCCAGCCTCCTTGCCAACGCCCCCTTTCCCTCTCCCCCTCCCGCTCGGCGCTGACC CCCCATCCCCACCCCCGTGGGAACACTGGGAGCCTGCACTCCACAGACCCTCTCCTT GCCTCTTCCCTCAGCCTCCGCTCCCCGCCCTCTTCCCGGCCCAGGGCGCCG GCCCACCCTTCCCTCCGCCGCCCCCCGGCCGCGGGGAGGACATGGCCGCGCACAG GCCGGTGGAATGGGTCCAGGCCGTGGTCAGCCGCTTCGACGAGCAGCTTCCAATAA AAACAGGACAGCAGAACACACATACCAAAGTCAGTACTGAGCACAACAAGGAATGTC TAATCAATATTTCCAAATACAAGTTTTCTTTGGTTATAAGCGGCCTCACTACTATTTTAA AGAATGTTAACTATATGAGAATATTTGGAGAAGCTGCTGAAAAAAATTTATATCTCTCT CAGTTGATTATATTGGATACACTGGAAAAATGTCTTGCTGGGCAACCAAAGGACACAA TGAGATTAGATGAAACGATGCTGGTCAAACAGTTGCTGCCAGAAATCTGCCATTTTCT TCACACCTGTCGTGAAGGAAACCAGCATGCAGCTGAACTTCGGAATTCTGCCTCTGG GGTTTTATTTTCTCTCAGCTGCAACAACTTCAATGCAGTCTTTAGTCGCATTTCTACCA GGTTACAGGAATTAACTGTTCAGAAGACAATGTTGATGTTCATGATATAGAATTG TTACAGTATATCAATGTGGATTGTGCAAAATTAAAACGACTCCTGAAGGAAACAGCAT TTAAATTTAAAGCCCTAAAGAAGGTTGCGCAGTTATAAATAGCCTGGAAAA GGCATTTTGGAACTGGGTAGAAAATTATCCAGATGAATTTACAAAACTGTACCAGATC CCACAGACTGATATGGCTGAATGTGCAGAAAAGCTATTTGACTTGGTGGATGGTTTTG CTGAAAGCACCAAACGTAAAGCAGCAGTTTGGCCACTACAAATCATTCTCCTTATCTT GTGTCCAGAAATAATCCAGGATATATCCAAAGACGTGGTTGATGAAAACAACATGAAT AAGAAGTTATTTCTGGACAGTCTACGAAAAGCTCTTGCTGGCCATGGAGGAAGTAGG CAGCTGACAGAAAGTGCTGCAATTGCCTGTGTCAAACTGTGTAAAGCAAGTACTTACA TCAATTGGGAAGATAACTCTGTCATTTTCCTACTTGTTCAGTCCATGGTGGTTGATCTT AAGAACCTGCTTTTTAATCCAAGTAAGCCATTCTCAAGAGGCAGTCAGCCTGCAGATG TGGATCTAATGATTGACTGCCTTGTTTCTTGCTTTCGTATAAGCCCTCACAACAACCAA CACTTTAAGATCTGCCTGGCTCAGAATTCACCTTCTACATTTCACTATGTGCTGGTAAA TTCACTCCATCGAATCATCACCAATTCCGCATTGGTGGCCTAAGATTGATGCT GTGTATTGTCACTCGGTTGAACTTCGAAATATGTTTGGTGAAACACTTCATAAAGCAGT GCAAGGTTGTGGAGCACACCCAGCAATACGAATGGCACCGAGTCTTACATTTAAAGA AAAAGTAACAAGCCTTAAATTTAAAGAAAAACCTACAGACCTGGAGACAAGAAGCTAT AAGTATCTTCTCTTGTCCATGGTGAAACTAATTCATGCAGCTCCAAAGCTCTTGCTTTG TAATCCAAGAAAACAGGGGCCCGAAACCCAAGGCAGTACAGCAGAATTACAGG GCTCGTCCAACTGGTCCCTCAGTCACACATGCCAGAGATTGCTCAGGAAGCAATGGA GGCTCTGCTGGTTCTTCATCAGTTAGATAGCATTGATTTGTGGAATCCTGATGCTCCT GTAGAAACATTTTGGGAGATTAGCTCACAAATGCTTTTTTACATCTGCAAGAAATTAAC TAGTCATCAAATGCTTAGTAGCACAGAAATTCTCAAGTGGTTGCGGGAAATATTGATC TGCAGGAATAAATTTCTTCTTAAAAATAAGCAGATAGAAGTTCCTGTCACTTTC

1 -allele diseases l l monoallelic mutations may be responsible for dominant or X-linked disorders new random mutations are the rule with an unpredictable pattern of distribution

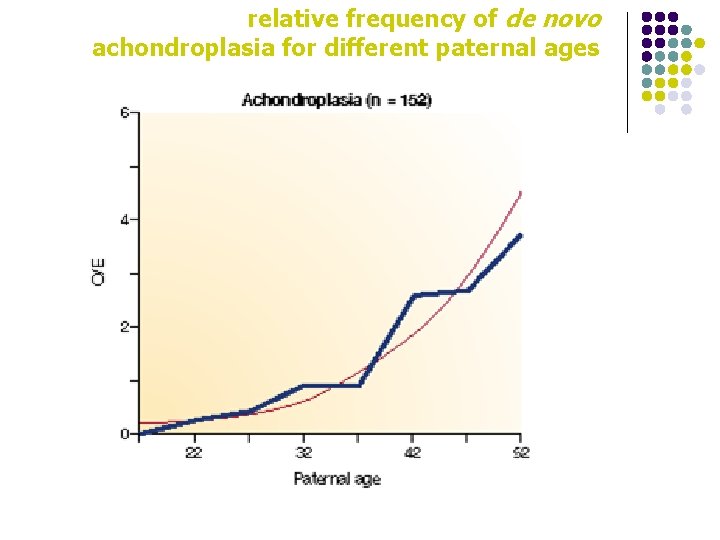
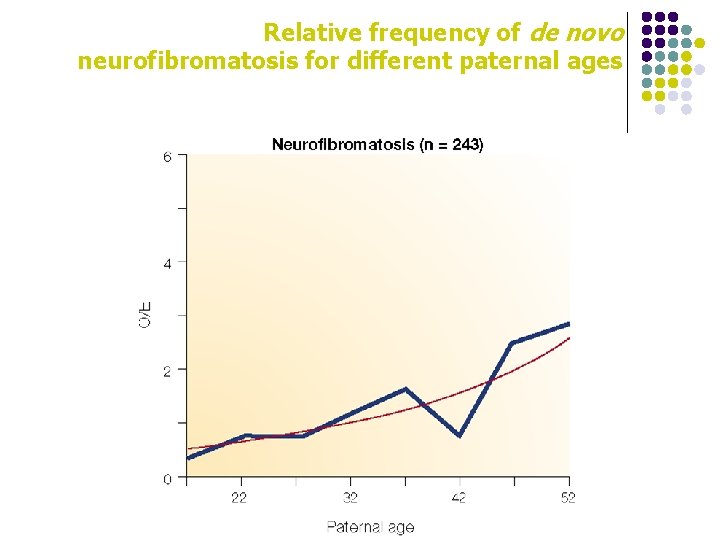
gender effect in mutations l l For mutations other than point mutations, sex biases in the mutation rate are very variable Small deletions are more frequent in females Germline base substitution mutations occur more frequently in males than in females, especially in older males Point mutations at some loci occur almost exclusively in males, whereas others occur ten times more than in females

relative frequency of de novo achondroplasia for different paternal ages

Relative frequency of de novo neurofibromatosis for different paternal ages

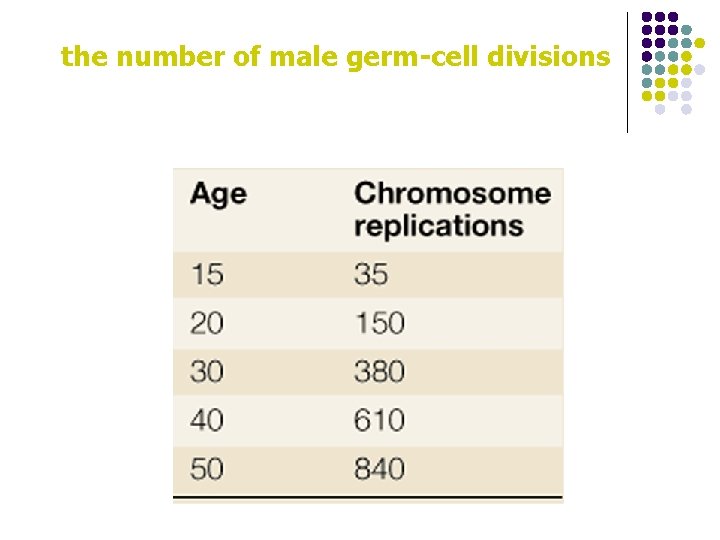
the number of male germ-cell divisions


2 -allele diseases l novel mutations are rare, usually mutations have a long history (100 -1000 generations) l mutations have an ethnical signature with a predictable pattern of distribution and frequency l biallelic mutations may be responsible for autosomal recessive disorders l polymorphisms and private variants are more easily discriminated vs true mutations

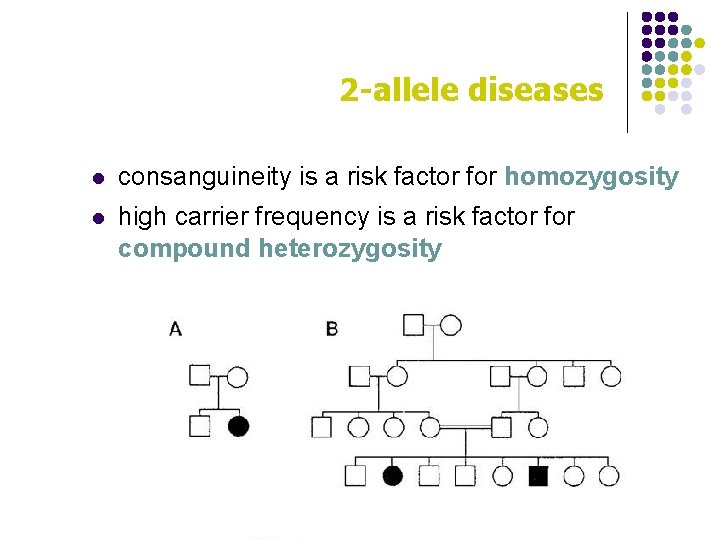
2 -allele diseases l consanguineity is a risk factor for homozygosity l high carrier frequency is a risk factor for compound heterozygosity

The effect of an allele l null or amorph = no product l hypomorph = reduced amount / activity l hypermorph = increased amount / activity l neomorph = novel product / activity l antimorph = antagonistic product / activity

Mutation detection l mutation scanning l l or resequencing methods for identifying previously unknown mutations genotyping l methods for scoring previously known mutations or single nucleotide polymorphisms (SNPs)

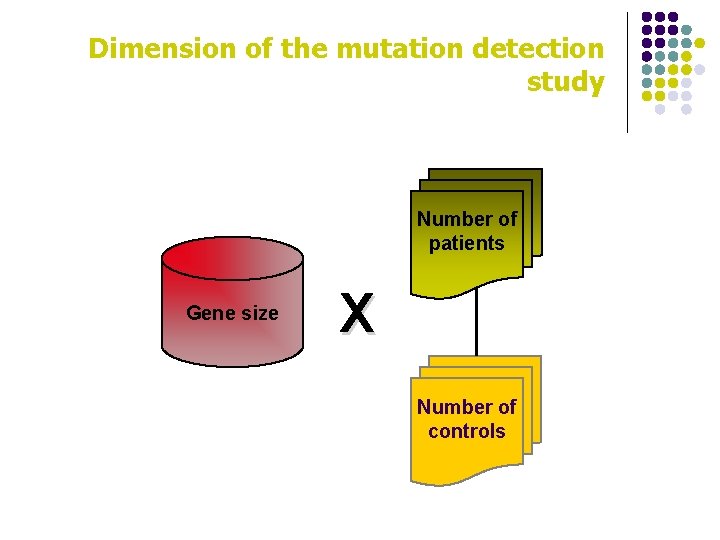
Key questions for mutation detection strategy l expected mutations are monoallelic or biallelic? l is the gene well recognized for that disease? l is the mutation pattern known? (deletion, dup, small mutations, etc. ) l which is the complexity of the gene? l how many patients must be examined? l how many controls should be examined? l how many mutations and how many variations have already been identified in this gene? l are there more members of the same gene family (or pseudogenes) in the genome?

Dimension of the mutation detection study Number of patients Gene size X Number of controls

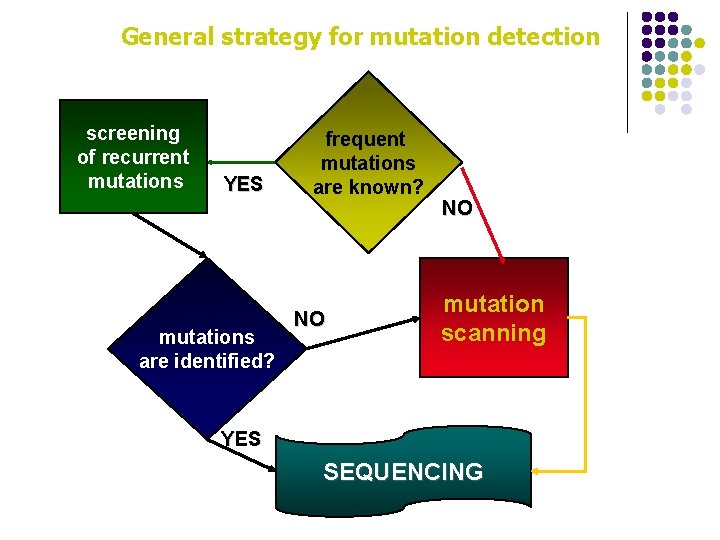
General strategy for mutation detection screening of recurrent mutations YES mutations are identified? frequent mutations are known? NO NO mutation scanning YES SEQUENCING

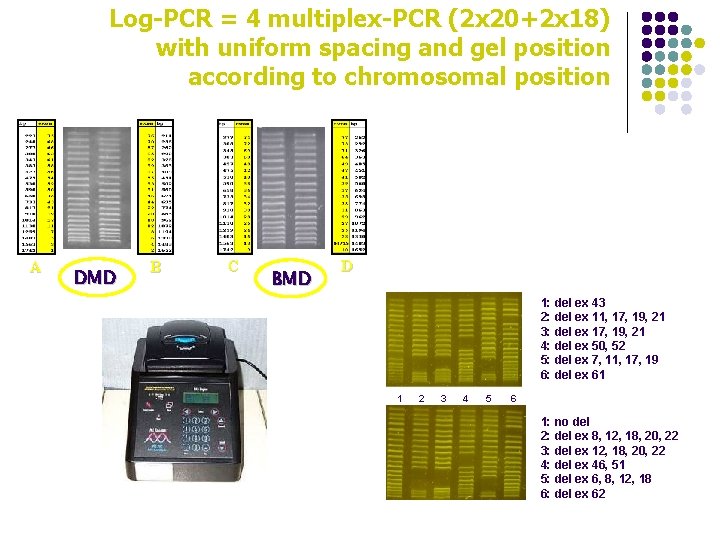
Log-PCR = 4 multiplex-PCR (2 x 20+2 x 18) with uniform spacing and gel position according to chromosomal position A DMD B C BMD D 1: del ex 43 2: del ex 11, 17, 19, 21 3: del ex 17, 19, 21 4: del ex 50, 52 5: del ex 7, 11, 17, 19 6: del ex 61 1 2 3 4 5 6 1: no del 2: del ex 8, 12, 18, 20, 22 3: del ex 12, 18, 20, 22 4: del ex 46, 51 5: del ex 6, 8, 12, 18 6: del ex 62

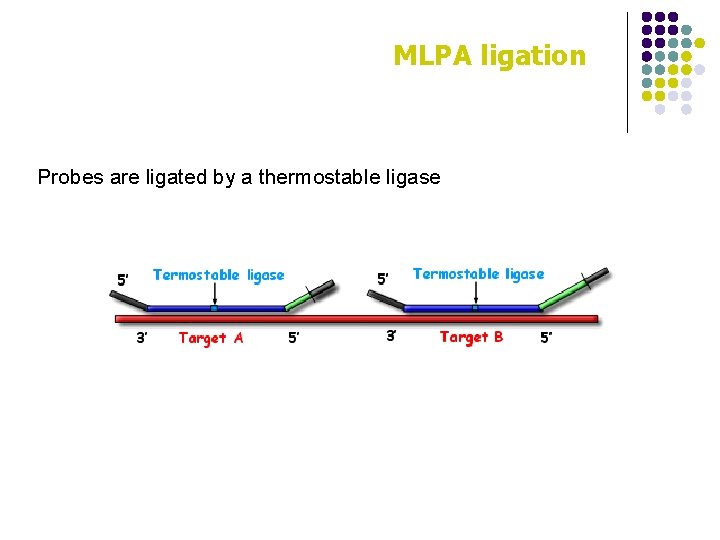
MLPA ligation Probes are ligated by a thermostable ligase

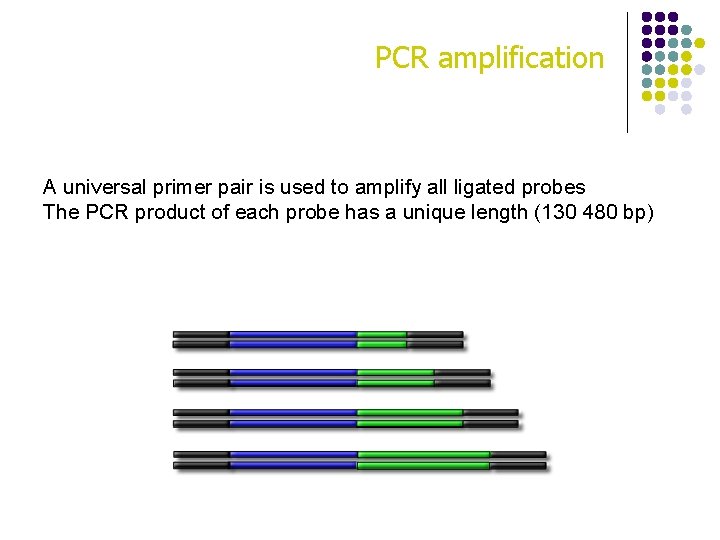
PCR amplification A universal primer pair is used to amplify all ligated probes The PCR product of each probe has a unique length (130 480 bp)

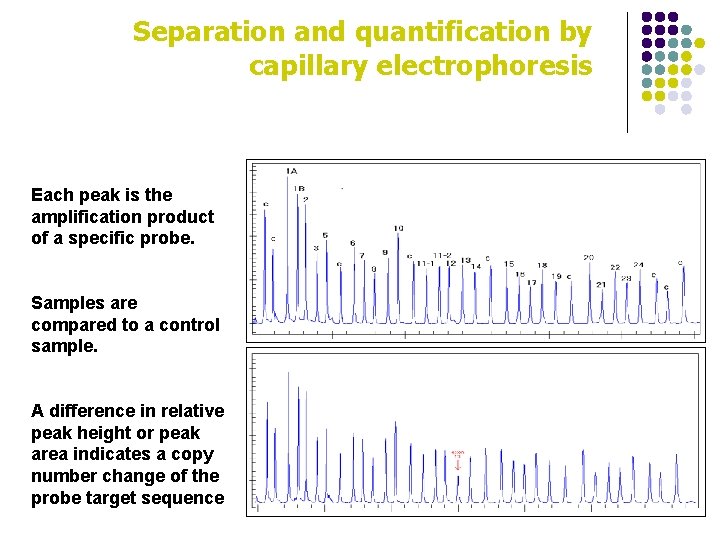
Separation and quantification by capillary electrophoresis Each peak is the amplification product of a specific probe. Samples are compared to a control sample. A difference in relative peak height or peak area indicates a copy number change of the probe target sequence

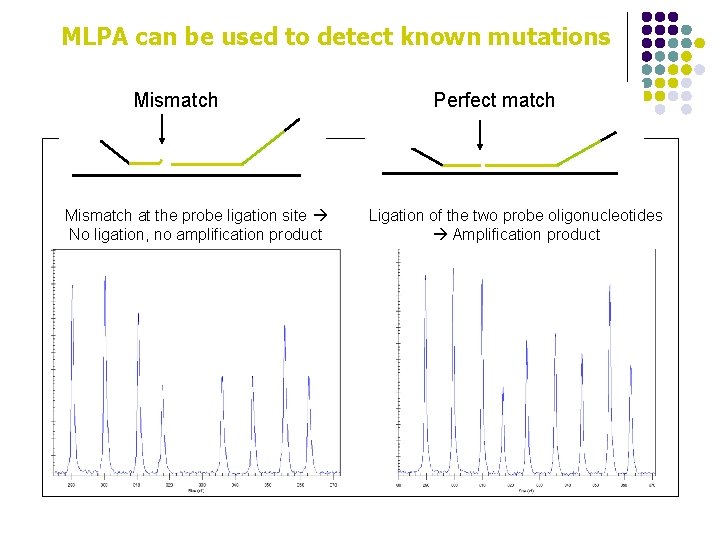
MLPA can be used to detect known mutations Mismatch at the probe ligation site No ligation, no amplification product Perfect match Ligation of the two probe oligonucleotides Amplification product

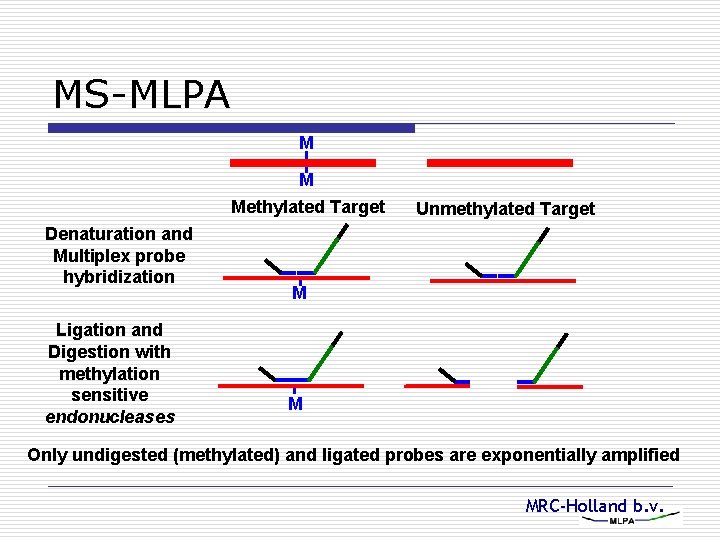
MS-MLPA M M Methylated Target Denaturation and Multiplex probe hybridization Ligation and Digestion with methylation sensitive endonucleases Unmethylated Target M M Only undigested (methylated) and ligated probes are exponentially amplified MRC-Holland b. v.

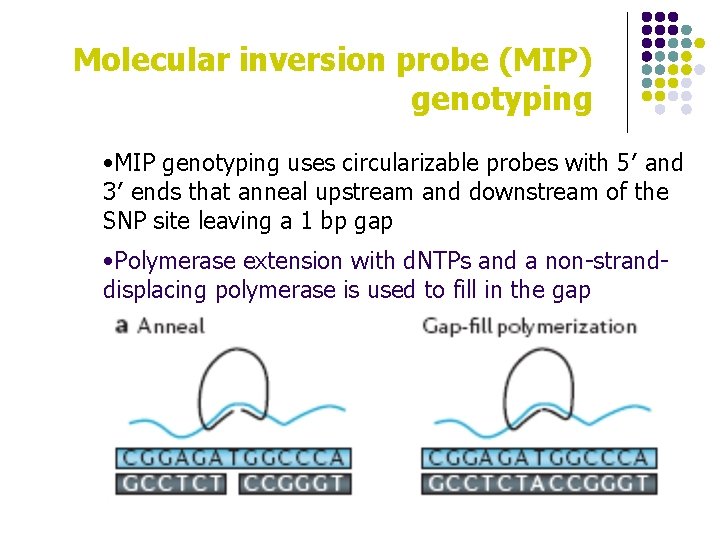
Molecular inversion probe (MIP) genotyping • MIP genotyping uses circularizable probes with 5′ and 3′ ends that anneal upstream and downstream of the SNP site leaving a 1 bp gap • Polymerase extension with d. NTPs and a non-stranddisplacing polymerase is used to fill in the gap

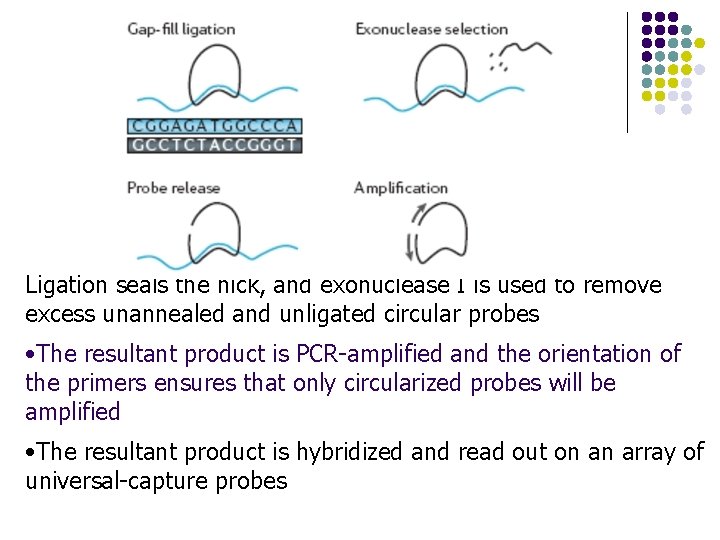
Ligation seals the nick, and exonuclease I is used to remove excess unannealed and unligated circular probes • The resultant product is PCR-amplified and the orientation of the primers ensures that only circularized probes will be amplified • The resultant product is hybridized and read out on an array of universal-capture probes

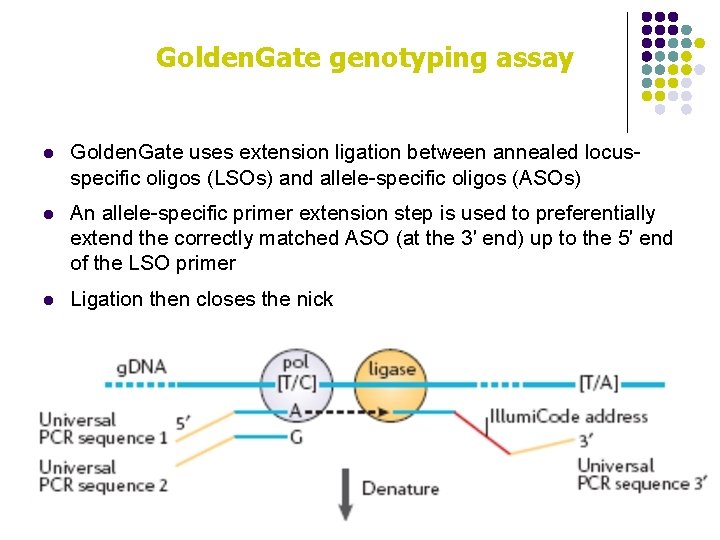
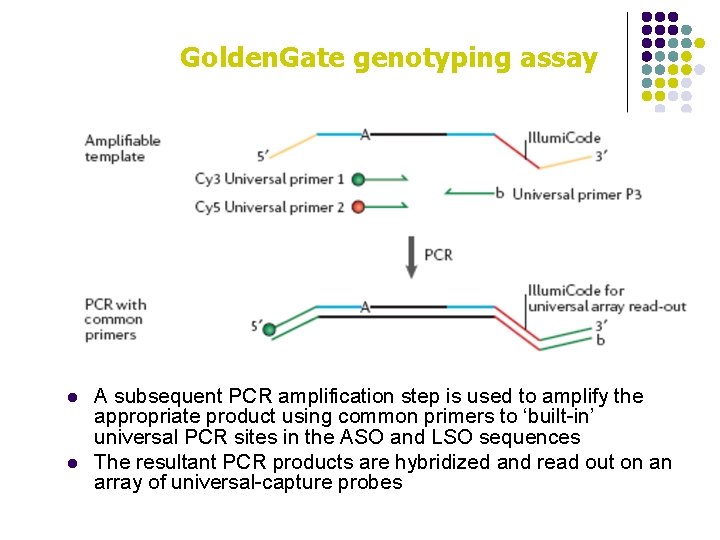
Golden. Gate genotyping assay l Golden. Gate uses extension ligation between annealed locusspecific oligos (LSOs) and allele-specific oligos (ASOs) l An allele-specific primer extension step is used to preferentially extend the correctly matched ASO (at the 3′ end) up to the 5′ end of the LSO primer l Ligation then closes the nick

Golden. Gate genotyping assay l l A subsequent PCR amplification step is used to amplify the appropriate product using common primers to ‘built-in’ universal PCR sites in the ASO and LSO sequences The resultant PCR products are hybridized and read out on an array of universal-capture probes

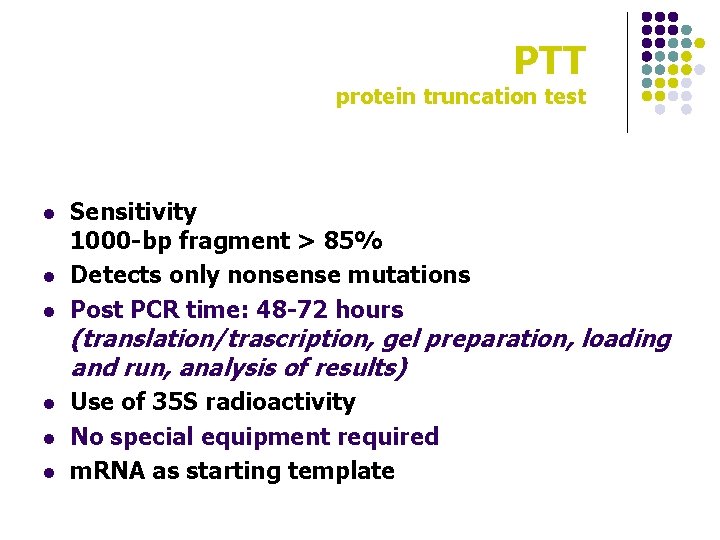
PTT protein truncation test l l l Sensitivity 1000 -bp fragment > 85% Detects only nonsense mutations Post PCR time: 48 -72 hours (translation/trascription, gel preparation, loading and run, analysis of results) Use of 35 S radioactivity No special equipment required m. RNA as starting template



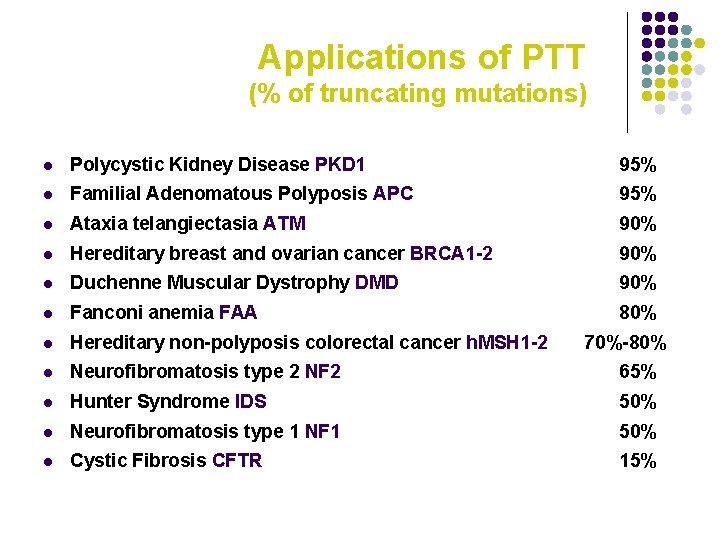
Applications of PTT (% of truncating mutations) l Polycystic Kidney Disease PKD 1 95% l Familial Adenomatous Polyposis APC 95% l Ataxia telangiectasia ATM 90% l Hereditary breast and ovarian cancer BRCA 1 -2 90% l Duchenne Muscular Dystrophy DMD 90% l Fanconi anemia FAA 80% l Hereditary non-polyposis colorectal cancer h. MSH 1 -2 l Neurofibromatosis type 2 NF 2 65% l Hunter Syndrome IDS 50% l Neurofibromatosis type 1 NF 1 50% l Cystic Fibrosis CFTR 15% 70%-80%

SSCP

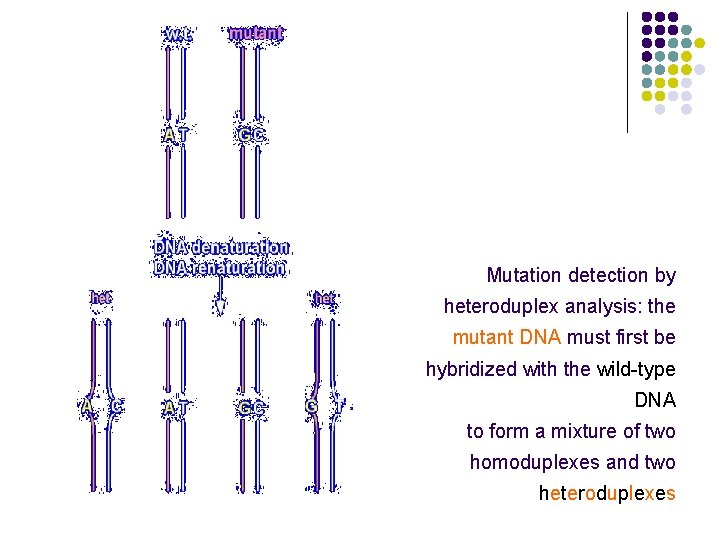
Mutation detection by heteroduplex analysis: the mutant DNA must first be hybridized with the wild-type DNA to form a mixture of two homoduplexes and two heteroduplexes

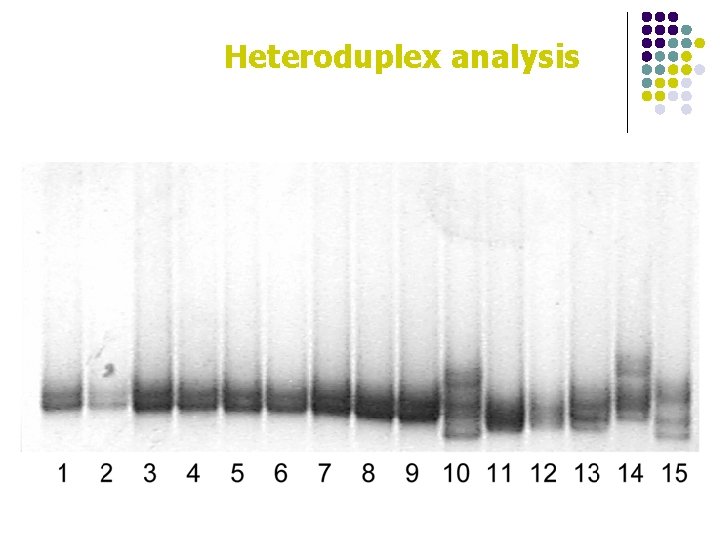
Heteroduplex analysis

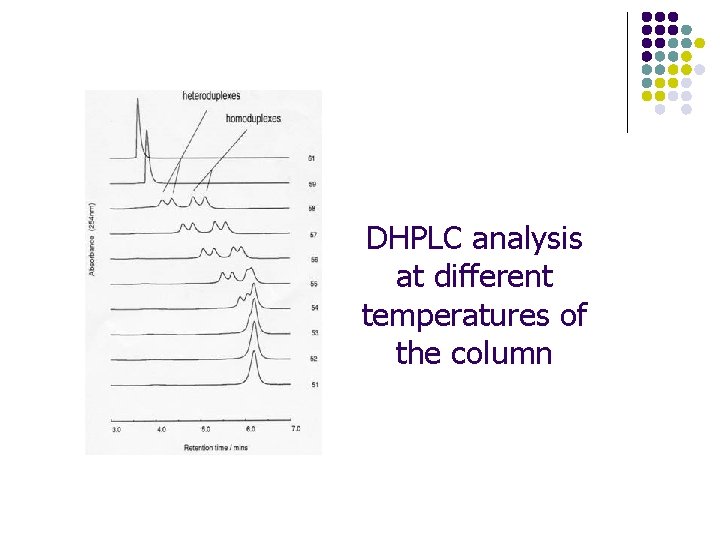
DHPLC denaturing HPLC from Transgenomic

DHPLC analysis at different temperatures of the column

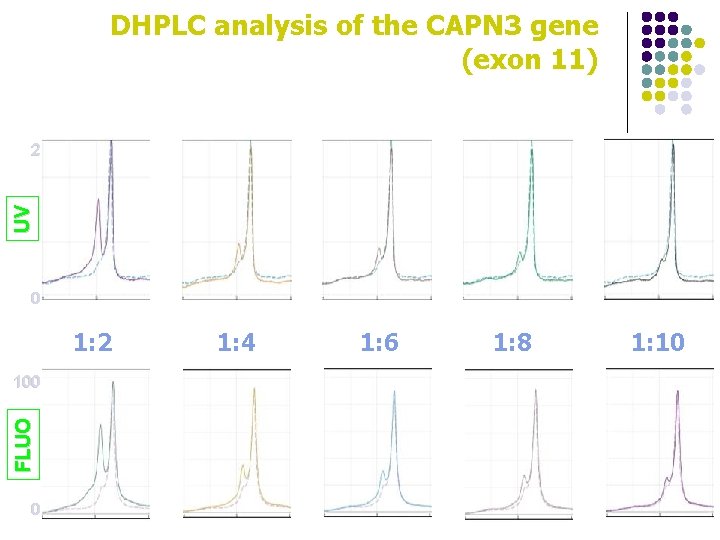
DHPLC analysis of the CAPN 3 gene (exon 11) UV 2 0 1: 2 FLUO 100 0 1: 4 1: 6 1: 8 1: 10

Sequencing artifacts FALSE POSITIVE (specificity) lwhen searching for heterozygous DNA differences there a number of potential mutations, together with sequence artifacts, compressions and differences in peak intensities that must be re-checked with additional primers and costs FALSE NEGATIVE (sensitivity) lloss of information farther away or closer to the primer ldoes not detect a minority of mutant molecules in a wildtype environment

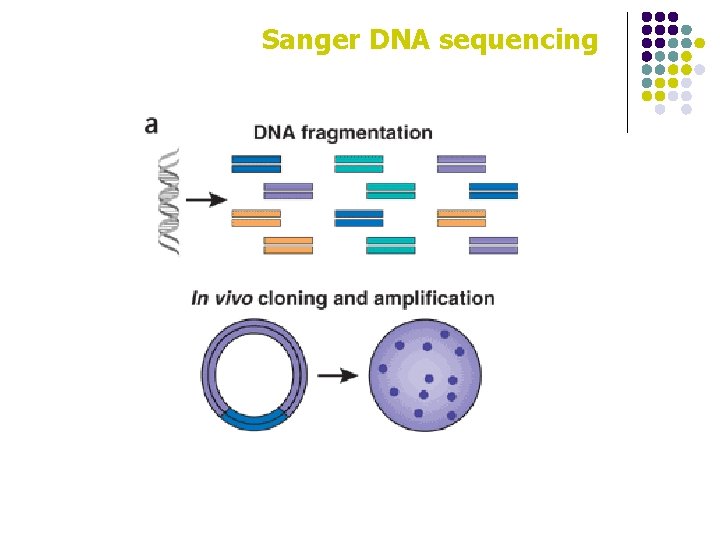
Sanger DNA sequencing


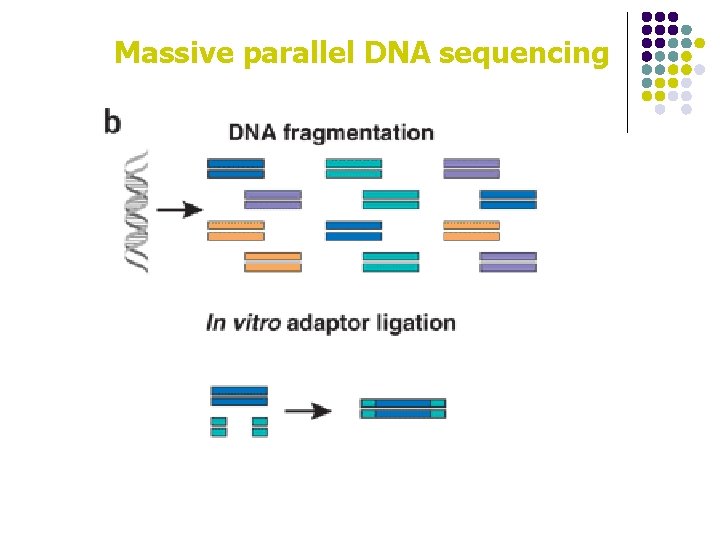
Massive parallel DNA sequencing

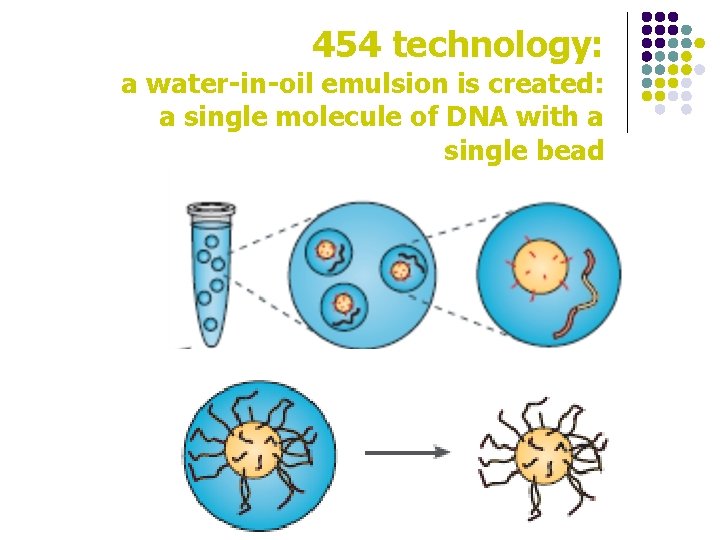
454 technology: a water-in-oil emulsion is created: a single molecule of DNA with a single bead

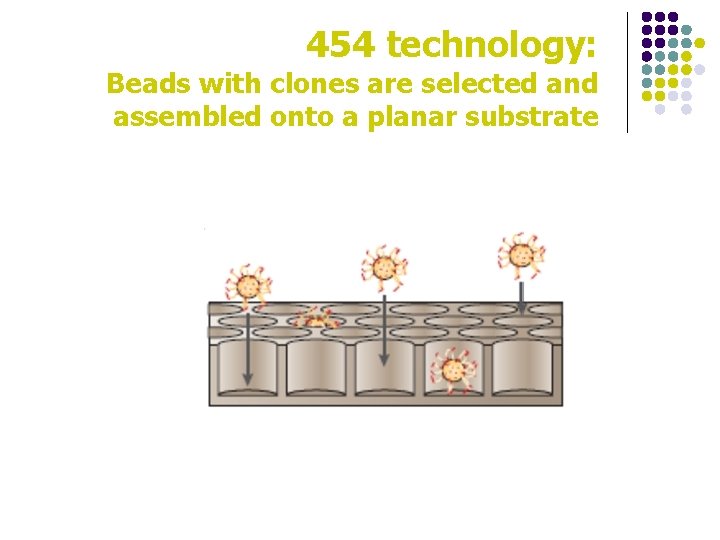
454 technology: Beads with clones are selected and assembled onto a planar substrate


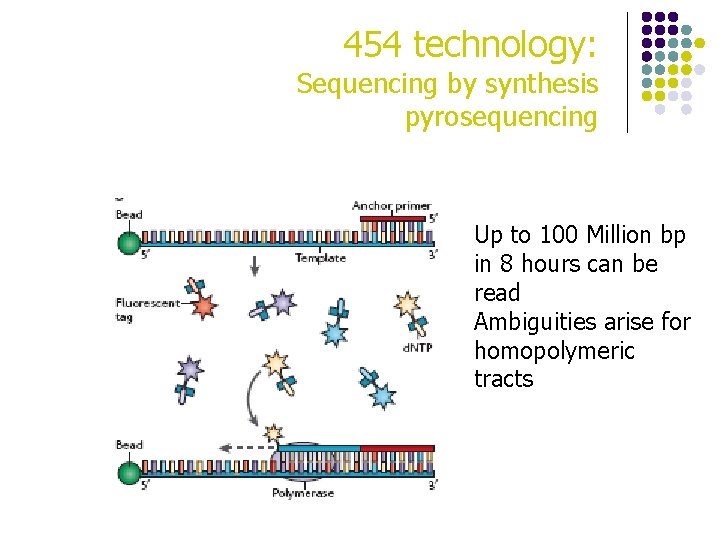
454 technology: Sequencing by synthesis pyrosequencing Up to 100 Million bp in 8 hours can be read Ambiguities arise for homopolymeric tracts

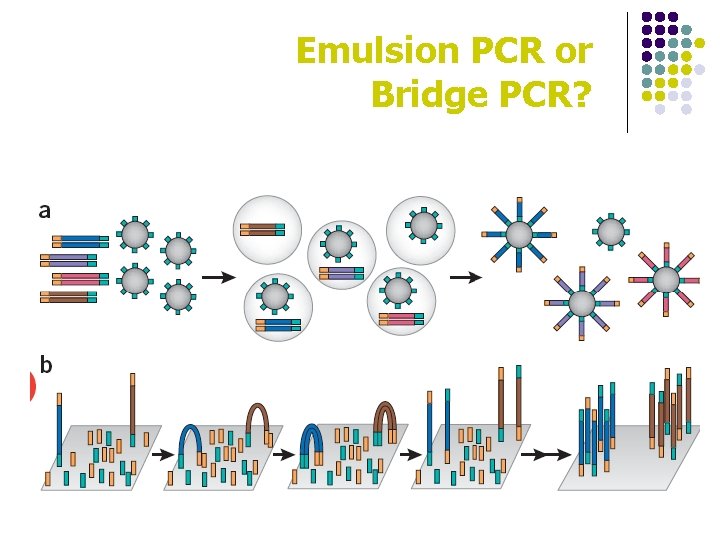
Emulsion PCR or Bridge PCR?



7. 4 x coverage 234 runs 24. 5 billions bp

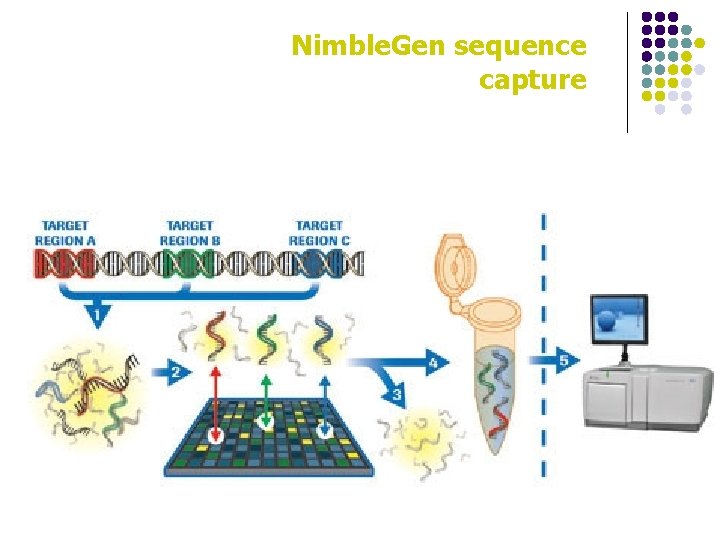
Nimble. Gen sequence capture

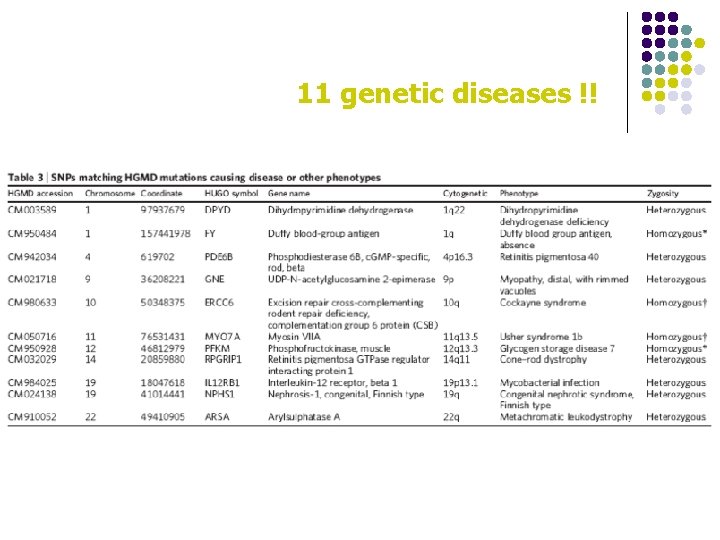
11 genetic diseases !!
 Nigro & nigro pc
Nigro & nigro pc Acrostic poem teamwork
Acrostic poem teamwork Elementi di fisica meccanica e termodinamica mazzoldi
Elementi di fisica meccanica e termodinamica mazzoldi Mutazione genica
Mutazione genica Mutazioni puntiformi
Mutazioni puntiformi Mutazione missenso
Mutazione missenso Miotonico significato
Miotonico significato Mutazioni puntiformi
Mutazioni puntiformi Allopoliploidia
Allopoliploidia Mutazioni genetiche
Mutazioni genetiche Mutazioni missenso
Mutazioni missenso Sindrome di turner immagini
Sindrome di turner immagini Mutazioni genetiche
Mutazioni genetiche Animazione
Animazione Ringraziamenti tesi
Ringraziamenti tesi Tecniche di purificazione
Tecniche di purificazione Tecniche investigative carabinieri
Tecniche investigative carabinieri Tecniche associate al pensiero computazionale:
Tecniche associate al pensiero computazionale: Casa di palma umberto riva
Casa di palma umberto riva Il testo narrativo
Il testo narrativo Tecniche di rappresentazione dello spazio
Tecniche di rappresentazione dello spazio Estrazione separazione miscugli
Estrazione separazione miscugli Tipi di presentazione dei personaggi
Tipi di presentazione dei personaggi Struttura di un testo
Struttura di un testo Le 5 c della comunicazione
Le 5 c della comunicazione Metodi e tecniche del servizio sociale slide
Metodi e tecniche del servizio sociale slide Forme della durata narrativa
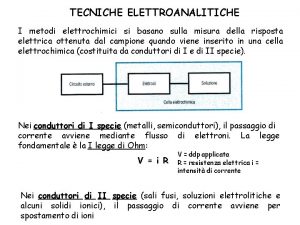
Forme della durata narrativa Tecniche analitiche di laboratorio
Tecniche analitiche di laboratorio Sezioni di solidi con piani inclinati
Sezioni di solidi con piani inclinati Tecniche traduzione inglese
Tecniche traduzione inglese Tecniche di conduzione dei gruppi slide
Tecniche di conduzione dei gruppi slide Schema base del testo
Schema base del testo Migi jodan
Migi jodan Misure tecniche, organizzative e procedurali
Misure tecniche, organizzative e procedurali Prompting fading shaping modeling chaining
Prompting fading shaping modeling chaining Tecniche di catalogazione
Tecniche di catalogazione Lo stile di un testo narrativo
Lo stile di un testo narrativo Sequenze nei testi
Sequenze nei testi Metodi di separazione dei miscugli esempi
Metodi di separazione dei miscugli esempi Tecniche pittoriche moderne
Tecniche pittoriche moderne Ciclo dei vinti
Ciclo dei vinti Teorie e tecniche della televisione
Teorie e tecniche della televisione Pensiero e poetica svevo
Pensiero e poetica svevo Tecniche di socializzazione
Tecniche di socializzazione Tecniche di purificazione
Tecniche di purificazione Ombra di un cono
Ombra di un cono Potenziometria
Potenziometria Comunicazione organizzativa aziendale
Comunicazione organizzativa aziendale Linguaggi e tecniche comunicative non verbali
Linguaggi e tecniche comunicative non verbali Vincenzo pisciuneri
Vincenzo pisciuneri Vincenzo civerchio
Vincenzo civerchio Vincenzo vagnoni
Vincenzo vagnoni