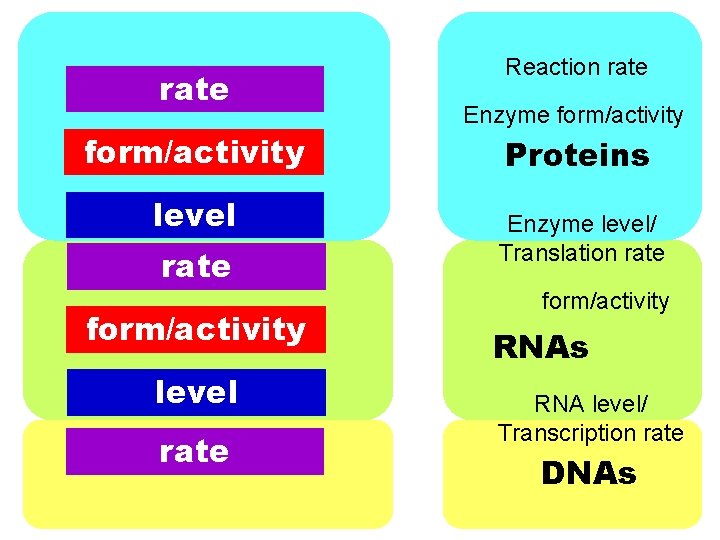
rate formactivity level rate Reaction rate Enzyme formactivity
- Slides: 44

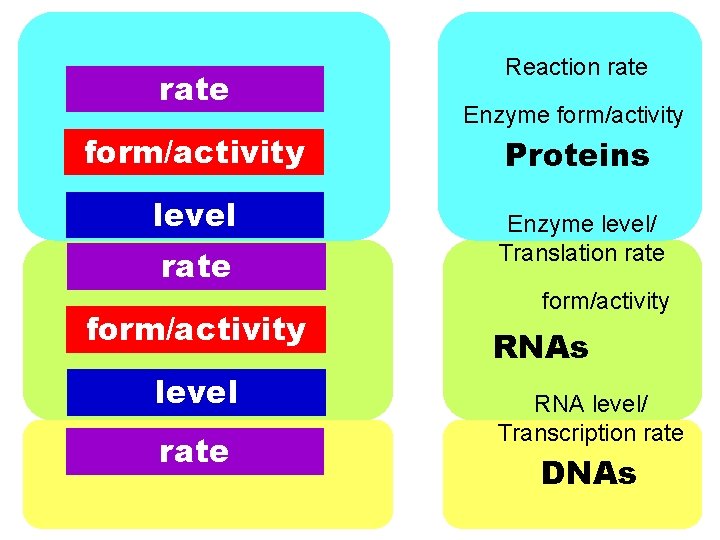
rate form/activity level rate Reaction rate Enzyme form/activity Proteins Enzyme level/ Translation rate form/activity RNAs RNA level/ Transcription rate DNAs

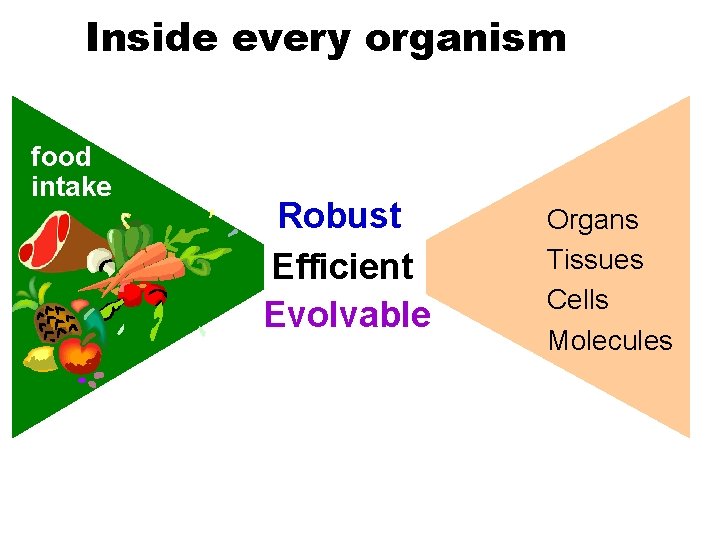
Inside every organism food intake Robust Efficient Evolvable Organs Tissues Cells Molecules

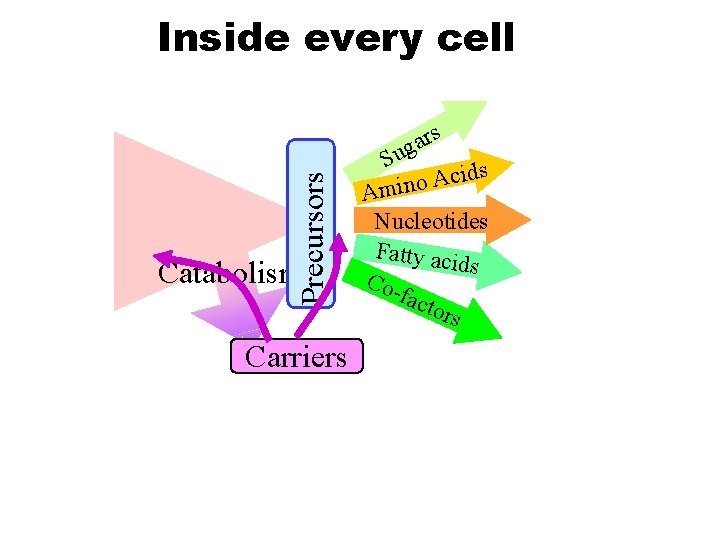
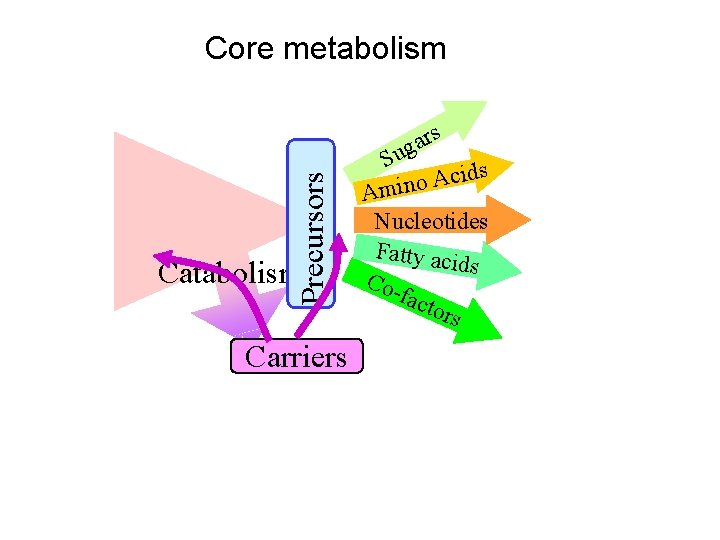
Precursors Inside every cell Catabolism Carriers rs a g Su ds i c A o Amin Nucleotides Fatty aci ds Cofact ors

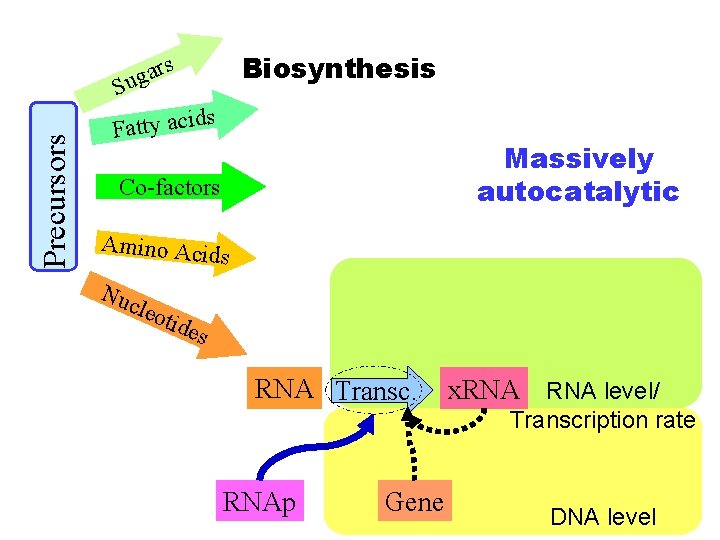
Biosynthesis s Precursors ar g u S Fatty acid s Massively autocatalytic Co-factors Amino Acid s Nuc leot ides RNA Transc. RNAp Gene x. RNA level/ Transcription rate DNA level

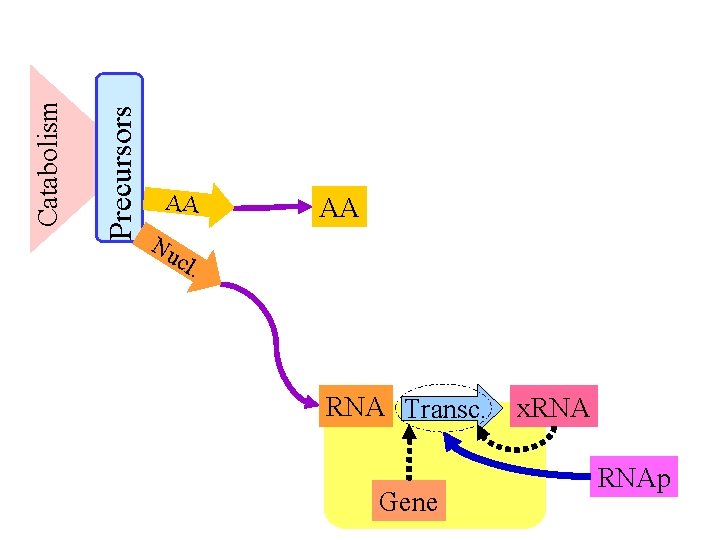
Precursors Catabolism AA Nu cl AA . RNA Transc. Gene x. RNAp

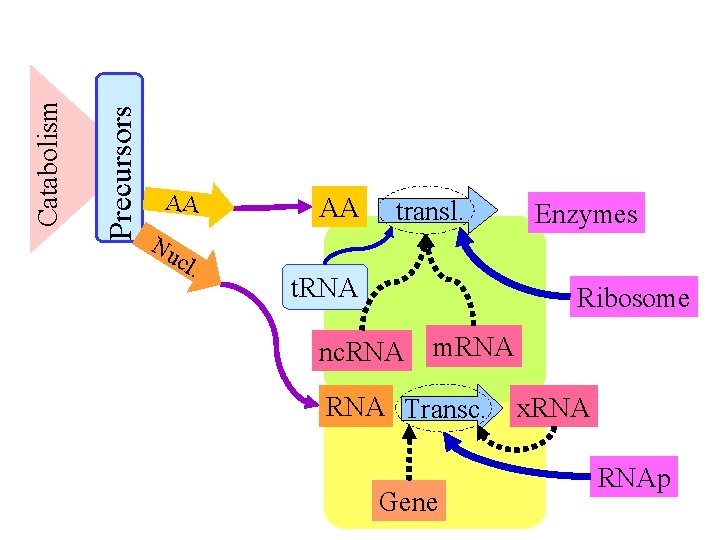
Precursors Catabolism AA Nu cl . AA transl. t. RNA Enzymes Ribosome nc. RNA m. RNA Transc. Gene x. RNAp

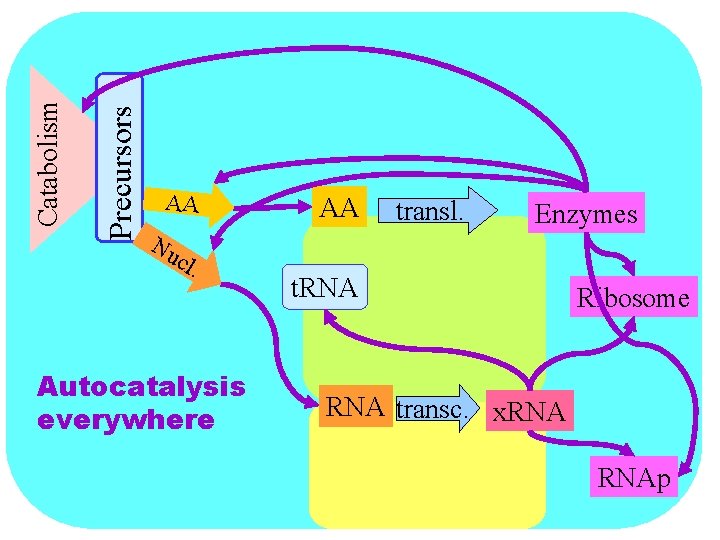
Precursors Catabolism AA Nu cl . Autocatalysis everywhere AA transl. Enzymes t. RNA Ribosome RNA transc. x. RNAp

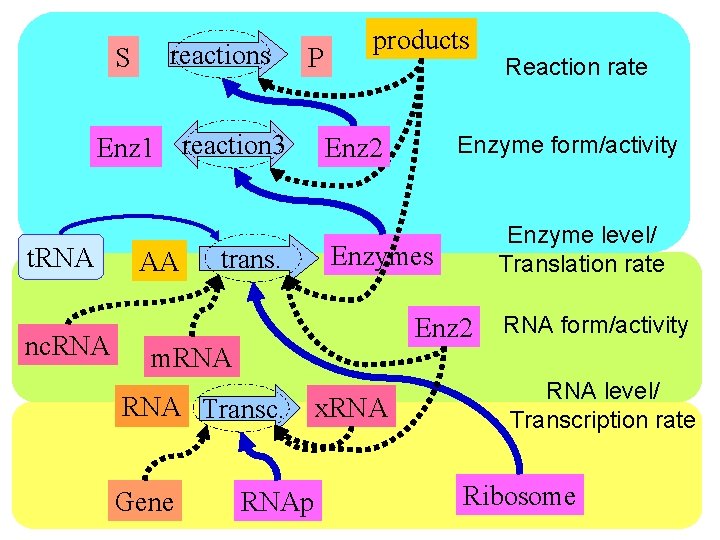
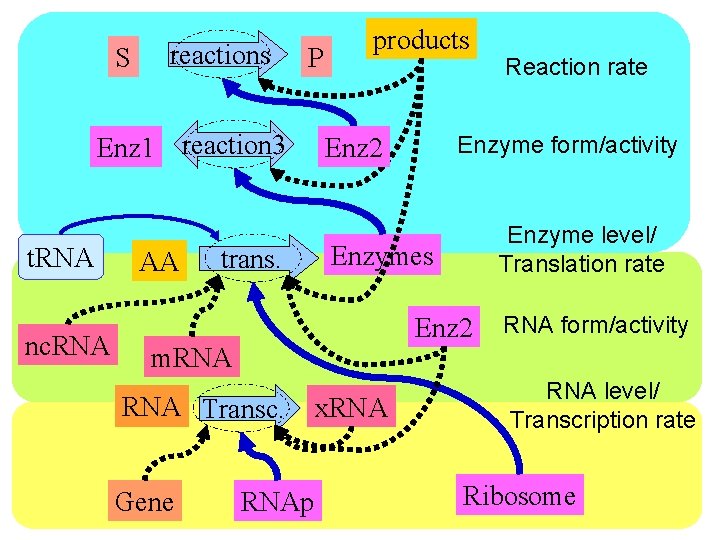
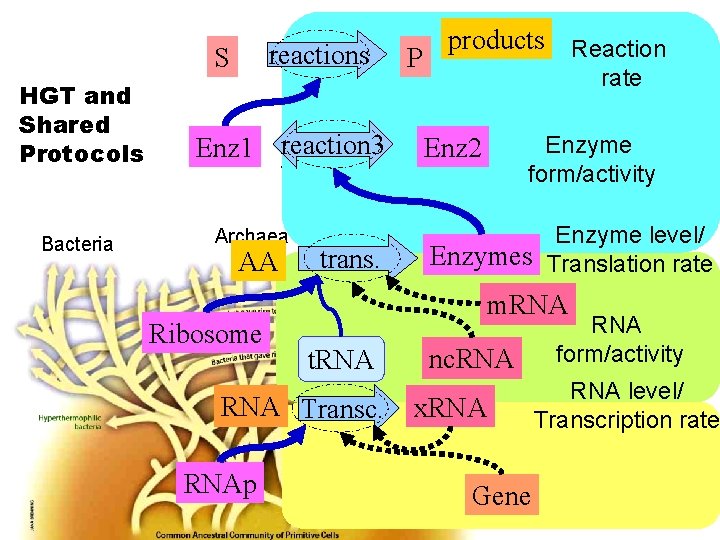
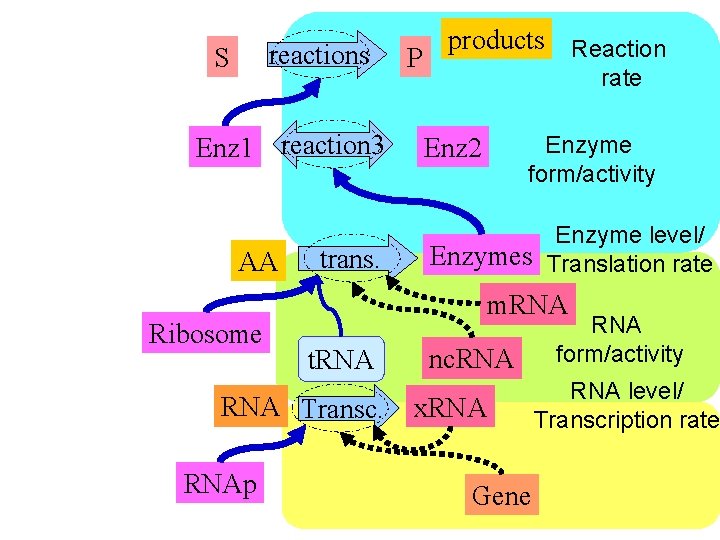
S reactions P Enz 1 reaction 3 t. RNA nc. RNA AA trans. products Enzyme level/ Translation rate Enzymes Enz 2 RNA Transc. Gene Enzyme form/activity Enz 2 m. RNAp x. RNA Reaction rate RNA form/activity RNA level/ Transcription rate Ribosome

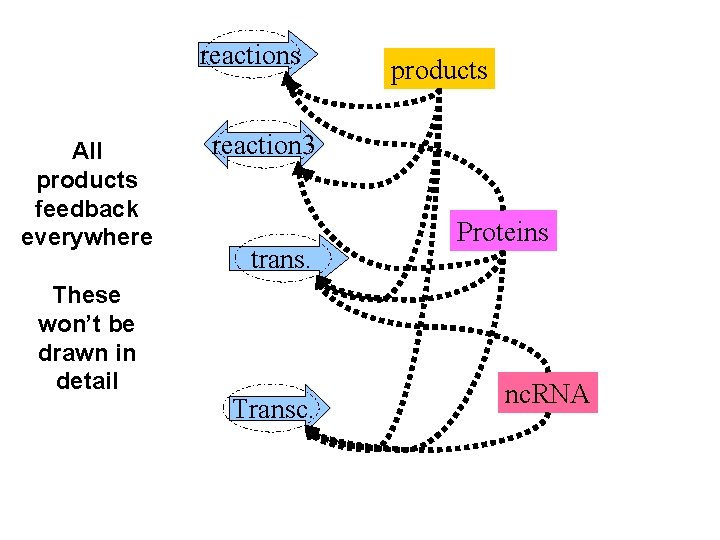
reactions All products feedback everywhere These won’t be drawn in detail products reaction 3 trans. Transc. Proteins nc. RNA

S reactions P Enz 1 reaction 3 t. RNA nc. RNA AA trans. products Enzyme level/ Translation rate Enzymes Enz 2 RNA Transc. Gene Enzyme form/activity Enz 2 m. RNAp x. RNA Reaction rate RNA form/activity RNA level/ Transcription rate Ribosome

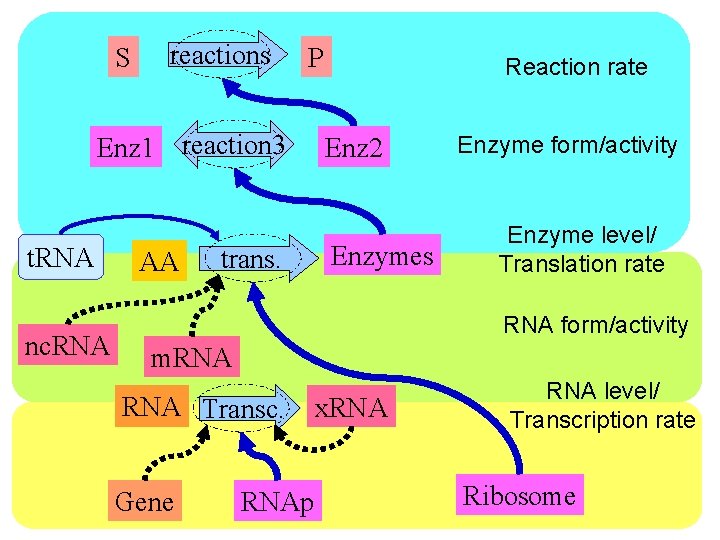
S reactions P Enz 1 reaction 3 t. RNA nc. RNA AA trans. Reaction rate Enz 2 Enzymes Enzyme form/activity Enzyme level/ Translation rate RNA form/activity m. RNA Transc. Gene RNAp x. RNA level/ Transcription rate Ribosome

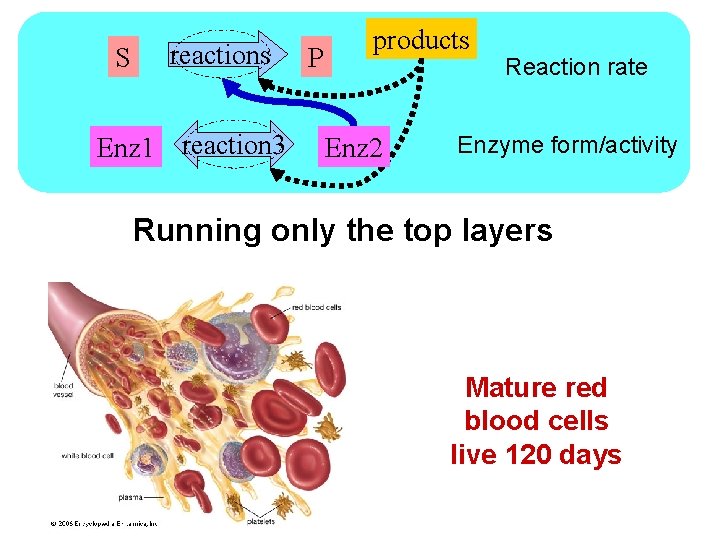
S reactions Enz 1 reaction 3 P products Enz 2 Reaction rate Enzyme form/activity Running only the top layers Mature red blood cells live 120 days

Diverse Application Diverse application s and genomes Genome Diverse

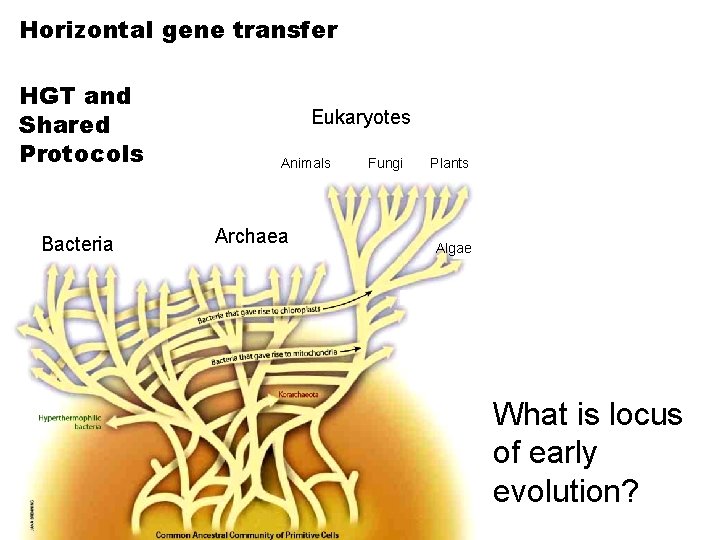
Horizontal gene transfer HGT and Shared Protocols Bacteria Eukaryotes Animals Archaea Fungi Plants Algae What is locus of early evolution?

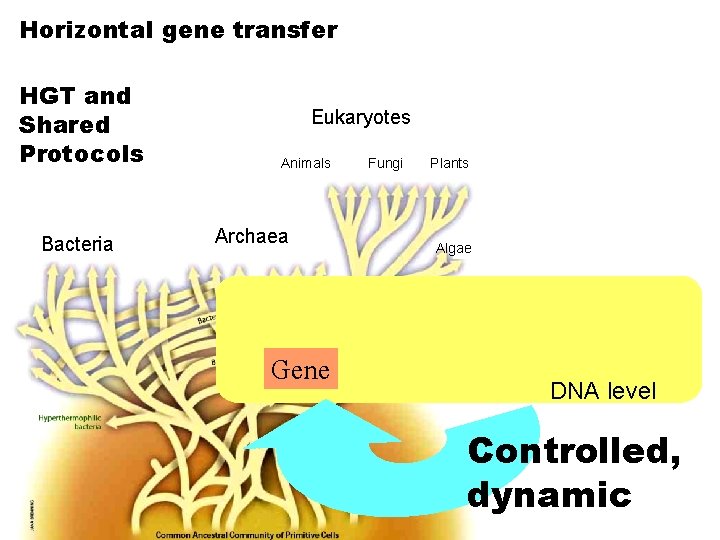
Horizontal gene transfer HGT and Shared Protocols Bacteria Eukaryotes Animals Archaea Gene Fungi Plants Algae DNA level Controlled, dynamic

reactions S HGT and Shared Protocols Bacteria products Reaction P rate Eukaryotes Enz 1 reaction 3 Enz 2 Animals Fungi Plants Archaea AA Ribosome trans. Enzyme level/ Enzymes Translation rate Algae m. RNA t. RNA Transc. RNAp Enzyme form/activity nc. RNA x. RNA Gene RNA form/activity RNA level/ Transcription rate

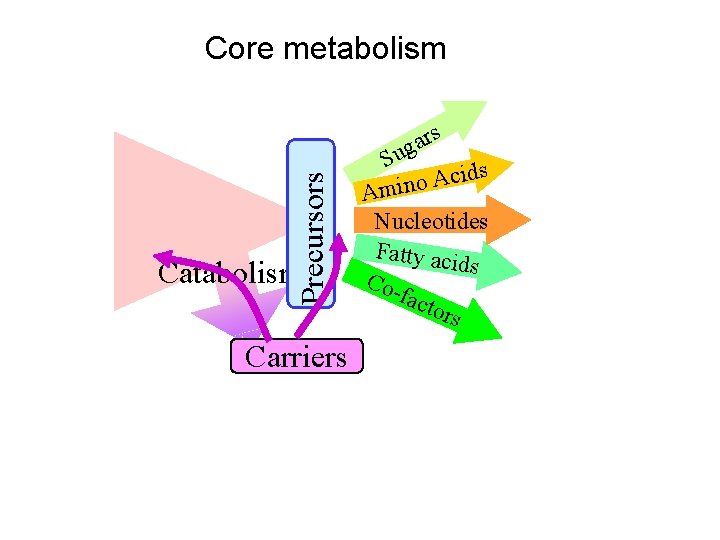
Precursors Core metabolism Carriers rs a g Su ds i c A o Amin Nucleotides Fatty aci ds Cofact ors

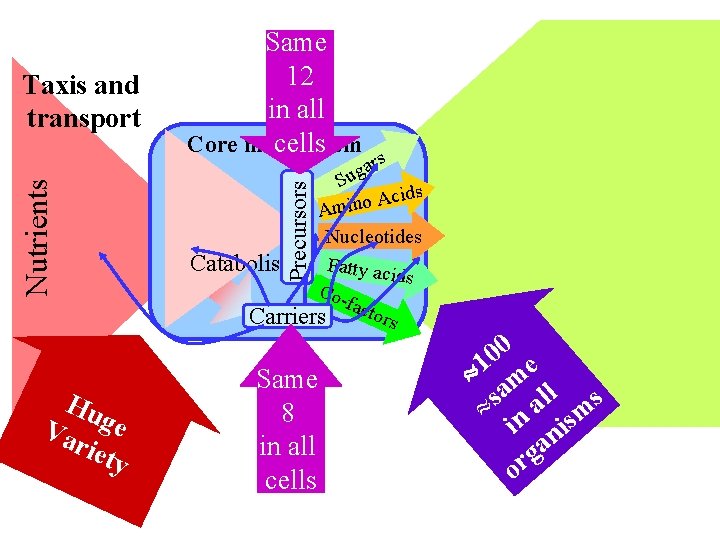
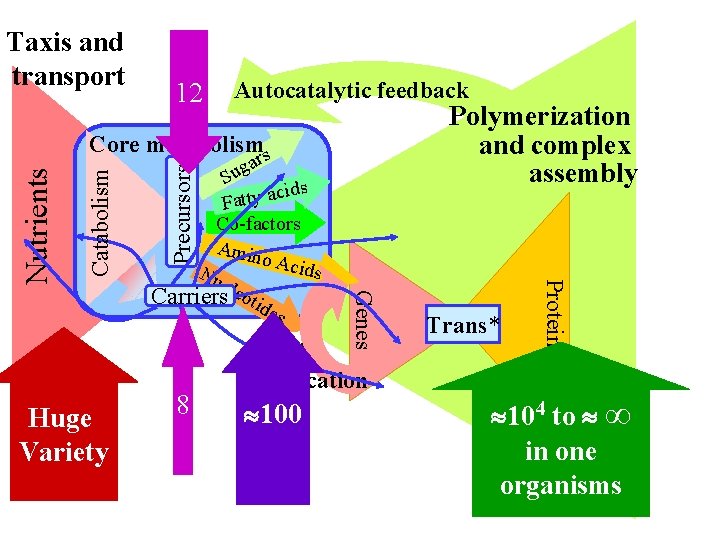
Hu Va ge riet y Precursors Nutrients Taxis and transport Same 12 in all Core metabolism cells rs a g Su cids A o n i m A Nucleotides Catabolism Fatty acid s Cofact ors Carriers Same 8 in all cells 0 0 1 me a ll s s » n a sm i ni a g or

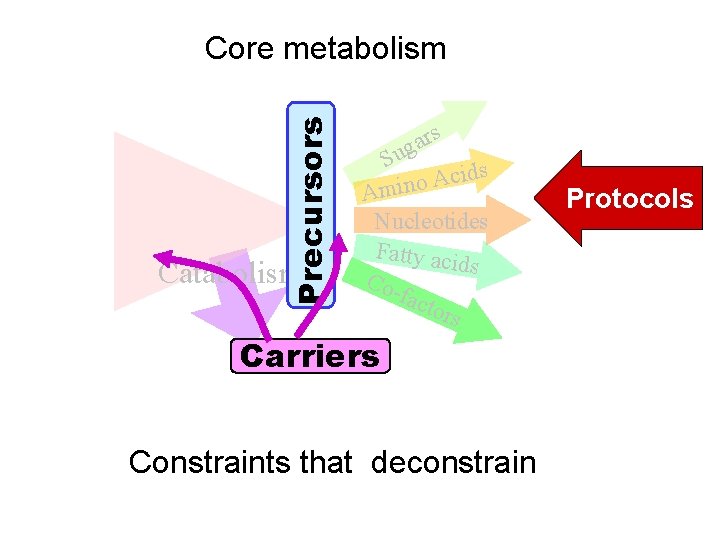
Precursors Core metabolism Catabolism rs a g Su ds i c A o Amin Nucleotides Fatty aci ds Cofact ors Carriers Constraints that deconstrain Protocols

Precursors Core metabolism Carriers rs a g Su ds i c A o Amin Nucleotides Fatty aci ds Cofact ors

Precursors Catabolism Carriers

Gly G 1 P G 6 P F 1 -6 BP Catabolism Gly 3 p 13 BPG ATP 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

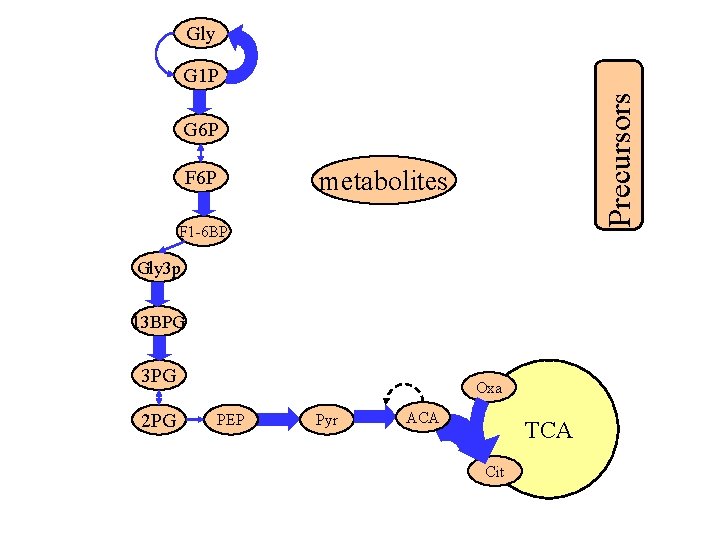
Gly Precursors G 1 P G 6 P metabolites F 6 P F 1 -6 BP Gly 3 p 13 BPG 3 PG 2 PG Oxa PEP Pyr ACA TCA Cit

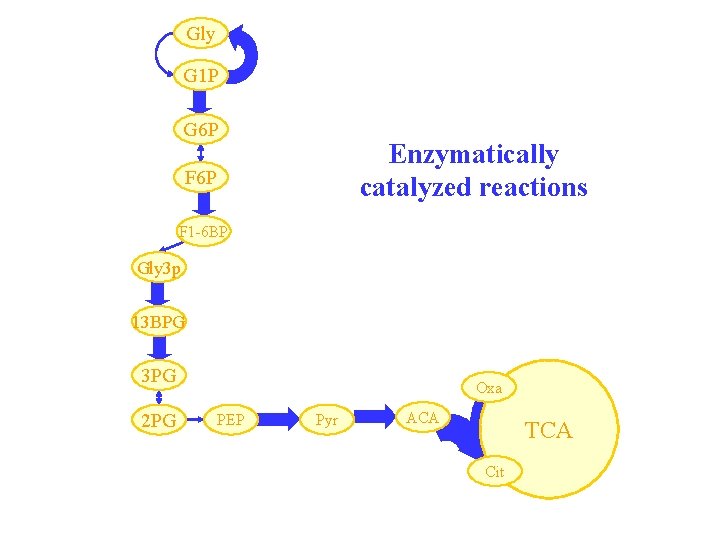
Gly G 1 P G 6 P Enzymatically catalyzed reactions F 6 P F 1 -6 BP Gly 3 p 13 BPG 3 PG 2 PG Oxa PEP Pyr ACA TCA Cit

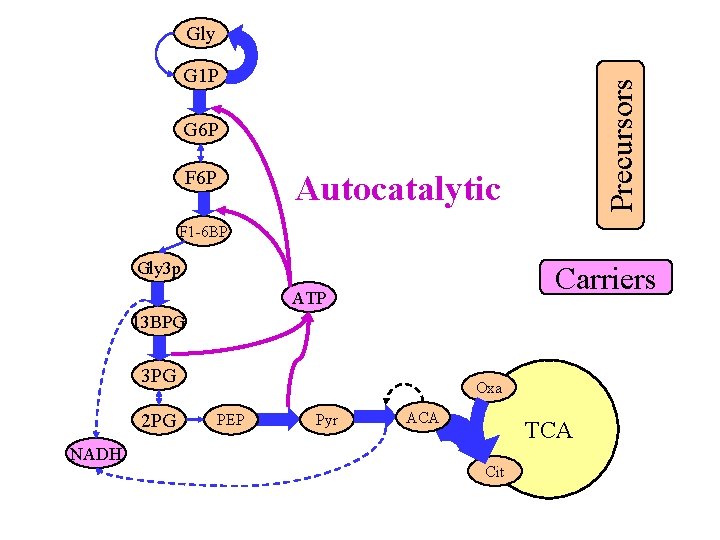
Gly Precursors G 1 P G 6 P F 6 P Autocatalytic F 1 -6 BP Gly 3 p Carriers ATP 13 BPG 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

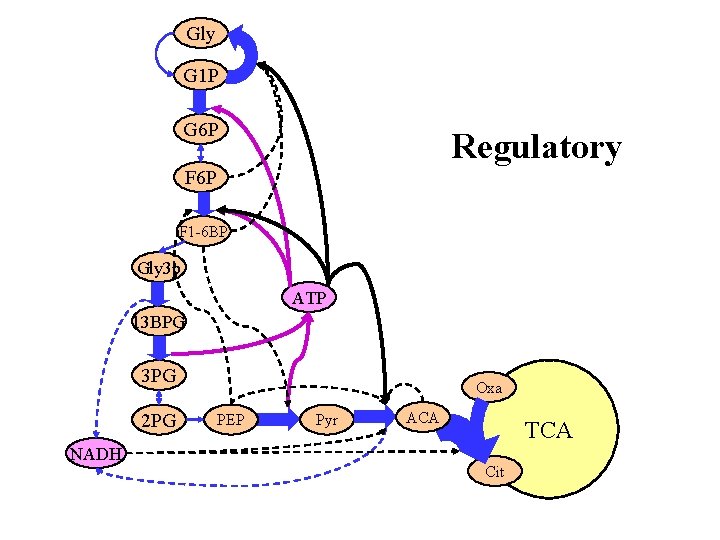
Gly G 1 P G 6 P Regulatory F 6 P F 1 -6 BP Gly 3 p ATP 13 BPG 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

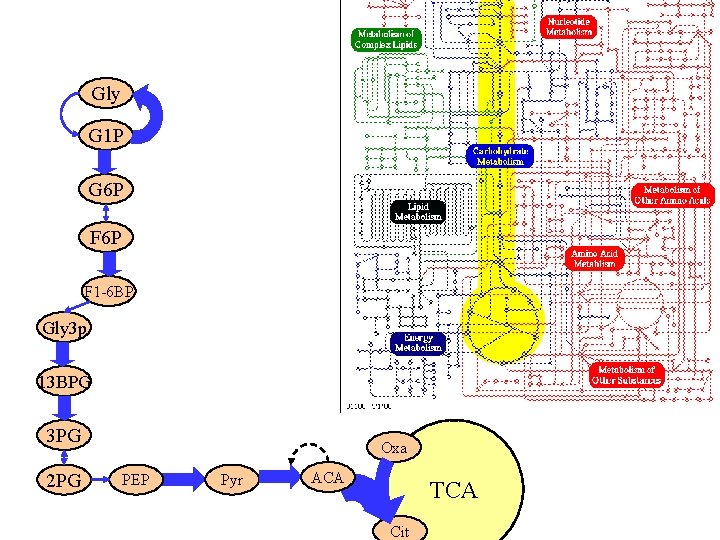
Gly G 1 P G 6 P F 1 -6 BP Gly 3 p 13 BPG 3 PG 2 PG Oxa PEP Pyr ACA TCA Cit

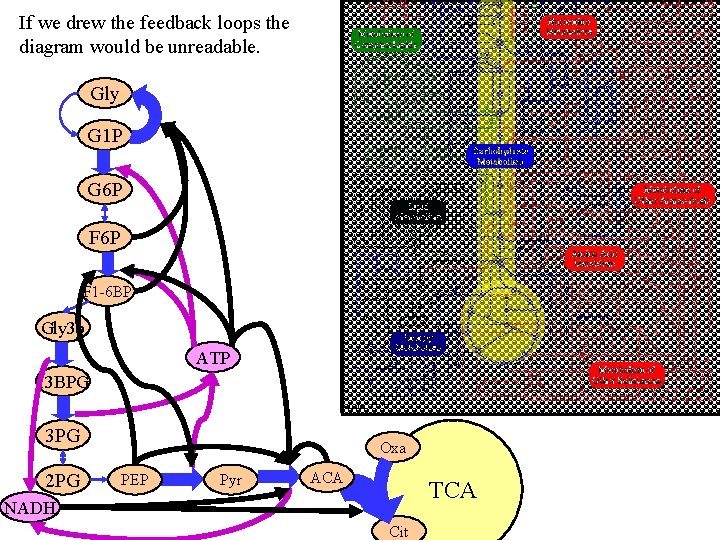
If we drew the feedback loops the diagram would be unreadable. Gly G 1 P G 6 P F 1 -6 BP Gly 3 p ATP 13 BPG 3 PG 2 PG Oxa PEP Pyr ACA TCA NADH Cit

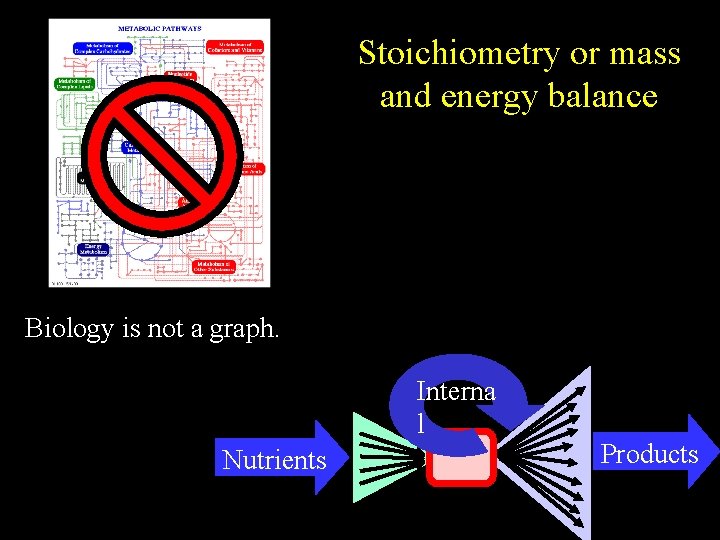
Stoichiometry or mass and energy balance Biology is not a graph. Interna l Nutrients Products

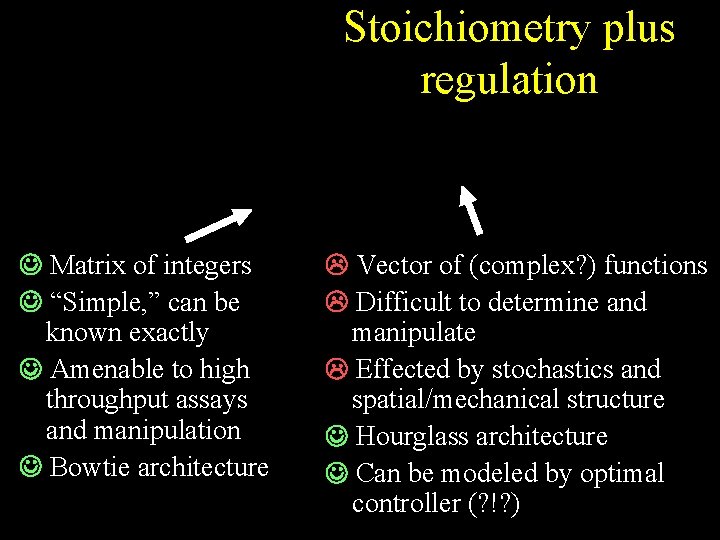
Stoichiometry plus regulation Matrix of integers “Simple, ” can be known exactly Amenable to high throughput assays and manipulation Bowtie architecture Vector of (complex? ) functions Difficult to determine and manipulate Effected by stochastics and spatial/mechanical structure Hourglass architecture Can be modeled by optimal controller (? !? )

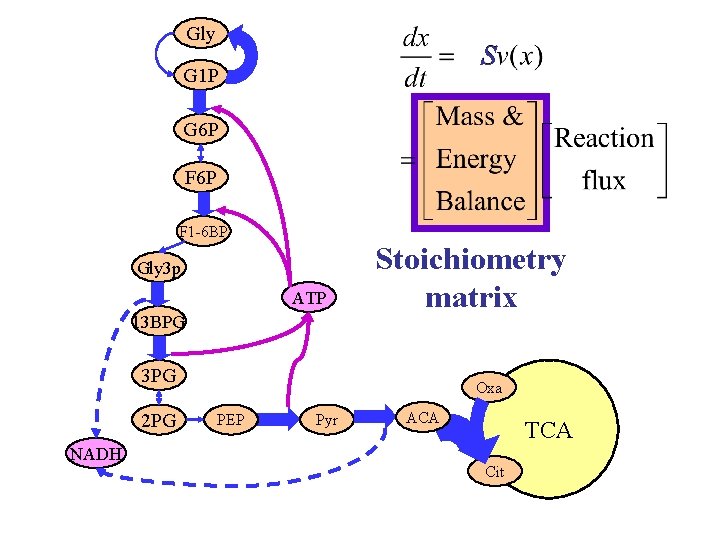
Gly S G 1 P G 6 P F 1 -6 BP Gly 3 p ATP 13 BPG Stoichiometry matrix 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

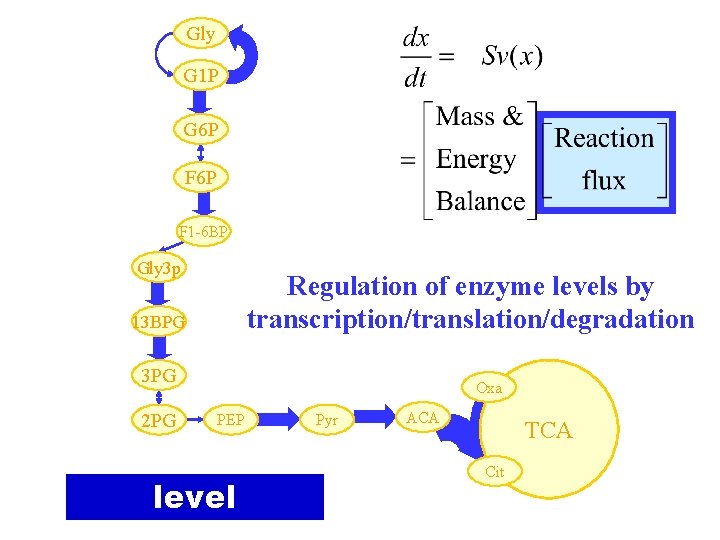
Gly G 1 P G 6 P F 1 -6 BP Gly 3 p Regulation of enzyme levels by transcription/translation/degradation 13 BPG 3 PG 2 PG Oxa PEP level Pyr ACA TCA Cit

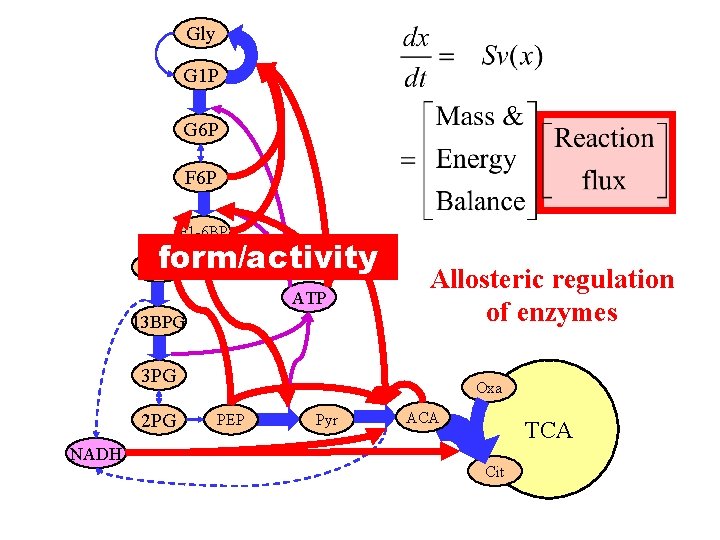
Gly G 1 P G 6 P F 1 -6 BP form/activity Gly 3 p ATP 13 BPG Allosteric regulation of enzymes 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

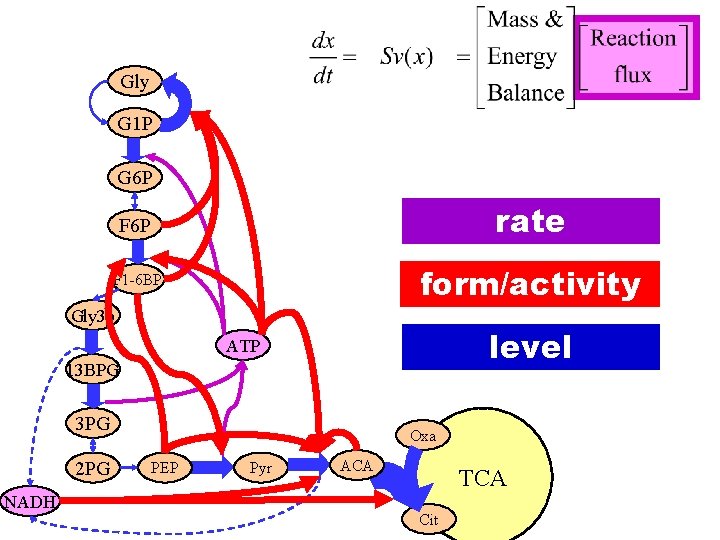
Gly G 1 P G 6 P F 6 P rate F 1 -6 BP form/activity Gly 3 p level ATP 13 BPG 3 PG 2 PG NADH Oxa PEP Pyr ACA TCA Cit

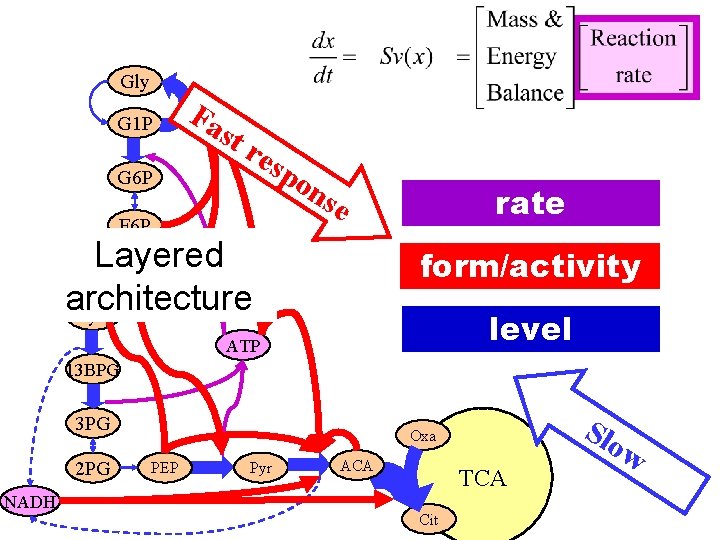
Gly G 1 P G 6 P Fa st res po F 6 P ns rate e Layered F 1 -6 BP architecture Gly 3 p form/activity level ATP 13 BPG 3 PG 2 PG NADH Slo Oxa PEP Pyr ACA TCA Cit w

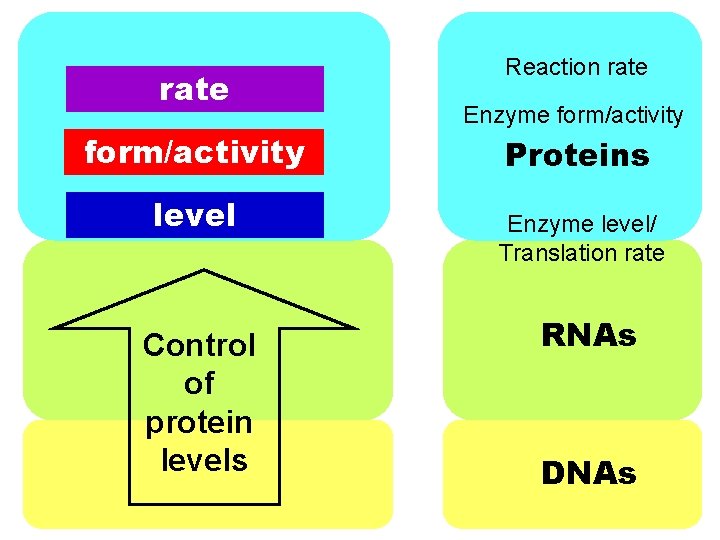
rate form/activity level Control of protein levels Reaction rate Enzyme form/activity Proteins Enzyme level/ Translation rate RNAs DNAs

rate form/activity level rate Reaction rate Enzyme form/activity Proteins Enzyme level/ Translation rate form/activity RNAs RNA level/ Transcription rate DNAs

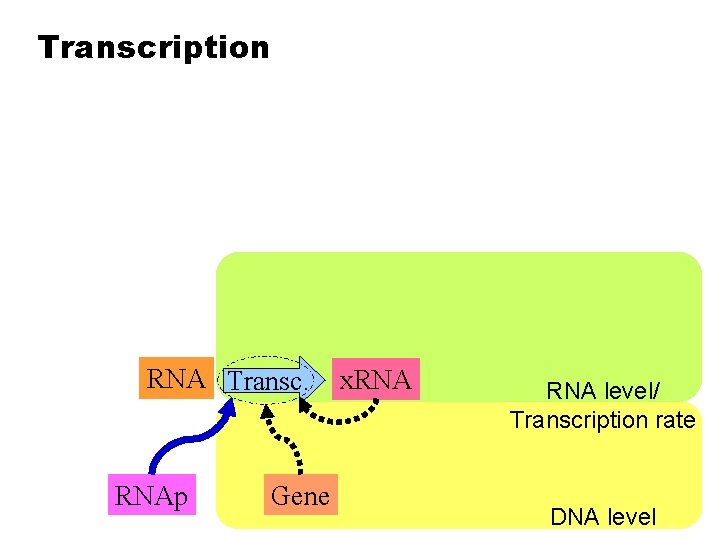
Transcription RNA Transc. RNAp Gene x. RNA level/ Transcription rate DNA level

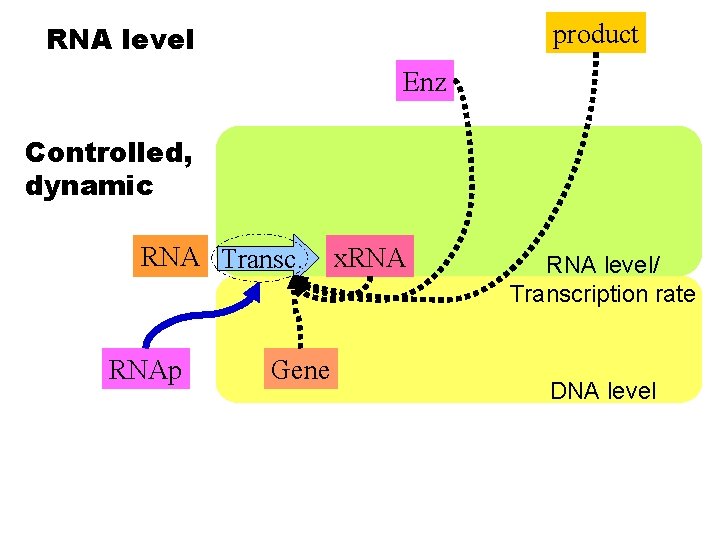
product RNA level Enz Controlled, dynamic RNA Transc. RNAp Gene x. RNA level/ Transcription rate DNA level

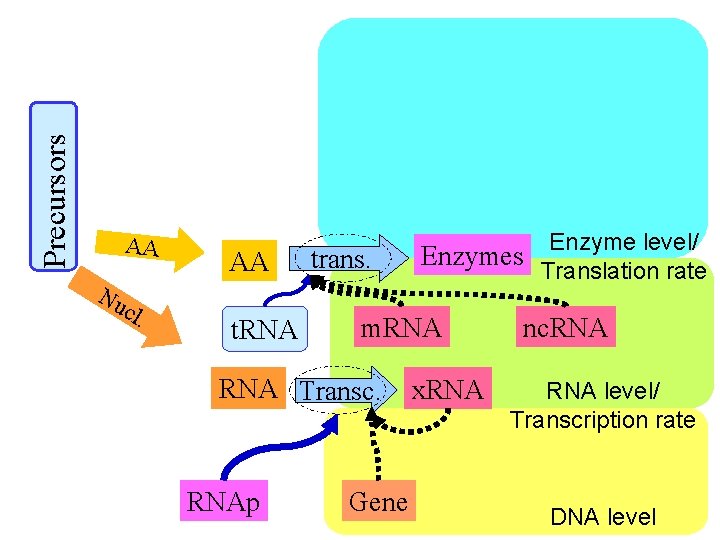
Precursors AA Nu cl . AA t. RNA trans. m. RNA Transc. RNAp Enzyme level/ Enzymes Translation rate Gene x. RNA nc. RNA level/ Transcription rate DNA level

reactions S Enz 1 reaction 3 AA Ribosome trans. rate Enzyme form/activity Enz 2 Enzyme level/ Enzymes Translation rate m. RNA t. RNA Transc. RNAp products Reaction P nc. RNA x. RNA Gene RNA form/activity RNA level/ Transcription rate

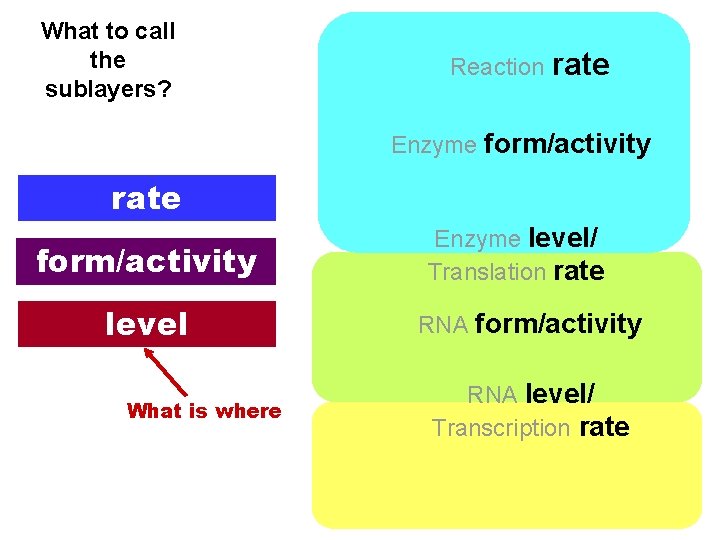
What to call the sublayers? Reaction rate Enzyme form/activity rate form/activity level What is where Enzyme level/ Translation rate RNA form/activity RNA level/ Transcription rate

12 Autocatalytic feedback Polymerization and complex assembly 8 Genes Huge Variety gar u S cids a y t t a F Co-factors Amin o Aci ds Nu cle Carriers otid es Precursors Catabolism Core metabolisms DNA replication 100 Trans* Proteins Nutrients Taxis and transport 104 to ∞ in one organisms

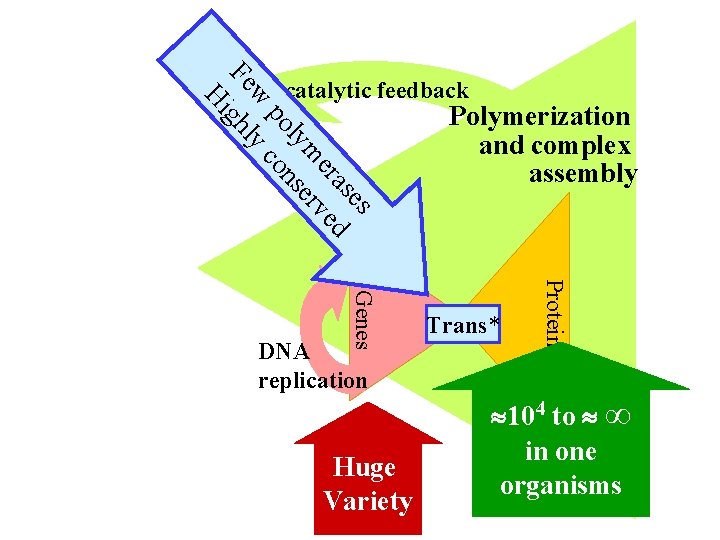
es as er ed m rv ly se po on w yc Fe ghl Hi Autocatalytic feedback Huge Variety Trans* Proteins Genes DNA replication Polymerization and complex assembly 104 to ∞ in one organisms