NEUTRON CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE TIM COMPLEXED



- Slides: 13

NEUTRON CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE (TIM) COMPLEXED WITH ITS INHIBITOR: PGH ESS Science Day – June 14 th Bénédicte LAFUMAT L. mexicana TIM dimer 1/11

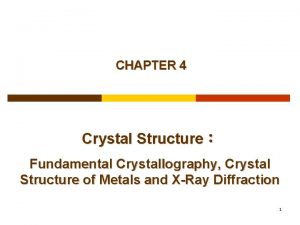
TRIOSEPHOSPHATE ISOMERASE (TIM) from the protozoan parasite Leishmania mexicana • H 95 Oxidoreductase activity E 167 PGH K 13 DHAP G 235 • G 3 P Homodimer : 27 k. Da S. U. 2/11

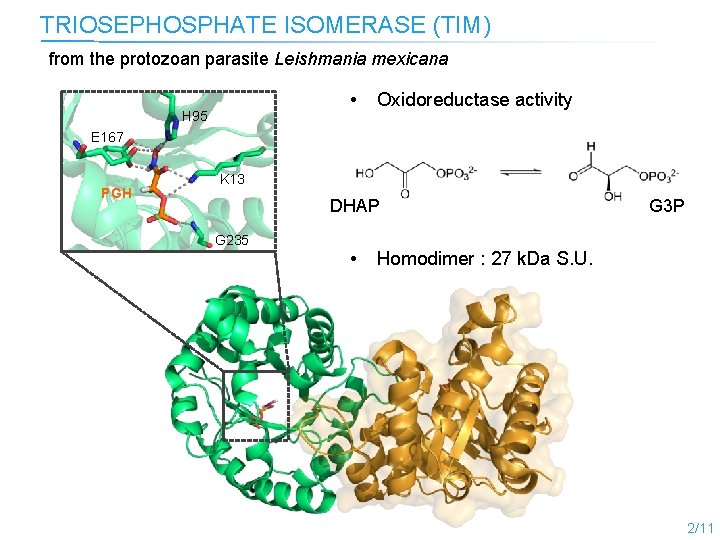
TIM ENZYMATIC MECHANISMS Di. Hydroxy. Acetone. Phosphate Glyceraldehyde-3 -Phosphate "classical" mechanism Acid-base catalysis: His 95/acid protonating the C 2 carbonyl oxygen and Glu 165/base grabbing the C 2 proton Low Barrier Hydrogen Bond mechanism Concerted deprotonation of the C 1 and protonation of the C 2 carbonyl oxygen 3/11

KEY QUESTION H 95 E 167 K 13 PGH G 235 Is TIM processing the “classical” or the “LBHB” enzymatic mechanism? Protonation state of the Glu 235, His 95 and Lys 13 and PGH inhibitor? 4/11

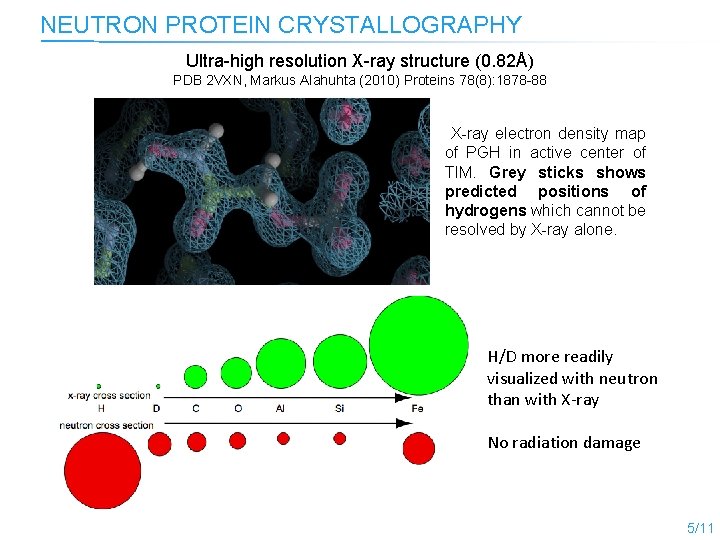
NEUTRON PROTEIN CRYSTALLOGRAPHY Ultra-high resolution X-ray structure (0. 82Å) PDB 2 VXN, Markus Alahuhta (2010) Proteins 78(8): 1878 -88 X-ray electron density map of PGH in active center of TIM. Grey sticks shows predicted positions of hydrogens which cannot be resolved by X-ray alone. H/D more readily visualized with neutron than with X-ray No radiation damage 5/11

TIM EXPRESSION, PURIFICATION & CRYSTALLIZATION Vinardas Keplas Ph. D student, Molecular Cell Biology Unit, Lund University LP 3 platform E. coli MDS 42 His-TIM deuterated minimal media M-9 Metal ion affinity chromatography His. Trap HP column His-Tag cleavage overnight TIM incubated with 3 C protease at 4°C Metal ion affinity chromatography His. Trap HP column Exclusion chromatography Hiload 16/600 Superdex 75 600 10 mg/m. L deuterated TIM 100 m. M Na. Cl 18 % w/v PEG 3350 Pure deuterated TIM Crystals growth from 1 to 3 weeks Microseeding in 200 µL drops 6/11

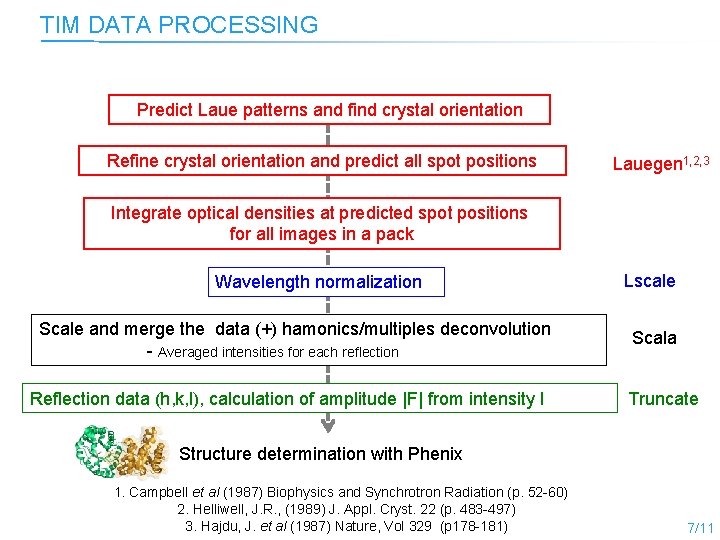
TIM DATA PROCESSING Predict Laue patterns and find crystal orientation Refine crystal orientation and predict all spot positions Lauegen 1, 2, 3 Integrate optical densities at predicted spot positions for all images in a pack Wavelength normalization Scale and merge the data (+) hamonics/multiples deconvolution - Averaged intensities for each reflection Reflection data (h, k, l), calculation of amplitude |F| from intensity I Lscale Scala Truncate Structure determination with Phenix 1. Campbell et al (1987) Biophysics and Synchrotron Radiation (p. 52 -60) 2. Helliwell, J. R. , (1989) J. Appl. Cryst. 22 (p. 483 -497) 3. Hajdu, J. et al (1987) Nature, Vol 329 (p 178 -181) 7/11

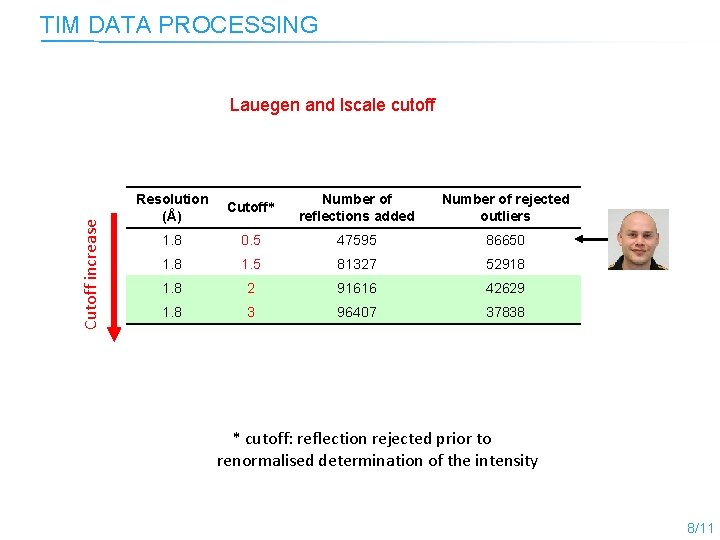
TIM DATA PROCESSING Cutoff increase Lauegen and lscale cutoff Resolution (Å) Cutoff* Number of reflections added Number of rejected outliers 1. 8 0. 5 47595 86650 1. 8 1. 5 81327 52918 1. 8 2 91616 42629 1. 8 3 96407 37838 * cutoff: reflection rejected prior to renormalised determination of the intensity 8/11

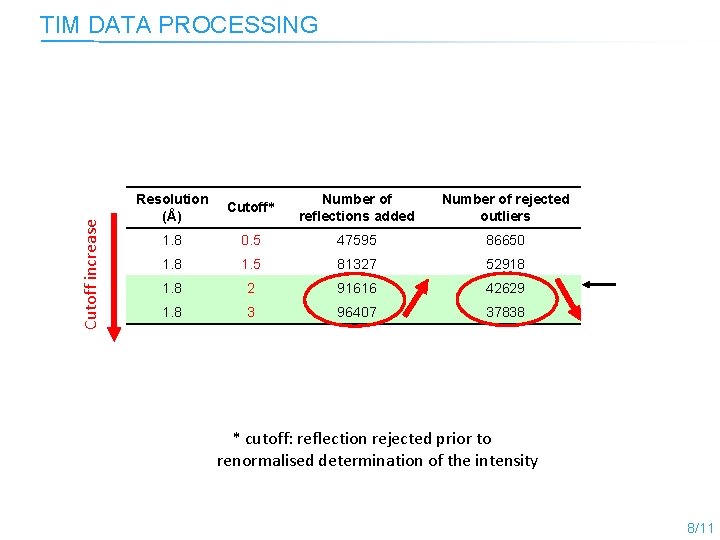
Cutoff increase TIM DATA PROCESSING Resolution (Å) Cutoff* Number of reflections added Number of rejected outliers 1. 8 0. 5 47595 86650 1. 8 1. 5 81327 52918 1. 8 2 91616 42629 1. 8 3 96407 37838 * cutoff: reflection rejected prior to renormalised determination of the intensity 8/11

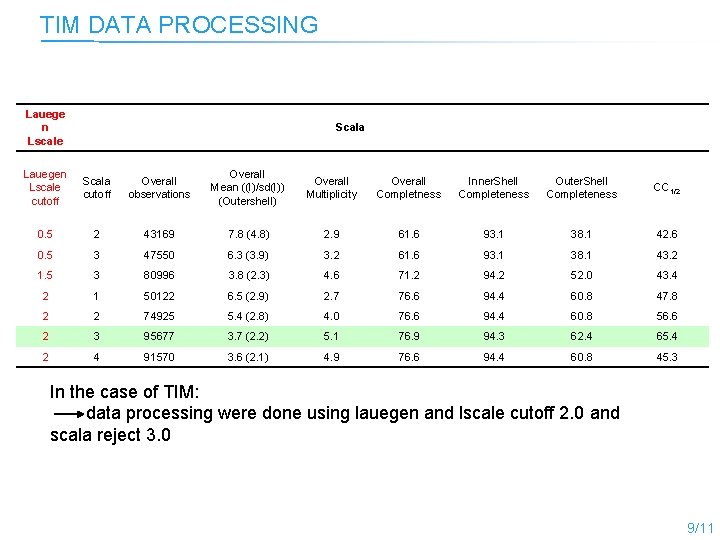
TIM DATA PROCESSING Lauege n Lscale Scala Lauegen Lscale cutoff Scala cutoff Overall observations Overall Mean ((I)/sd(I)) (Outershell) Overall Multiplicity Overall Completness Inner. Shell Completeness Outer. Shell Completeness CC 1/2 0. 5 2 43169 7. 8 (4. 8) 2. 9 61. 6 93. 1 38. 1 42. 6 0. 5 3 47550 6. 3 (3. 9) 3. 2 61. 6 93. 1 38. 1 43. 2 1. 5 3 80996 3. 8 (2. 3) 4. 6 71. 2 94. 2 52. 0 43. 4 2 1 50122 6. 5 (2. 9) 2. 7 76. 6 94. 4 60. 8 47. 8 2 2 74925 5. 4 (2. 8) 4. 0 76. 6 94. 4 60. 8 56. 6 2 3 95677 3. 7 (2. 2) 5. 1 76. 9 94. 3 62. 4 65. 4 2 4 91570 3. 6 (2. 1) 4. 9 76. 6 94. 4 60. 8 45. 3 In the case of TIM: data processing were done using lauegen and lscale cutoff 2. 0 and scala reject 3. 0 9/11

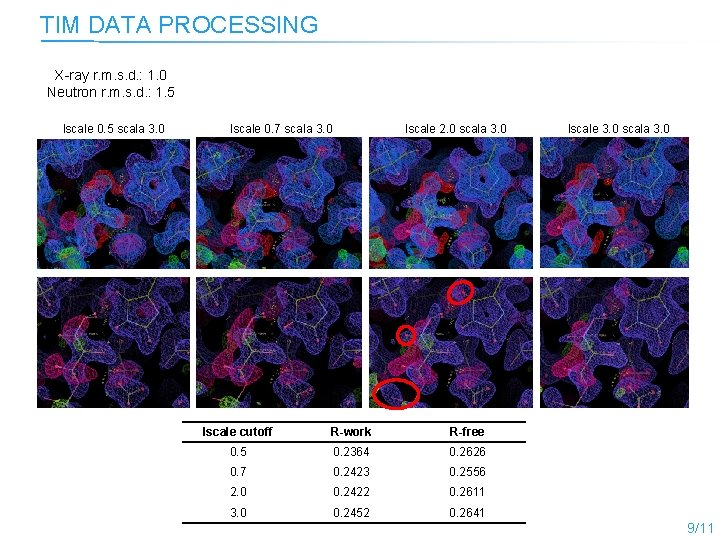
TIM DATA PROCESSING X-ray r. m. s. d. : 1. 0 Neutron r. m. s. d. : 1. 5 lscale 0. 5 scala 3. 0 lscale 0. 7 scala 3. 0 lscale 2. 0 scala 3. 0 lscale cutoff R-work R-free 0. 5 0. 2364 0. 2626 0. 7 0. 2423 0. 2556 2. 0 0. 2422 0. 2611 3. 0 0. 2452 0. 2641 lscale 3. 0 scala 3. 0 9/11

CONCLUSION & PERSPECTIVES 1. Improvement of the data treatment: Lauegen, lscale and scala cutoff 2. Need of new data processing software developements to: Initially better orient the crystal Nicolas Coquelle: https: //github. com/coquellen Find the best compromise to solve the structure Rejection cutoff of reflections 3. TIM structure present possible tautomerization of PGH inhibitor 10/11

ACKNOWLEDGEMENTS Esko Oksanen NMX Scientific Project Leader, ESS Vinardas Keplas Ph. D student, Molecular Cell Biology Unit, Lund University Claes von Wachenfeldt Senior lecturer, Molecular Cell Biology Unit, Lund University Valeria Bugris Research Associate, BRC (Szeged, HU) Matthew Blakeley Nicolas Coquelle LADI-III responsible, ILL (Grenoble, FR) LADI-III co-responsible, ILL (Grenoble, FR) Zoë Fisher Head of Deuteration Macromolecular Crystallization (DEMAX), ESS All the NMX, DEMAX and LP 3 group Wolfgang Knecht, LP 3 Manager Maria Gourdon, LP 3 scientist Annika Rogstam, LP 3 scientist Katarina Koruza, Ph. D student 11/11
 Triosephosphate isomerase
Triosephosphate isomerase Tim enzyme
Tim enzyme Cscl structure
Cscl structure Silicon unit cell
Silicon unit cell Titanium crystal structure
Titanium crystal structure Giant ionic lattice examples
Giant ionic lattice examples Miller indices examples
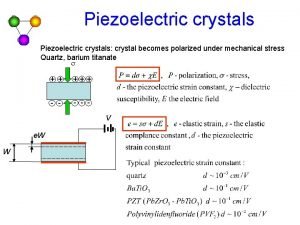
Miller indices examples Piezoelectric crystal atomic structure
Piezoelectric crystal atomic structure Basis in crystal structure
Basis in crystal structure Batio3 coordination number
Batio3 coordination number Structure of nacl crystal
Structure of nacl crystal Metallic crystal structure
Metallic crystal structure Atomic packing factor for bcc
Atomic packing factor for bcc Microwaves definition science
Microwaves definition science