Enzyme Regulation Regulation of Enzyme Activity Enzyme quantity



![Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of](https://slidetodoc.com/presentation_image/c25bd9fa20274ebb20749678cff1f894/image-7.jpg)

- Slides: 14

Enzyme Regulation

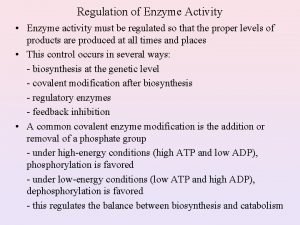
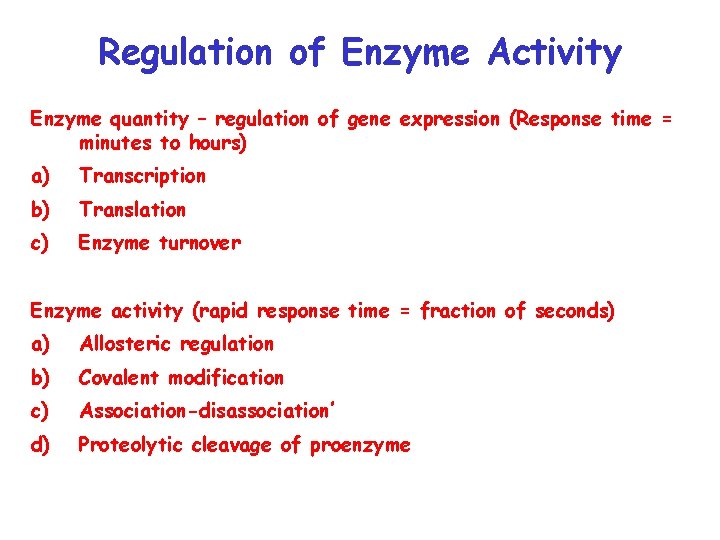
Regulation of Enzyme Activity Enzyme quantity – regulation of gene expression (Response time = minutes to hours) a) Transcription b) Translation c) Enzyme turnover Enzyme activity (rapid response time = fraction of seconds) a) Allosteric regulation b) Covalent modification c) Association-disassociation’ d) Proteolytic cleavage of proenzyme

Allosteric Regulation • End products are often inhibitors • Allosteric modulators bind to site other than the active site • Allosteric enzymes usually have 4 o structure • Vo vs [S] plots give sigmoidal curve for at least one substrate • Can remove allosteric site without effecting enzymatic action

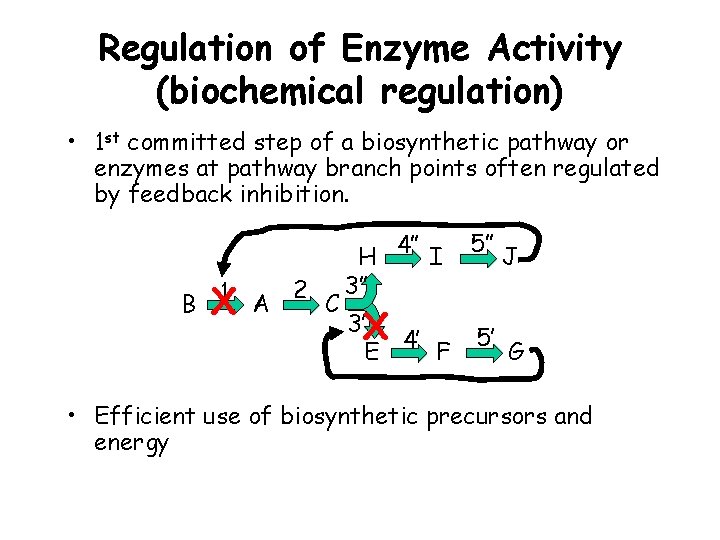
Regulation of Enzyme Activity (biochemical regulation) • 1 st committed step of a biosynthetic pathway or enzymes at pathway branch points often regulated by feedback inhibition. 1 A 2 C B X H 4” I 3” 3’X E 4’ F 5” 5’ J G • Efficient use of biosynthetic precursors and energy

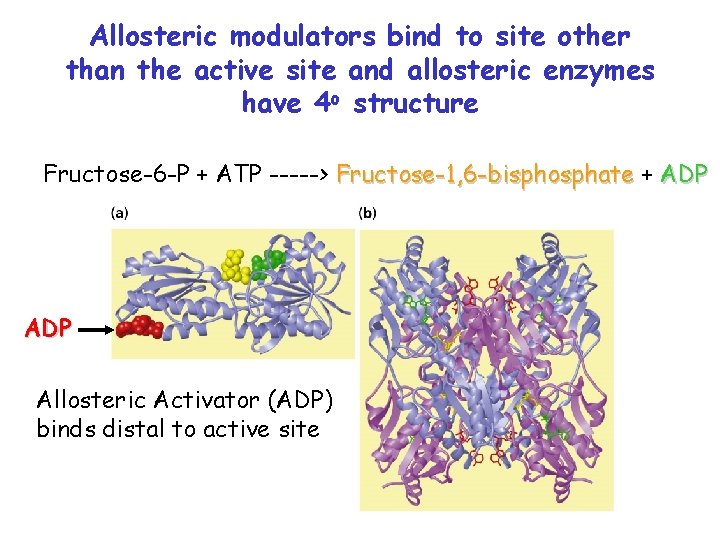
Phosphofructokinase( PFK) Fructose-6 -P + ATP -----> Fructose-1, 6 -bisphosphate + ADP • PFK catalyzes 1 st committed step in glycolysis (10 steps total) (Glucose + 2 ADP + 2 NAD+ + 2 Pi 2 pyruvate + 2 ATP + 2 NADH) • Phosphoenolpyruvate is an allosteric inhibitor of PFK • ADP is an allosteric activator of PFK

Allosteric modulators bind to site other than the active site and allosteric enzymes have 4 o structure Fructose-6 -P + ATP -----> Fructose-1, 6 -bisphosphate + ADP Allosteric Activator (ADP) binds distal to active site
![Vo vs S plots give sigmoidal curve for at least one substrate Binding of Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of](https://slidetodoc.com/presentation_image/c25bd9fa20274ebb20749678cff1f894/image-7.jpg)
Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of allosteric inhibitor or activator does not effect Vmax, but does alter Km Allosteric enzyme do not follow M-M kinetics

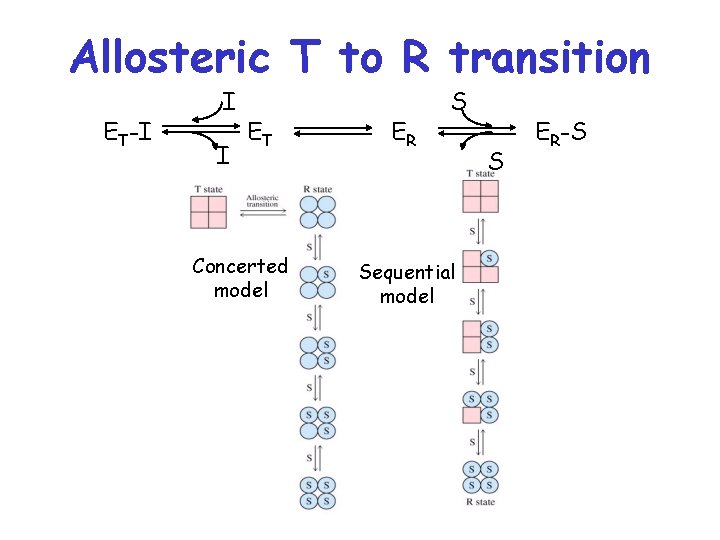
Allosteric T to R transition ET-I I I ET Concerted model ER S Sequential model S ER-S

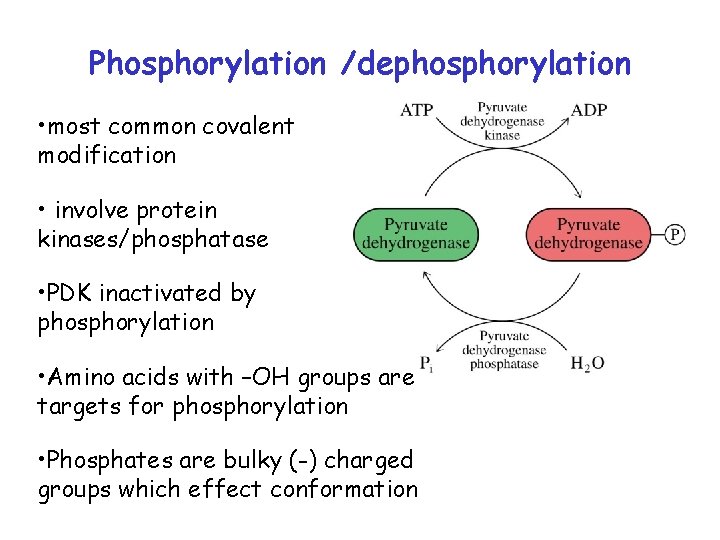
Covalent modification • Regulation by covalent modification is slower than allosteric regulation • Reversible • Require one enzyme for activation and one enzyme for inactivation • Covalent modification freezes enzyme T or R conformation

Phosphorylation /dephosphorylation • most common covalent modification • involve protein kinases/phosphatase • PDK inactivated by phosphorylation • Amino acids with –OH groups are targets for phosphorylation • Phosphates are bulky (-) charged groups which effect conformation

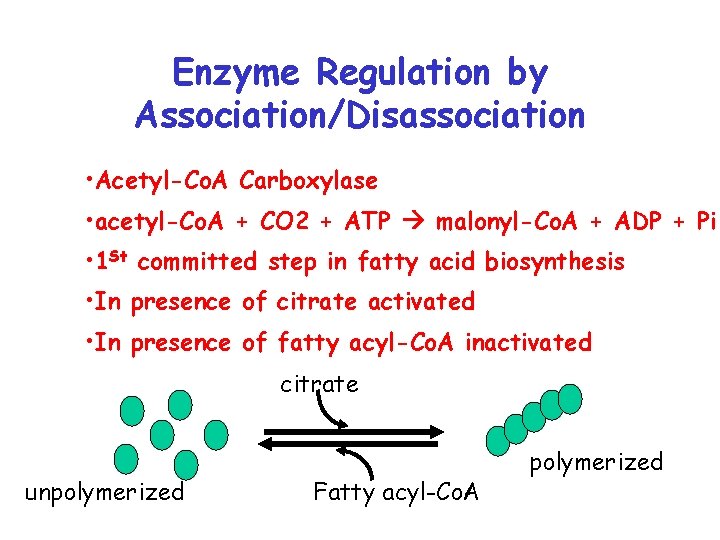
Enzyme Regulation by Association/Disassociation • Acetyl-Co. A Carboxylase • acetyl-Co. A + CO 2 + ATP malonyl-Co. A + ADP + Pi • 1 St committed step in fatty acid biosynthesis • In presence of citrate activated • In presence of fatty acyl-Co. A inactivated citrate unpolymerized Fatty acyl-Co. A polymerized

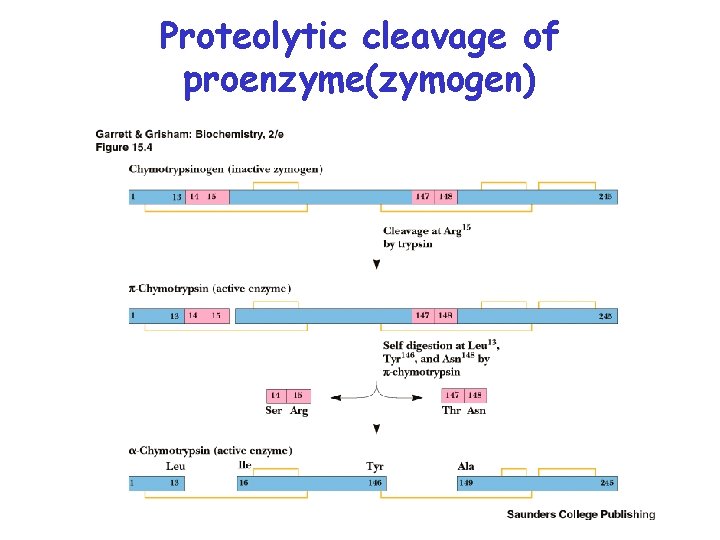
Proteolytic cleavage of proenzyme(zymogen)

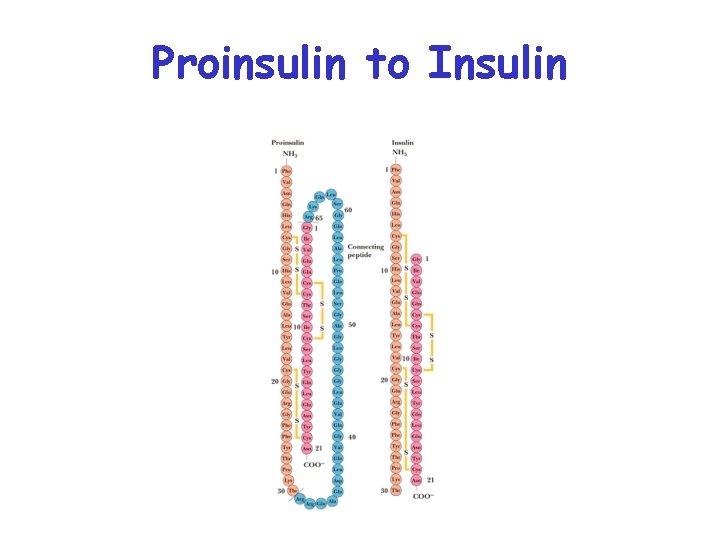
Proinsulin to Insulin

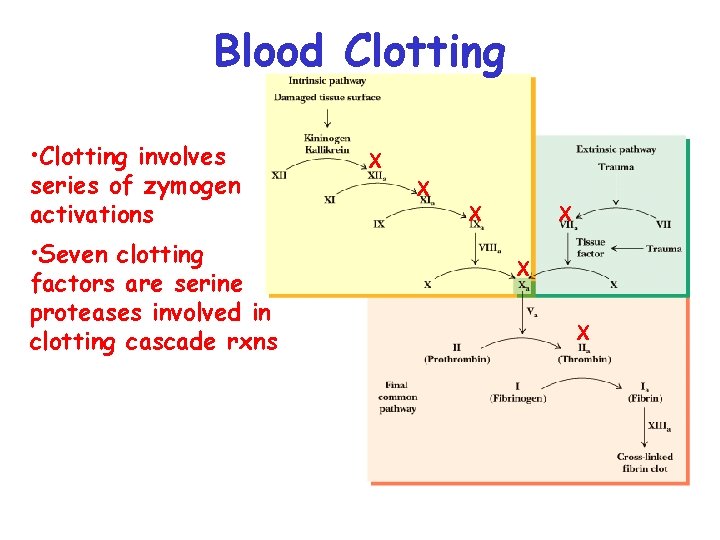
Blood Clotting • Clotting involves series of zymogen activations • Seven clotting factors are serine proteases involved in clotting cascade rxns X X X
 Covalently modulated enzymes
Covalently modulated enzymes Enzyme regulation
Enzyme regulation Vary inversely
Vary inversely All examples of vector quantity
All examples of vector quantity Scalar quantity
Scalar quantity Vector and scalar quantities
Vector and scalar quantities Quantity has magnitude only
Quantity has magnitude only Enzyme regulation
Enzyme regulation Thiamin
Thiamin Enzyme activity graph
Enzyme activity graph Factors affecting enzyme activity bbc bitesize
Factors affecting enzyme activity bbc bitesize Enzyme activity graph
Enzyme activity graph Enzyme cut-outs activity answer key
Enzyme cut-outs activity answer key Factors affecting enzyme activity slideshare
Factors affecting enzyme activity slideshare Enzymease
Enzymease