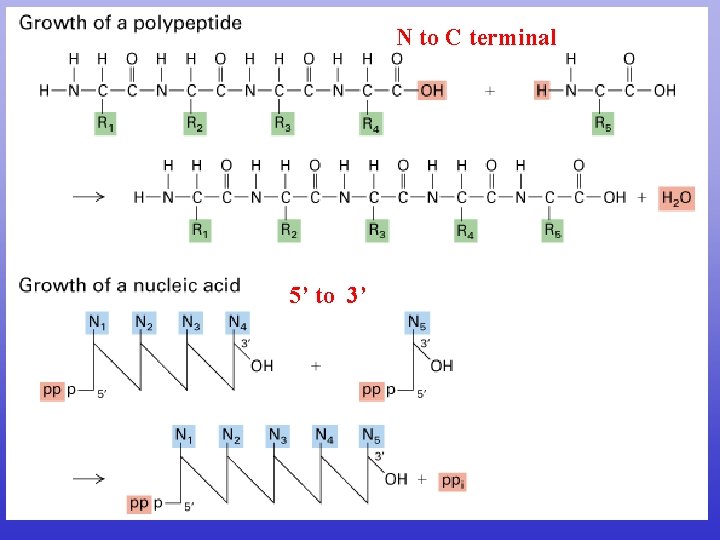
N to C terminal 5 to 3 CDS











































- Slides: 68






N to C terminal 5’ to 3’

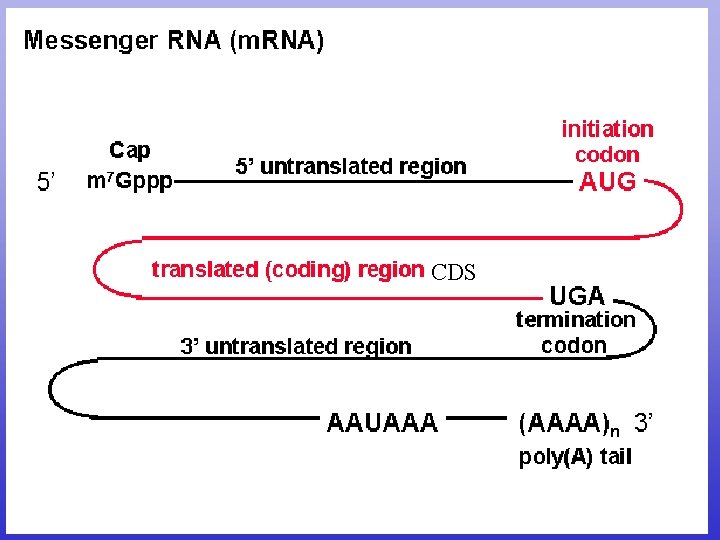
CDS






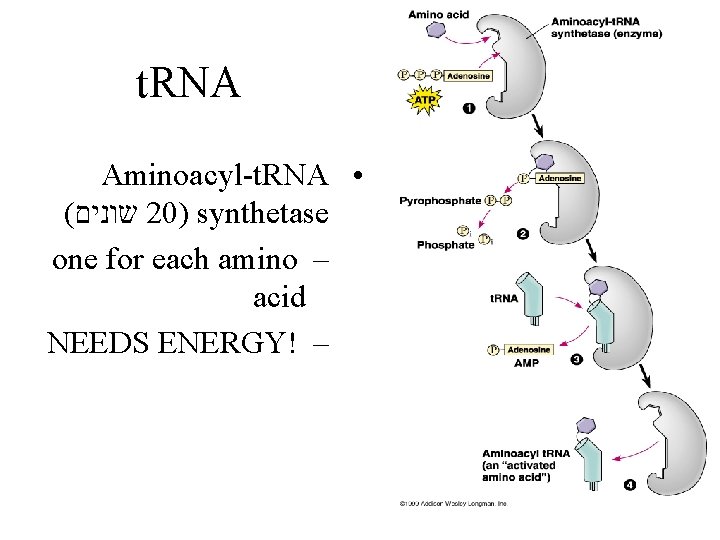
t. RNA Aminoacyl-t. RNA • ( שונים 20) synthetase one for each amino – acid NEEDS ENERGY! –






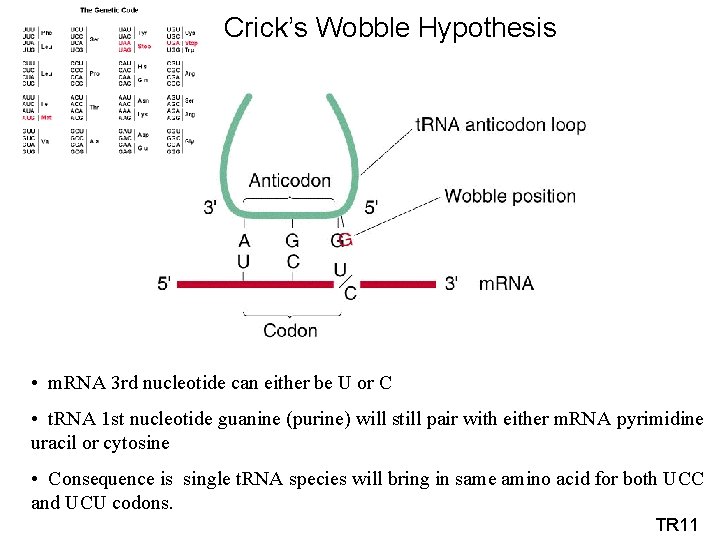
Crick’s Wobble Hypothesis • m. RNA 3 rd nucleotide can either be U or C • t. RNA 1 st nucleotide guanine (purine) will still pair with either m. RNA pyrimidine uracil or cytosine • Consequence is single t. RNA species will bring in same amino acid for both UCC and UCU codons. TR 11




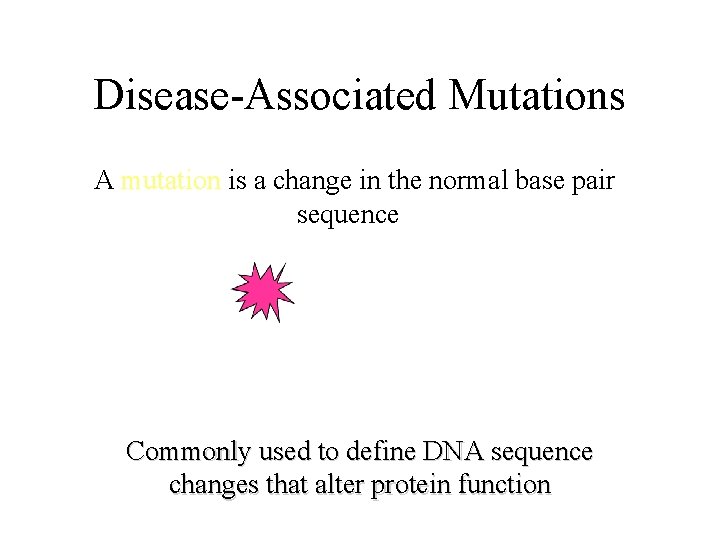
Disease-Associated Mutations A mutation is a change in the normal base pair sequence Commonly used to define DNA sequence changes that alter protein function

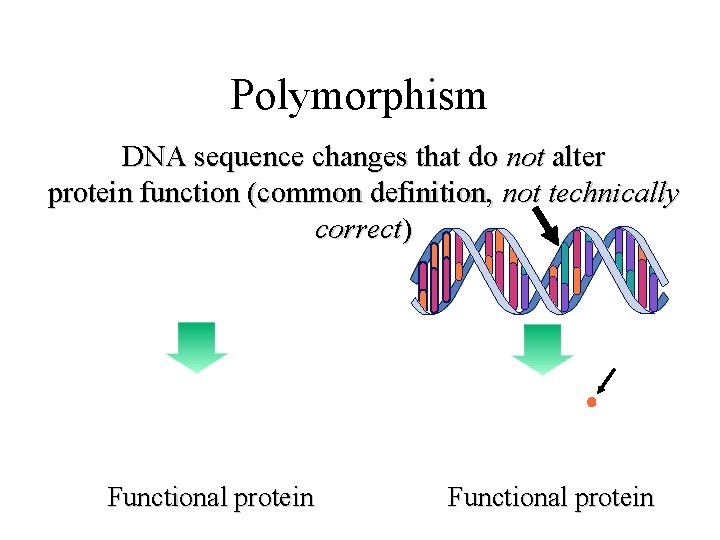
Polymorphism DNA sequence changes that do not alter protein function (common definition, not technically correct) Functional protein

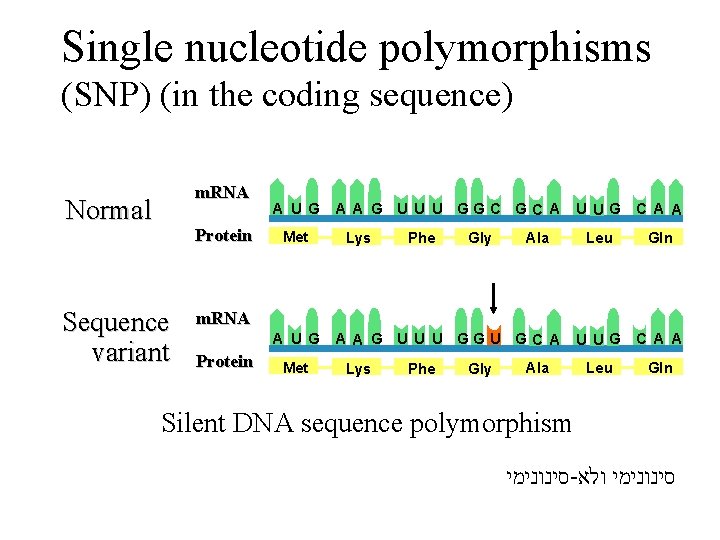
Single nucleotide polymorphisms (SNP) (in the coding sequence) m. RNA Normal Protein Sequence variant A U G Met A A G U U U GGC GC A U UG C A A Lys Phe Gly Ala Leu Gln m. RNA A U G Protein Met A A G U U U GGU GC A U UG C A A Lys Phe Gly Ala Leu Gln Silent DNA sequence polymorphism סינונימי - סינונימי ולא

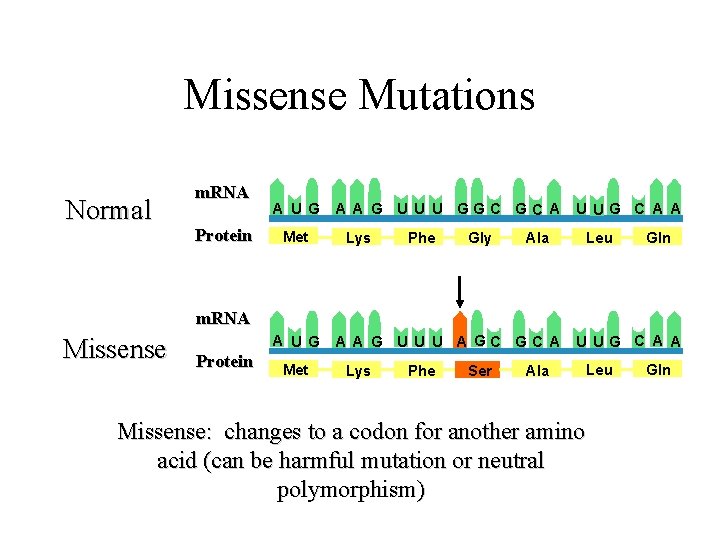
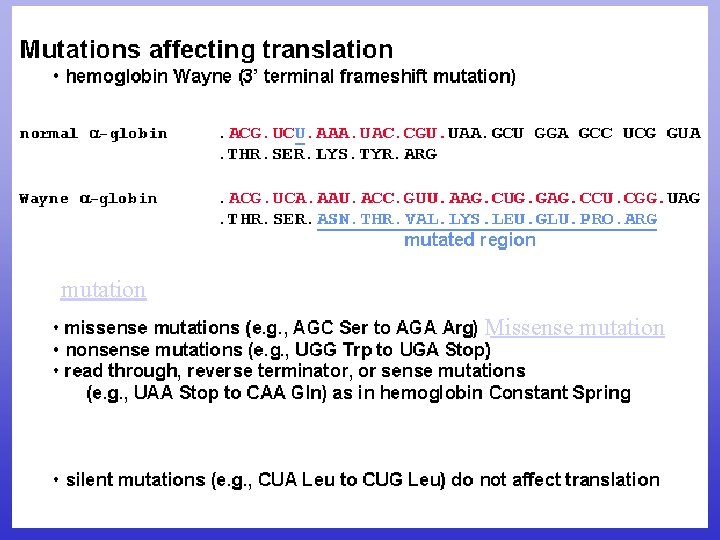
Missense Mutations Normal m. RNA Protein A U G Met A A G U U U GGC GC A U UG C A A Lys Phe Gly Ala Leu Gln m. RNA Missense A U G Protein Met A A G U U U AGC GC A U UG C A A Lys Phe Ser Ala Leu Missense: changes to a codon for another amino acid (can be harmful mutation or neutral polymorphism) Gln

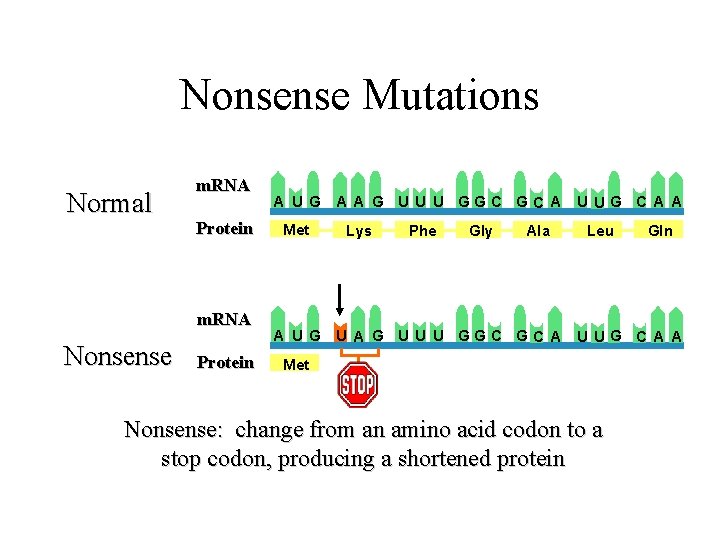
Nonsense Mutations Normal m. RNA Protein m. RNA Nonsense Protein A U G Met A U G A A G U U U GGC GC A U UG C A A Lys Phe Gly Ala Leu Gln U A G U U U GGC GC A U UG C A A Met Nonsense: change from an amino acid codon to a stop codon, producing a shortened protein

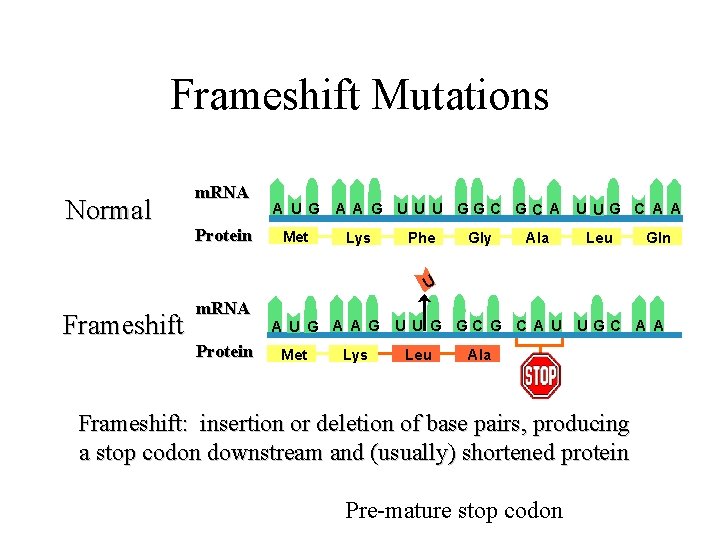
Frameshift Mutations Normal m. RNA Protein A U G Met A A G U U U GGC GC A U UG C A A Lys Phe Gly Ala Leu Gln U Frameshift m. RNA Protein A U G A A G U U G GC G C A U UGC A A Met Lys Leu Ala Frameshift: insertion or deletion of base pairs, producing a stop codon downstream and (usually) shortened protein Pre-mature stop codon

mutation Missense mutation






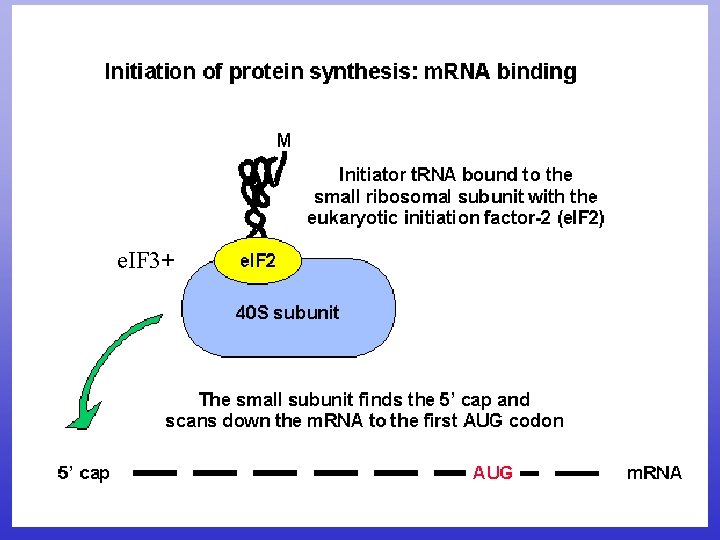
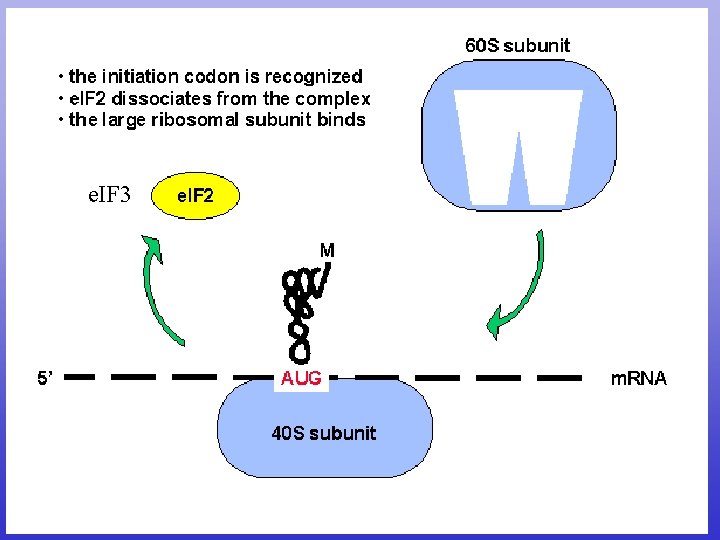
e. IF 3+

e. IF 3


Translation



סרט טרנסלציה Life cycle of m. RNA movie

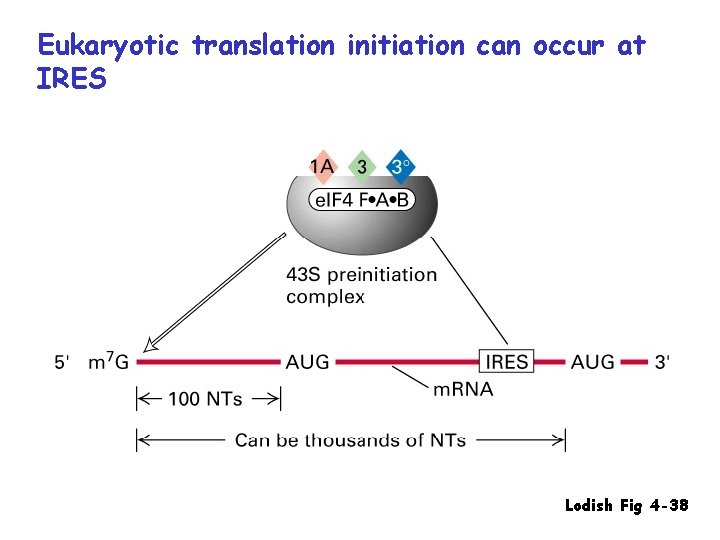
Eukaryotic translation initiation can occur at IRES Lodish Fig 4 -38

Ada Yonat Movie



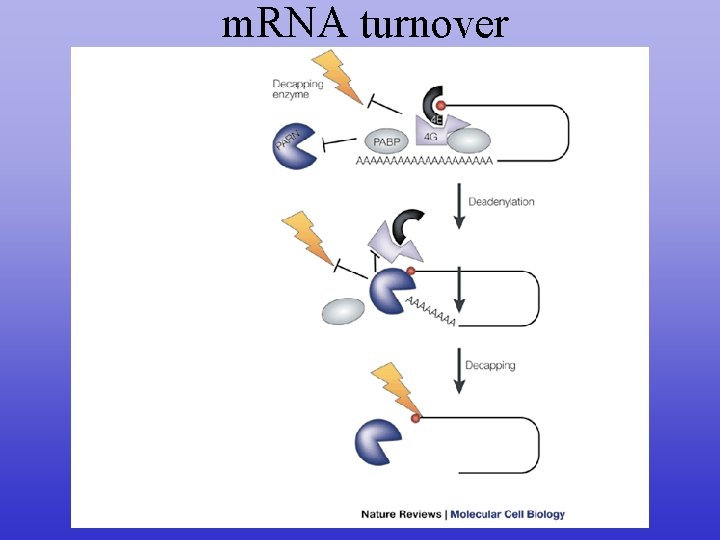
m. RNA turnover

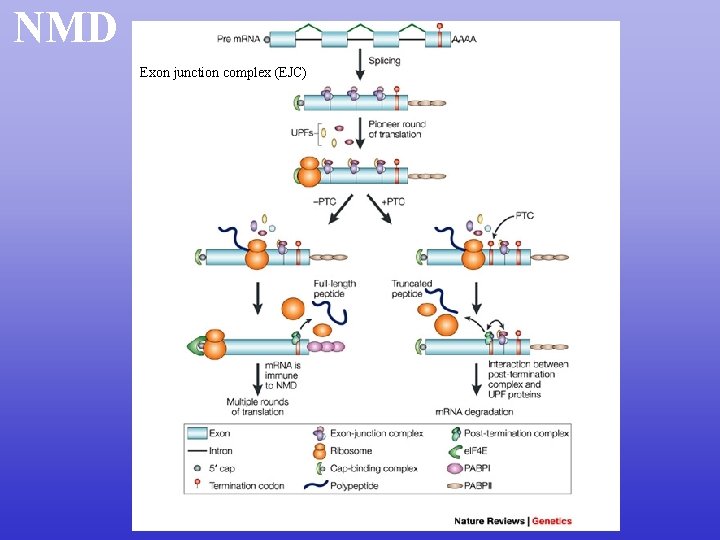
NMD Exon junction complex (EJC)

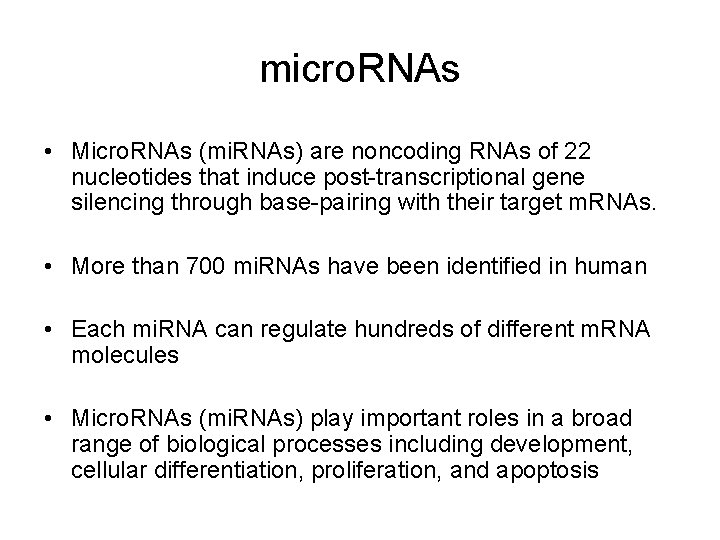
micro. RNAs • Micro. RNAs (mi. RNAs) are noncoding RNAs of 22 nucleotides that induce post-transcriptional gene silencing through base-pairing with their target m. RNAs. • More than 700 mi. RNAs have been identified in human • Each mi. RNA can regulate hundreds of different m. RNA molecules • Micro. RNAs (mi. RNAs) play important roles in a broad range of biological processes including development, cellular differentiation, proliferation, and apoptosis

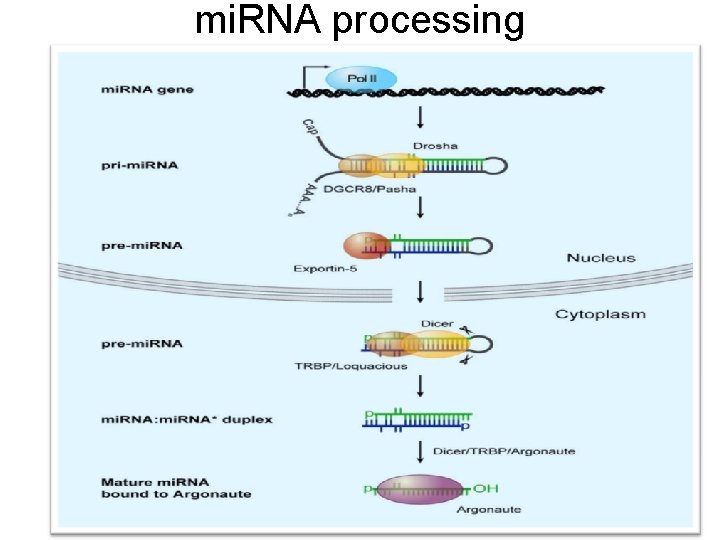
mi. RNA processing

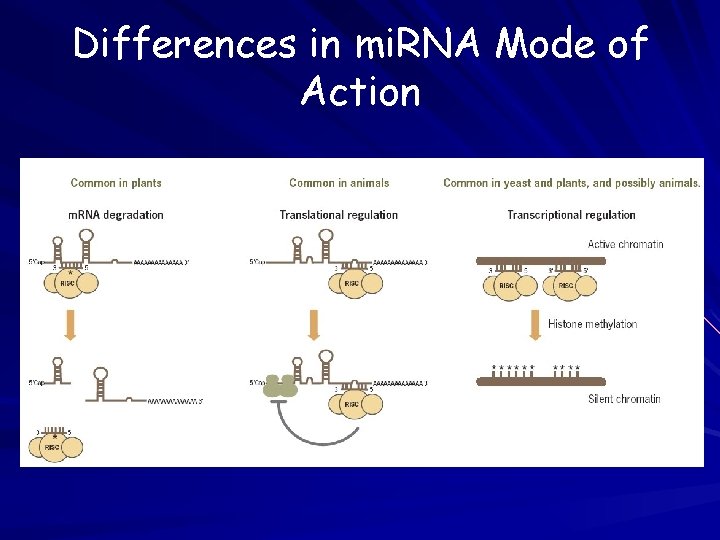
Differences in mi. RNA Mode of Action

mi. RNA Registry http: //www. sanger. ac. uk/Software/Rfa m/mirna/index. shtml Latest release contains 4584 predicted and verified mi. RNAs (May 2007) 321 predicted and 223 experimentally verified in Homo sapiens Mouse and human are highly conserved Human is not conserved with plants בקרה על התפתחות ומעורבות בסרטן





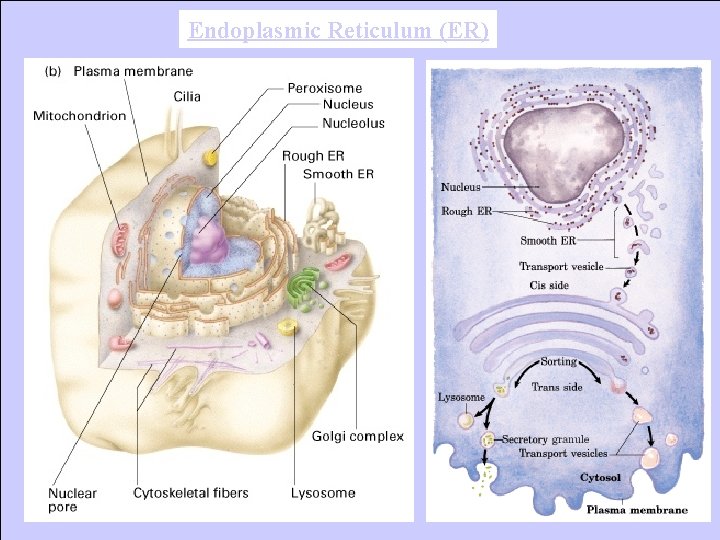
Endoplasmic Reticulum (ER)








FD - Familial Dysautonomia Riley-Day Syndrome Gil Ast

FD- Familial Dysautonomia • An autosomal recessive congenital neuropathy • Poor development and progressive degeneration of sensory and autonomic nervous system • Common in Ashkenazi Jewish population – carrier: 1: 30 (1: 18 in Polish Jews), live births: 1: 3, 600 • Symptoms: decreased sensitivity to pain and temperature, cardiovascular instability, gastrointestinal dysfunction, postural hypotension, and more • 50% of patients die before the age of 30

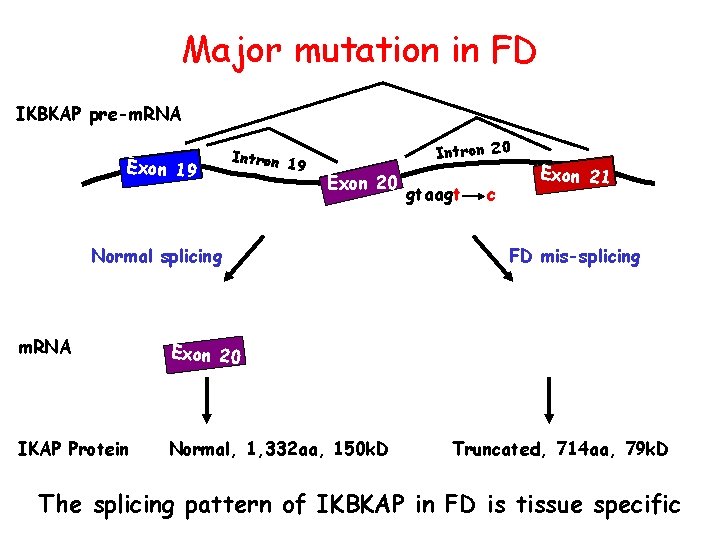
Major mutation in FD IKBKAP pre-m. RNA Exon 1 9 Intron 20 Exon 20 Normal splicing m. RNA Exon 20 IKAP Protein Normal, 1, 332 aa, 150 k. D gtaagt c E xon 21 FD mis-splicing Truncated, 714 aa, 79 k. D The splicing pattern of IKBKAP in FD is tissue specific

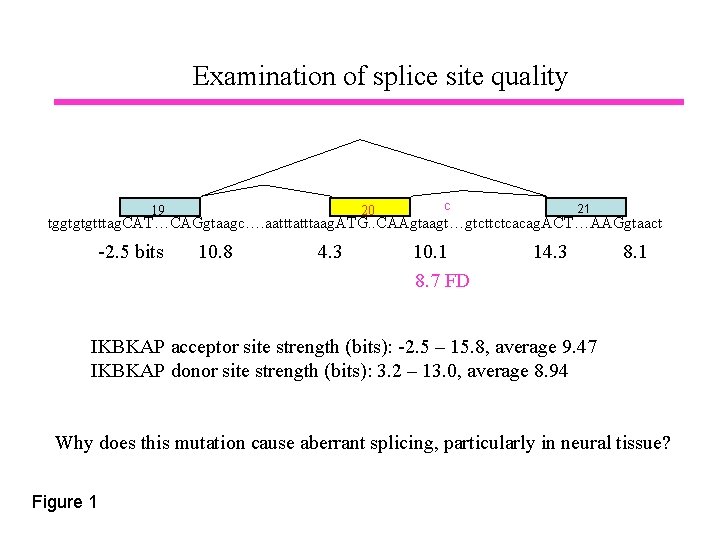
Examination of splice site quality 19 20 c 21 tggtgtgtttag. CAT…CAGgtaagc…. aatttaag. ATG. . CAAgtaagt…gtcttctcacag. ACT…AAGgtaact -2. 5 bits 10. 8 4. 3 10. 1 8. 7 FD 14. 3 8. 1 IKBKAP acceptor site strength (bits): -2. 5 – 15. 8, average 9. 47 IKBKAP donor site strength (bits): 3. 2 – 13. 0, average 8. 94 Why does this mutation cause aberrant splicing, particularly in neural tissue? Figure 1
