more regulating gene expression We looked at the







- Slides: 67

more regulating gene expression

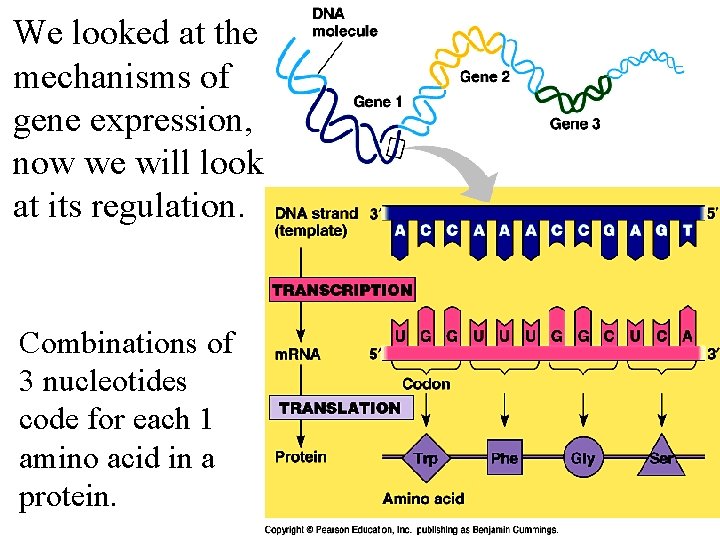
We looked at the mechanisms of gene expression, now we will look at its regulation. Combinations of 3 nucleotides code for each 1 amino acid in a protein.

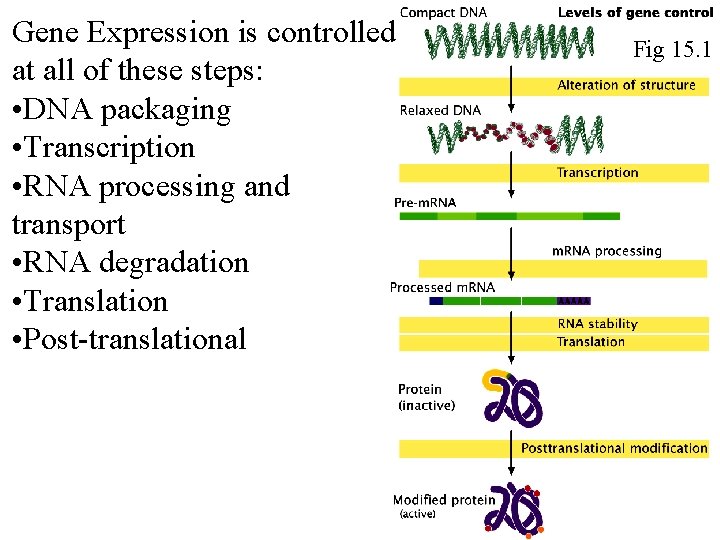
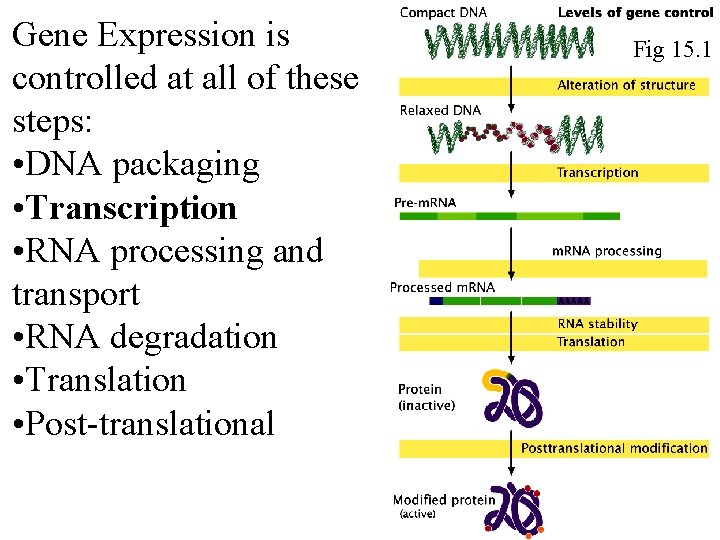
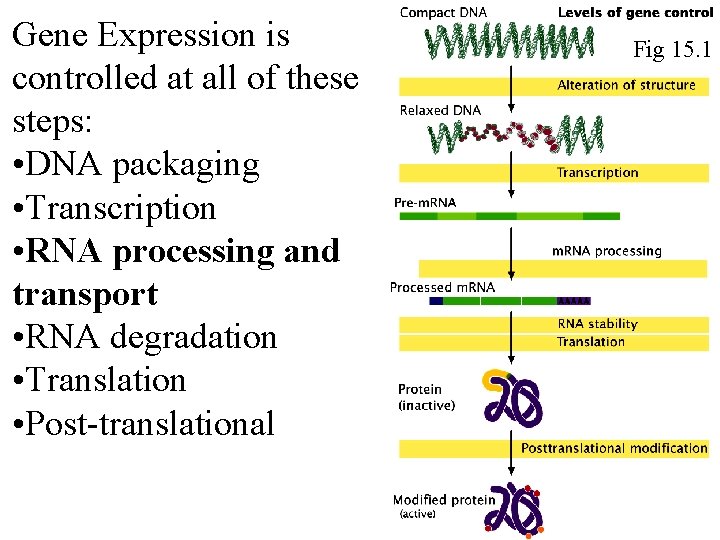
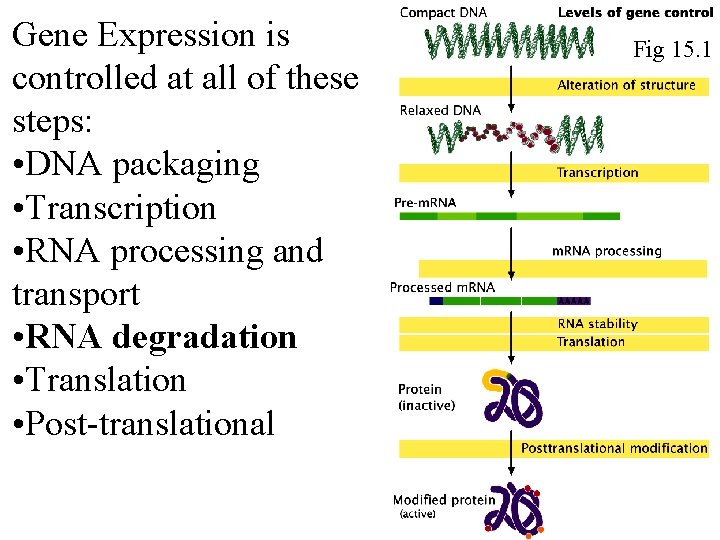
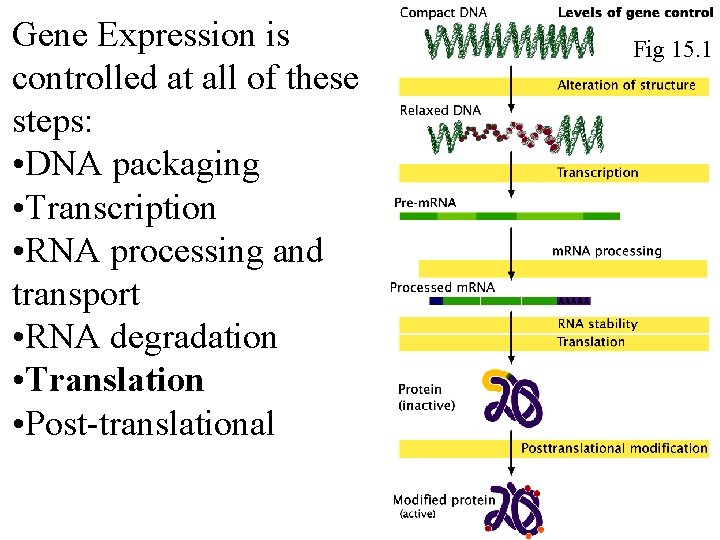
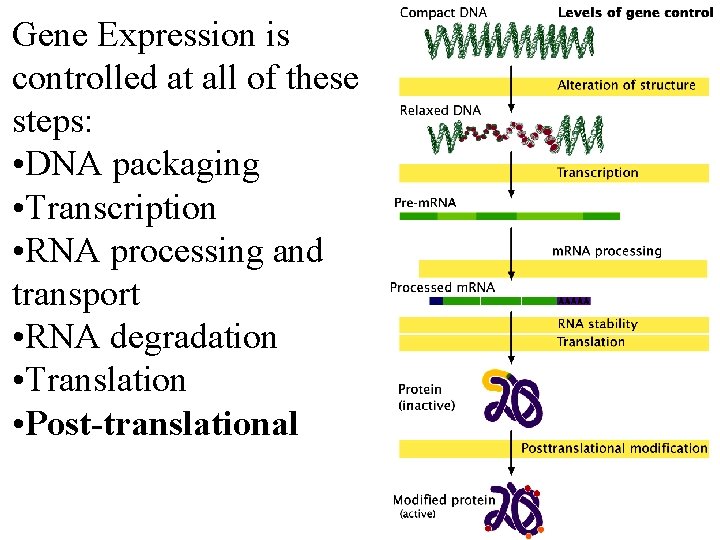
Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

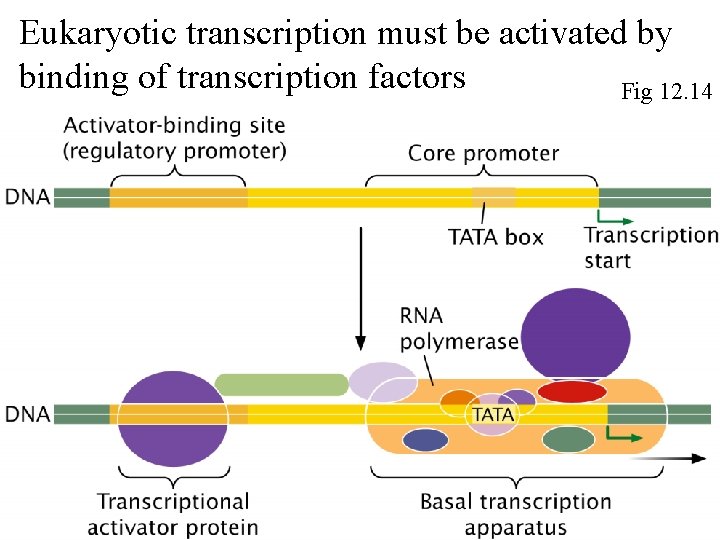
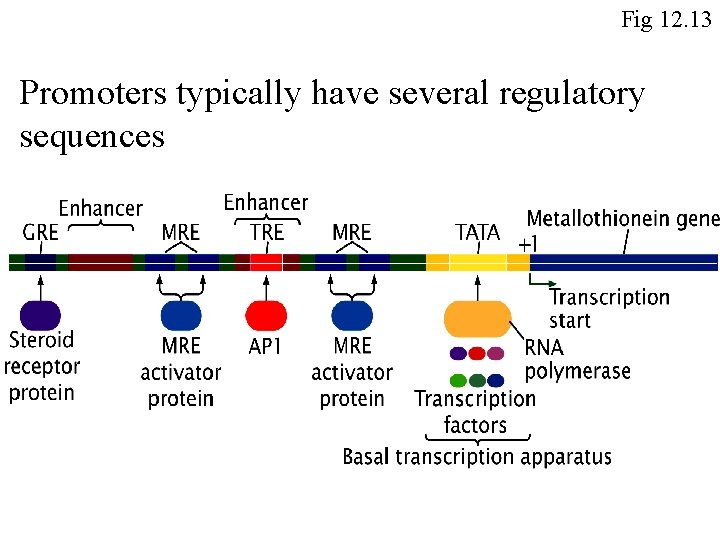
Eukaryotic transcription must be activated by binding of transcription factors Fig 12. 14

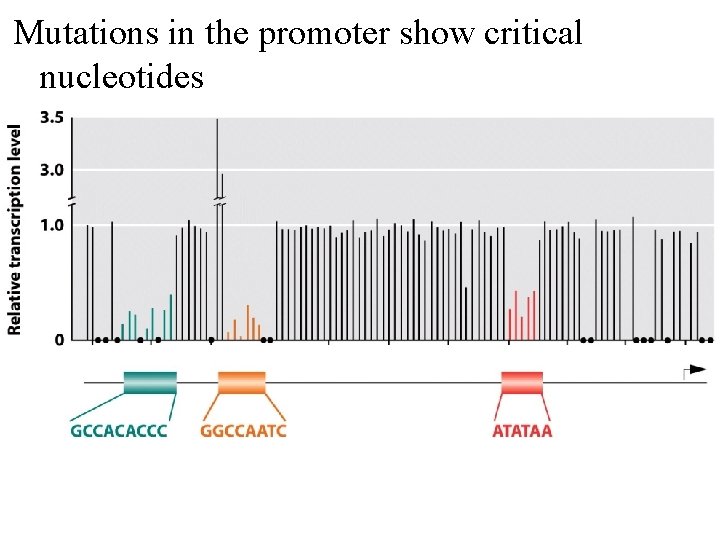
Mutations in the promoter show critical nucleotides

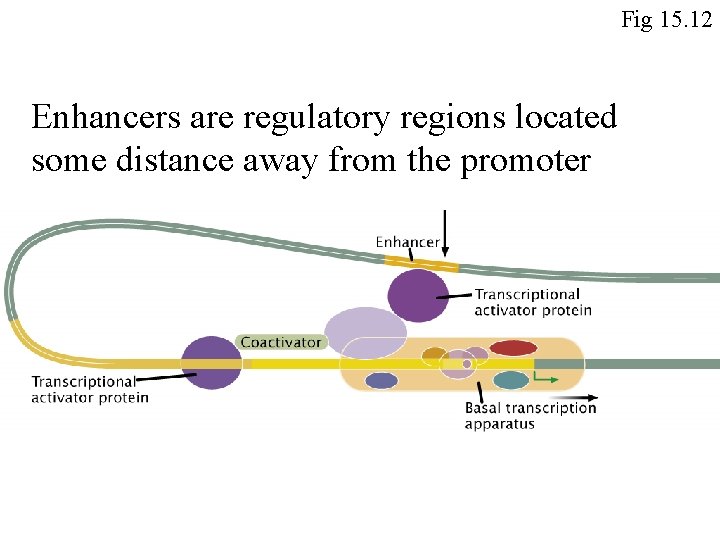
Fig 15. 12 Enhancers are regulatory regions located some distance away from the promoter

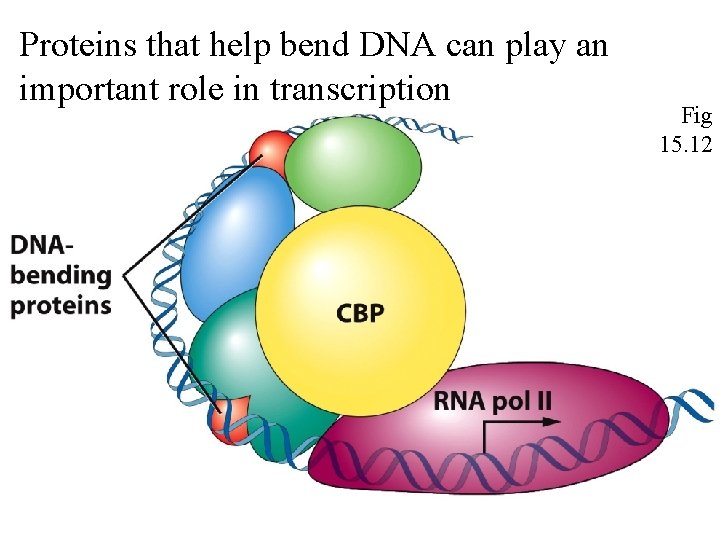
Proteins that help bend DNA can play an important role in transcription Fig 15. 12

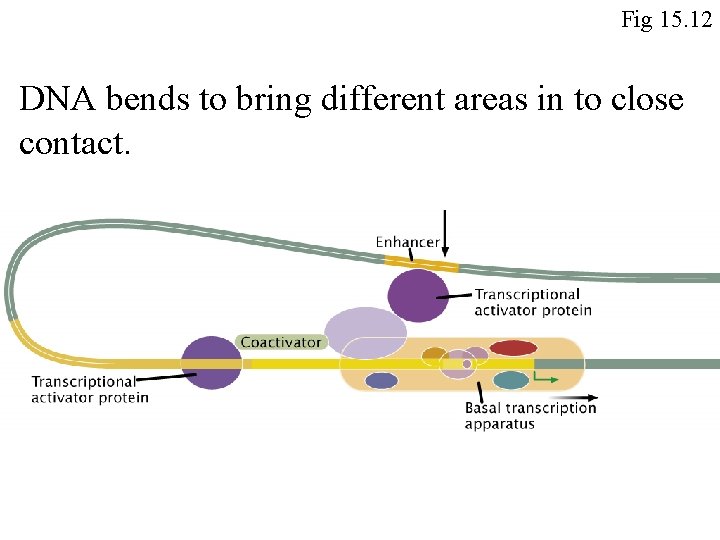
Fig 15. 12 DNA bends to bring different areas in to close contact.

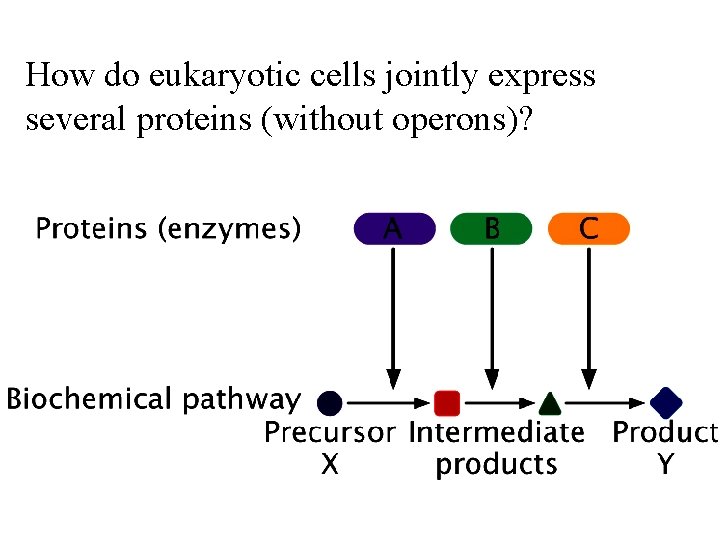
How do eukaryotic cells jointly express several proteins (without operons)?

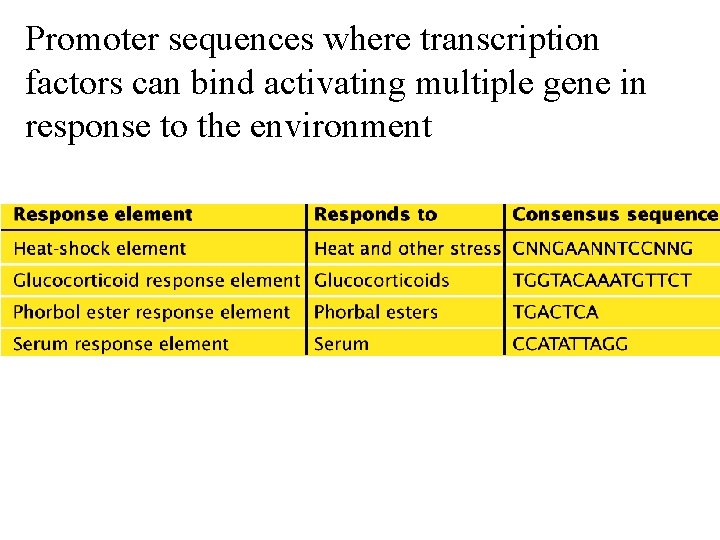
Promoter sequences where transcription factors can bind activating multiple gene in response to the environment

Fig 12. 13 Promoters typically have several regulatory sequences

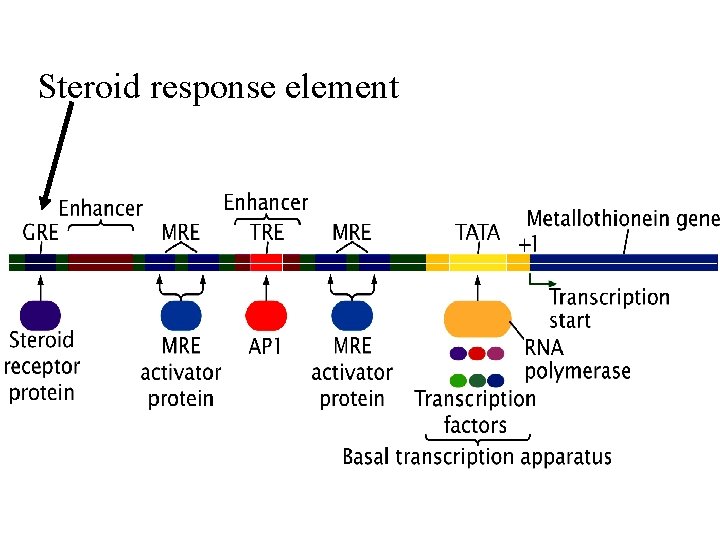
Steroid response element

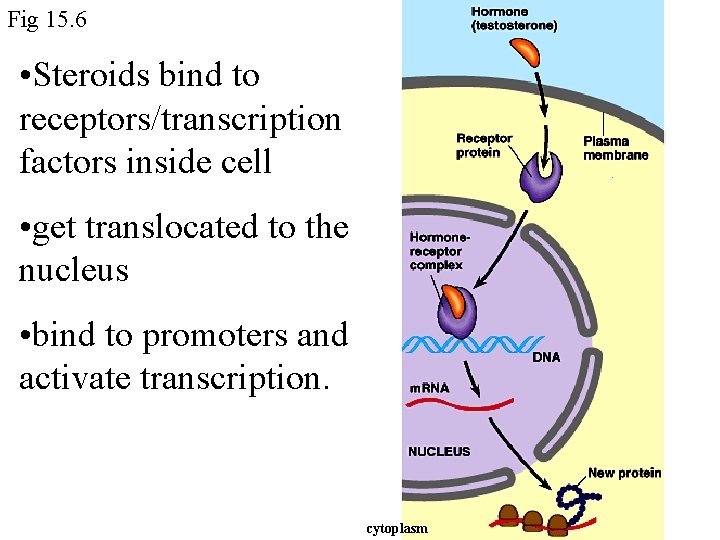
Fig 15. 6 • Steroids bind to receptors/transcription factors inside cell • get translocated to the nucleus • bind to promoters and activate transcription. cytoplasm

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

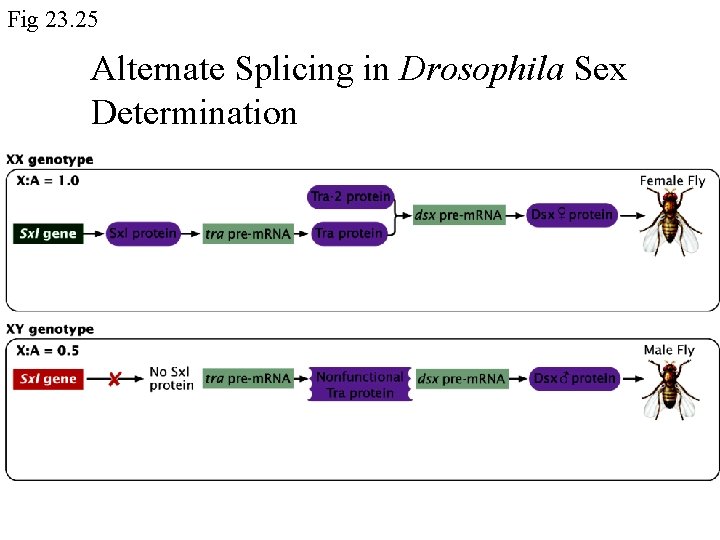
Fig 23. 25 Alternate Splicing in Drosophila Sex Determination

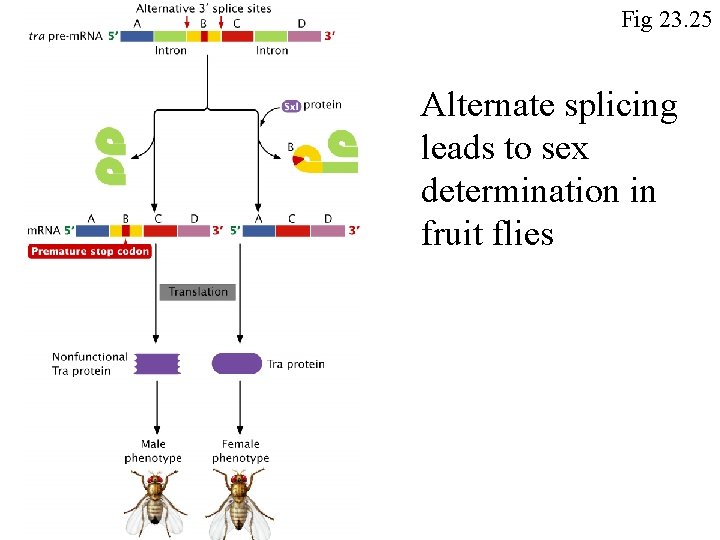
Fig 23. 25 Alternate splicing leads to sex determination in fruit flies

Mammalian m. RNA Splice-Isoform Selection Is Tightly Controlled Jennifer L. Chisa and David T. Burke Genetics, Vol. 175: 1079 -1087, March 2007 • Regulation of gene expression is often in response to a changing environment. • But how stable can alternative splicing be, and does it play a role in maintaining homeostasis?

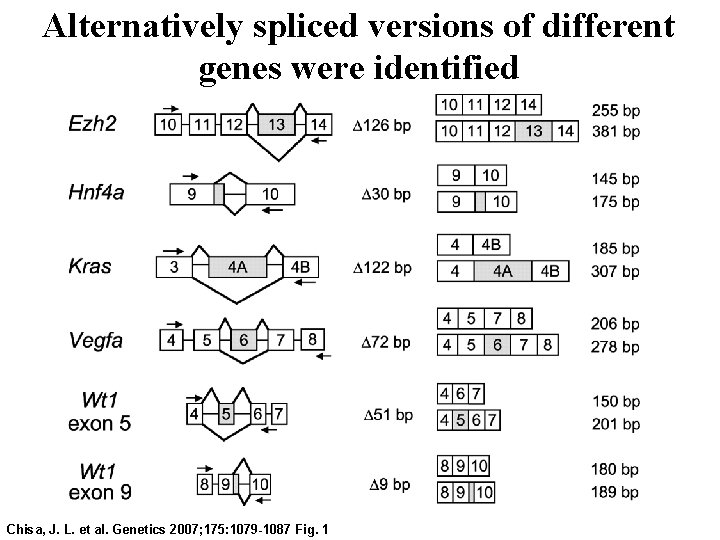
• Alternative splicing modifies at least half of all primary m. RNA transcripts in mammals. • More than one alternative splice isoform can be maintained concurrently in the steady state m. RNA pool of a single tissue or cell type, and changes in the ratios of isoforms have been associated with physiological variation and susceptibility to disease. • Splice isoforms with opposing functions can be generated; for example, different isoforms of Bcl-x have pro-apoptotic and anti-apoptotic function. Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 1

Alternatively spliced versions of different genes were identified Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 1

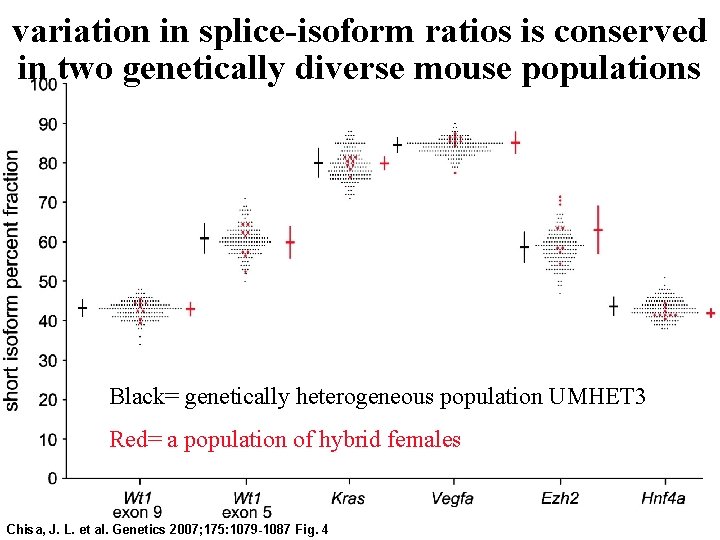
variation in splice-isoform ratios is conserved in two genetically diverse mouse populations Black= genetically heterogeneous population UMHET 3 Red= a population of hybrid females Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 4

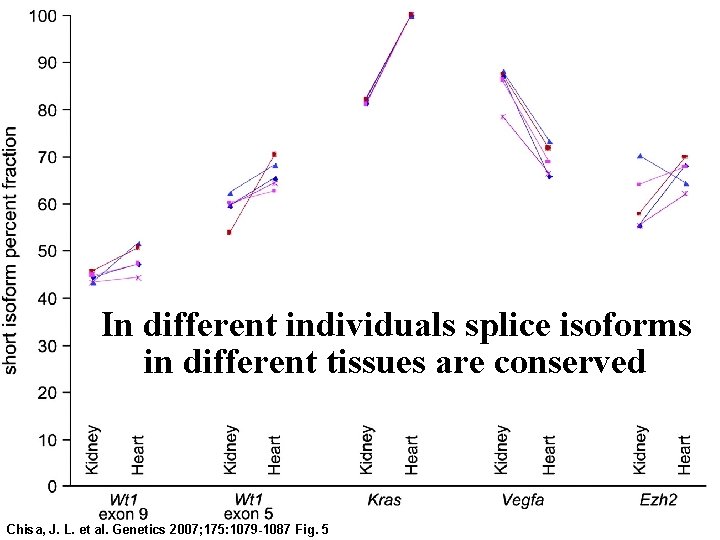
In different individuals splice isoforms in different tissues are conserved Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 5

Conclusions: • Alternate splicing for some genes is tightly regulated between different individuals. • Slight differences in alternative splicing may be indicative of abnormalities (disease).

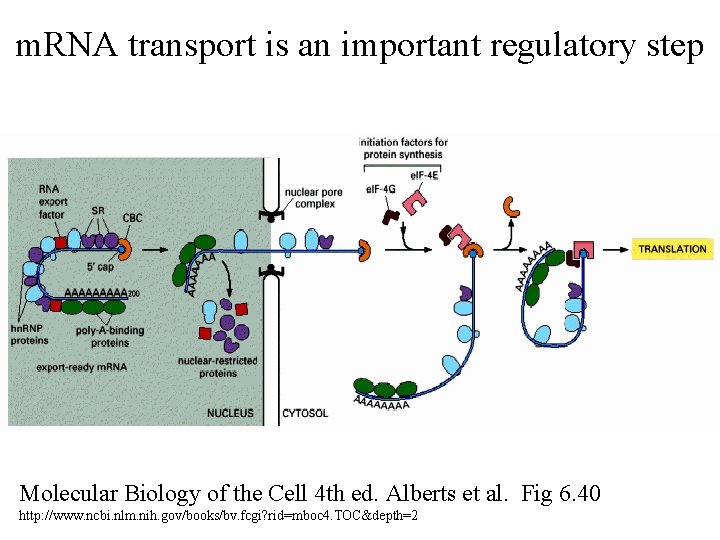
m. RNA transport is an important regulatory step Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 6. 40 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

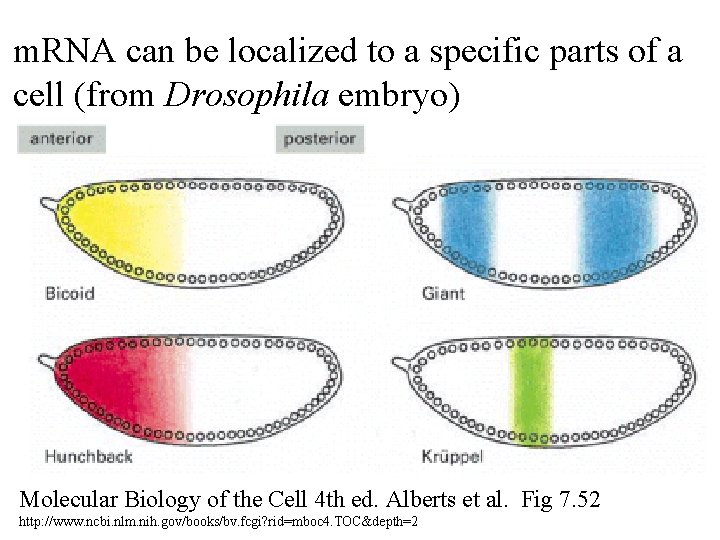
m. RNA can be localized to a specific parts of a cell (from Drosophila embryo) Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 52 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

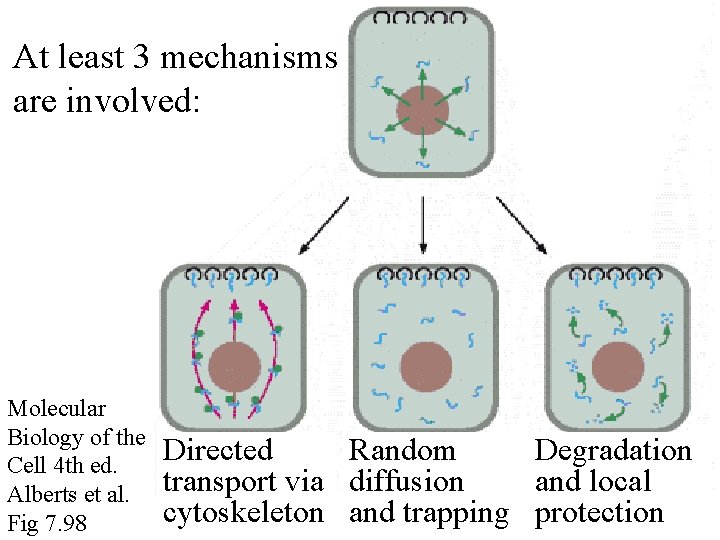
At least 3 mechanisms are involved: Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 98 Directed Random Degradation transport via diffusion and local cytoskeleton and trapping protection

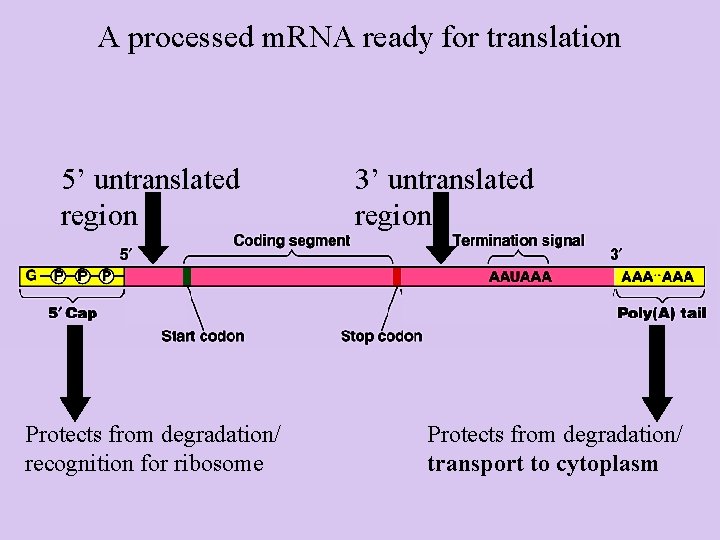
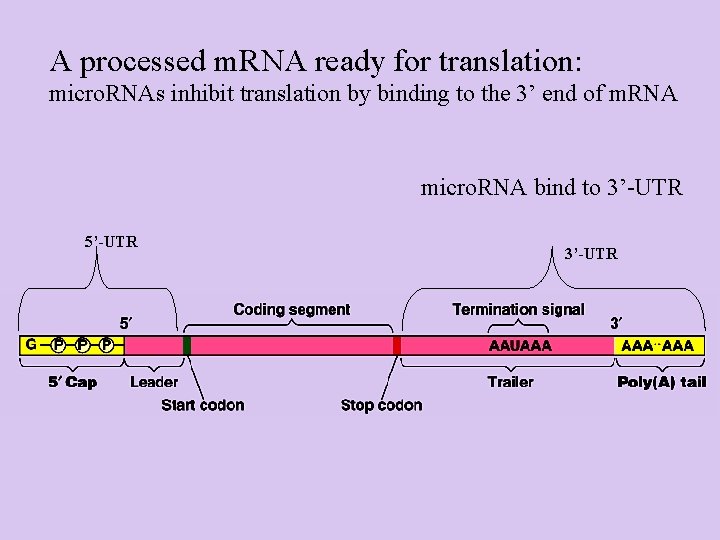
A processed m. RNA ready for translation 5’ untranslated region Protects from degradation/ recognition for ribosome 3’ untranslated region Protects from degradation/ transport to cytoplasm

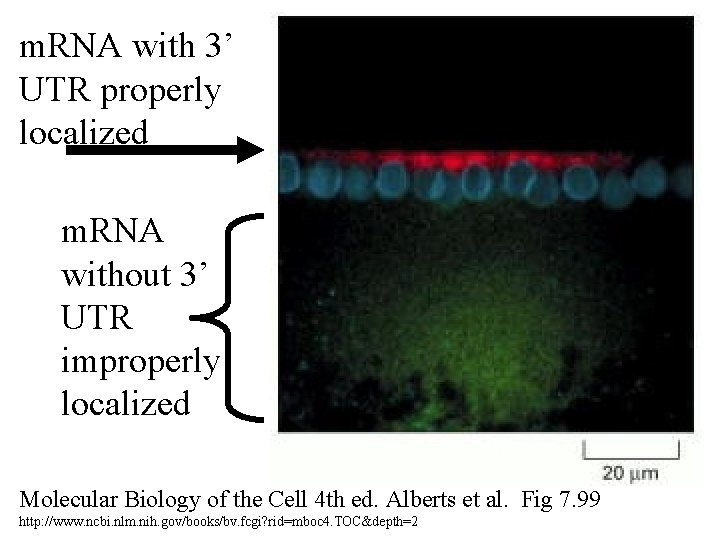
m. RNA with 3’ UTR properly localized m. RNA without 3’ UTR improperly localized Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 99 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

Seeds germinated underground begin growing in darkness then emerge into light and begin photosynthesis energy from seed energy from sun

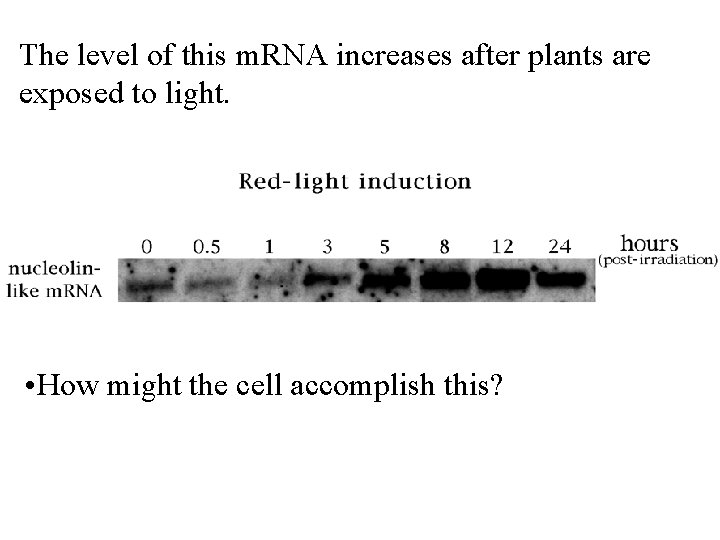
The level of this m. RNA increases after plants are exposed to light. • How might the cell accomplish this?

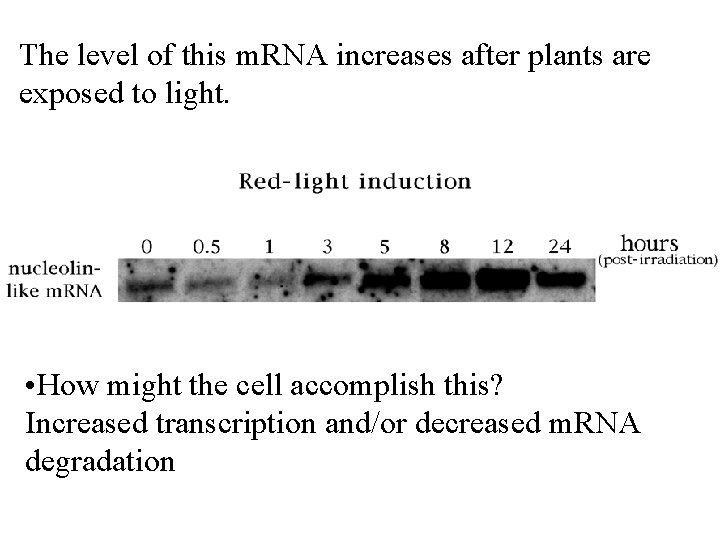
The level of this m. RNA increases after plants are exposed to light. • How might the cell accomplish this? Increased transcription and/or decreased m. RNA degradation

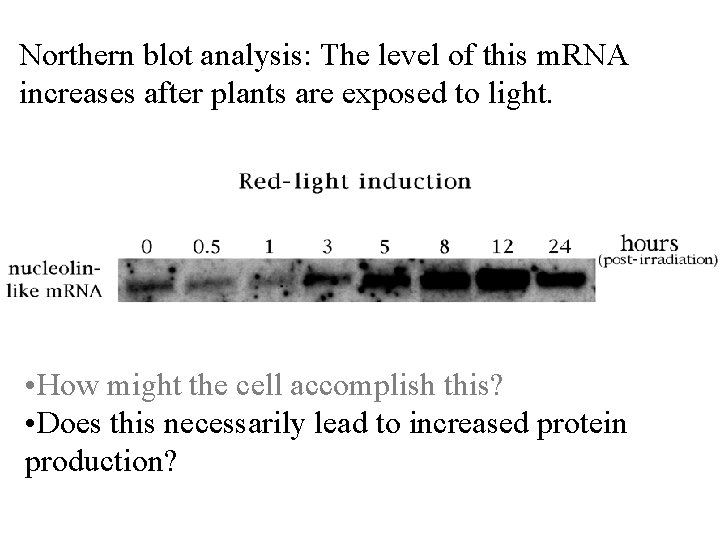
Northern blot analysis: The level of this m. RNA increases after plants are exposed to light. • How might the cell accomplish this? • Does this necessarily lead to increased protein production?

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

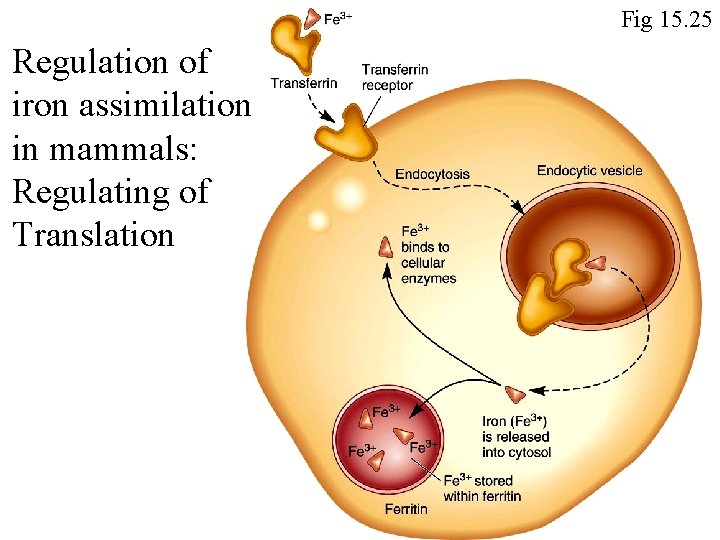
Fig 15. 25 Regulation of iron assimilation in mammals: Regulating of Translation

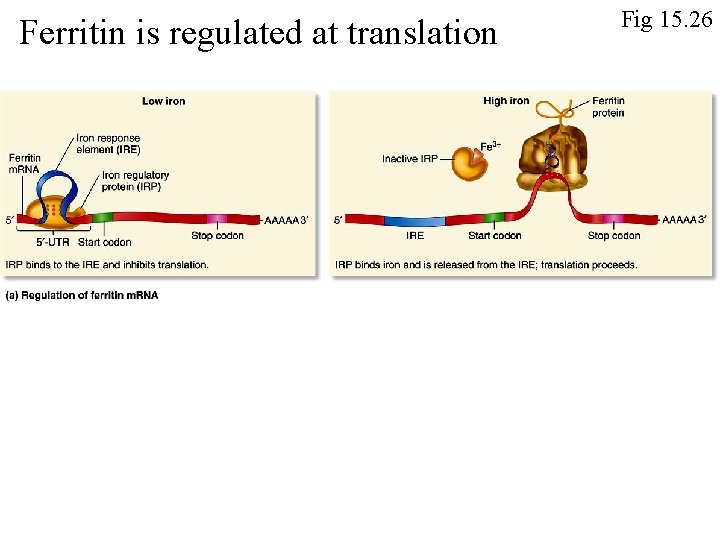
Ferritin is regulated at translation Fig 15. 26

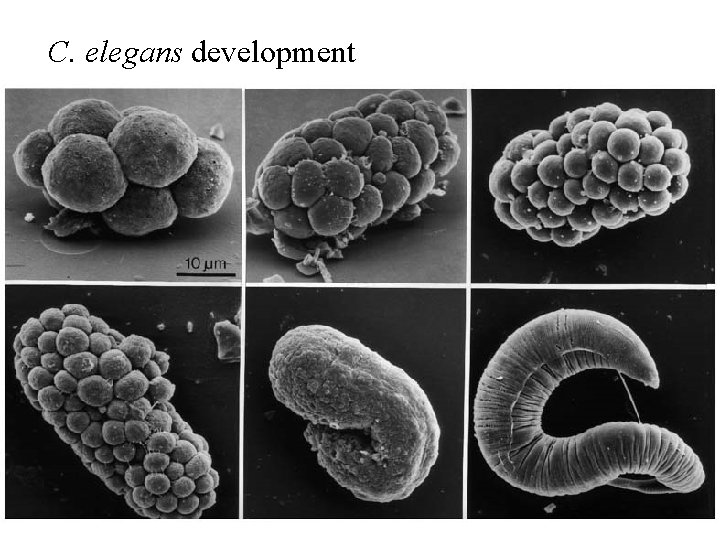
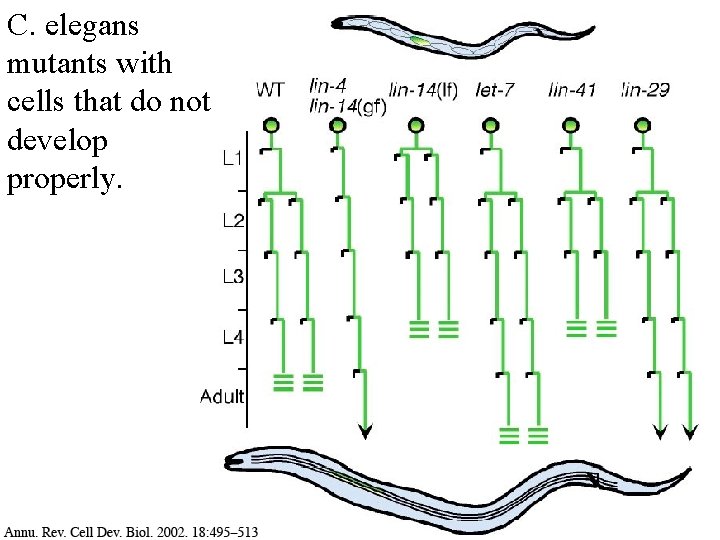
C. elegans is commonly used to study development

C. elegans development

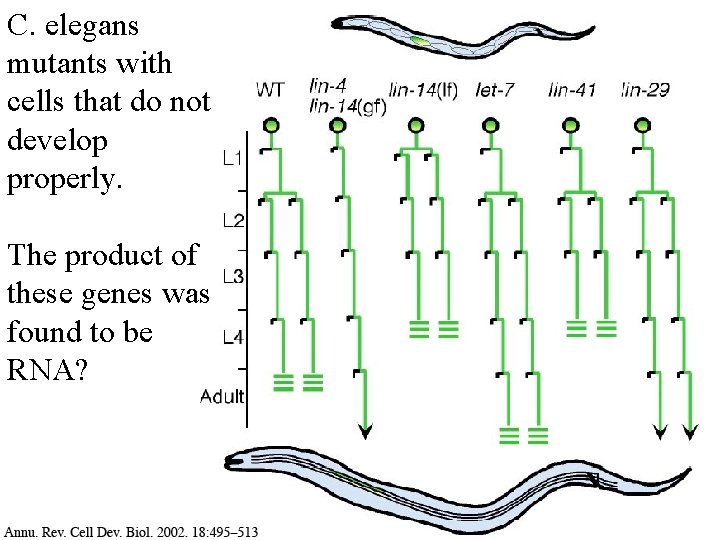
C. elegans mutants with cells that do not develop properly.

C. elegans mutants with cells that do not develop properly. The product of these genes was found to be RNA?

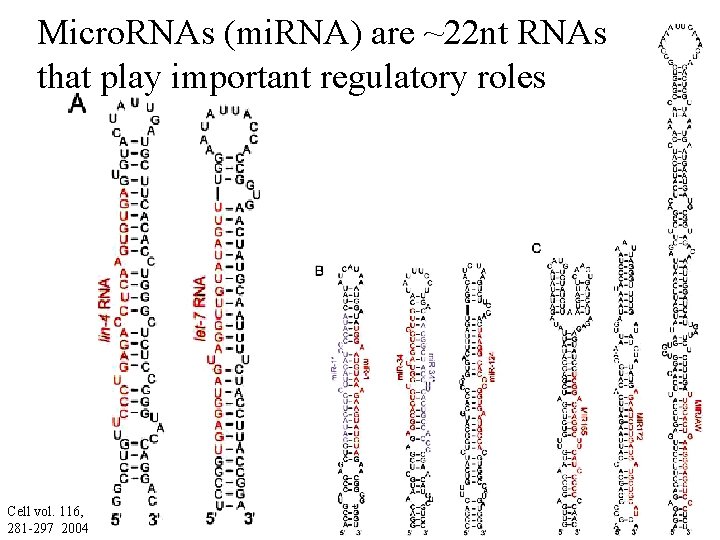
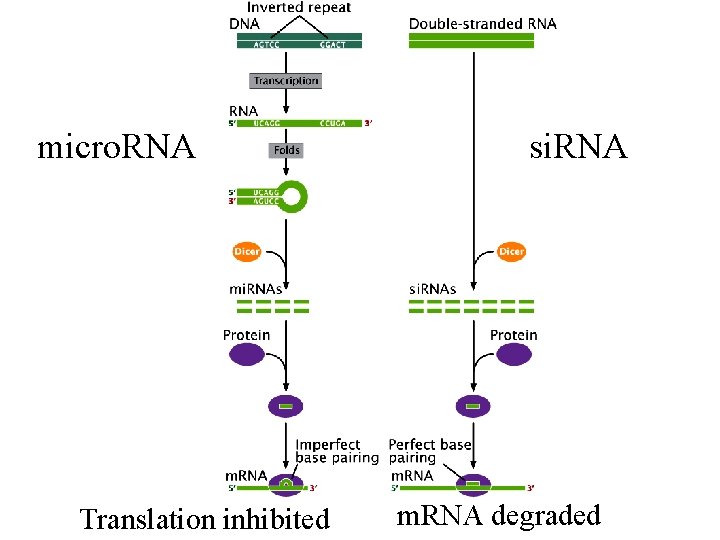
Micro. RNAs (mi. RNA) are ~22 nt RNAs that play important regulatory roles Cell vol. 116, 281 -297 2004

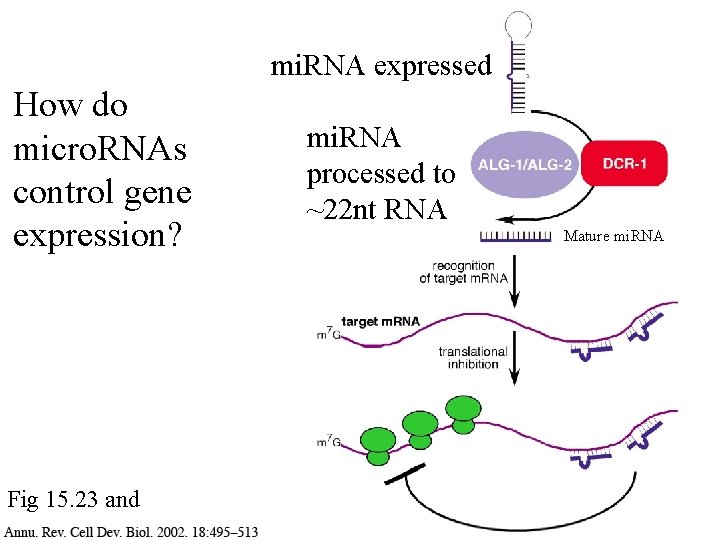
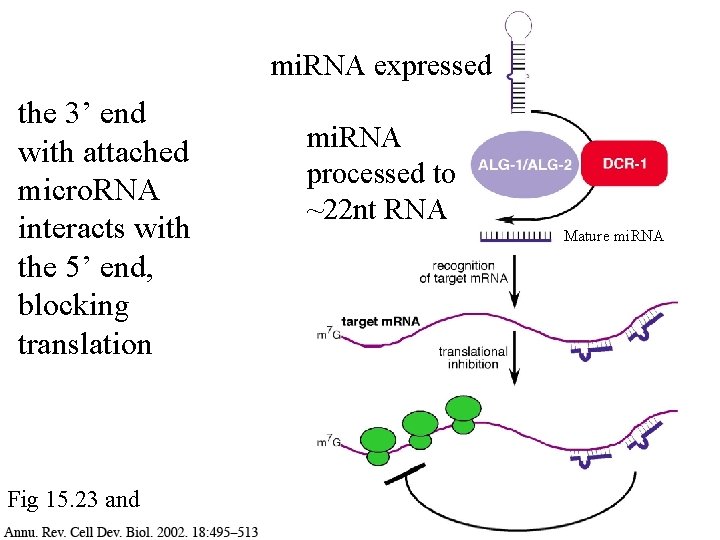
mi. RNA expressed How do micro. RNAs control gene expression? Fig 15. 23 and mi. RNA processed to ~22 nt RNA Mature mi. RNA

A processed m. RNA ready for translation: micro. RNAs inhibit translation by binding to the 3’ end of m. RNA micro. RNA bind to 3’-UTR 5’-UTR 3’-UTR

mi. RNA expressed the 3’ end with attached micro. RNA interacts with the 5’ end, blocking translation Fig 15. 23 and mi. RNA processed to ~22 nt RNA Mature mi. RNA

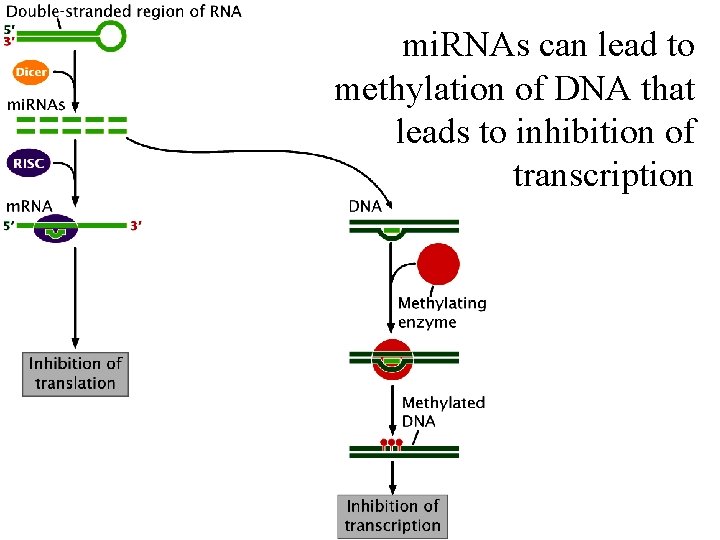
mi. RNAs can lead to methylation of DNA that leads to inhibition of transcription

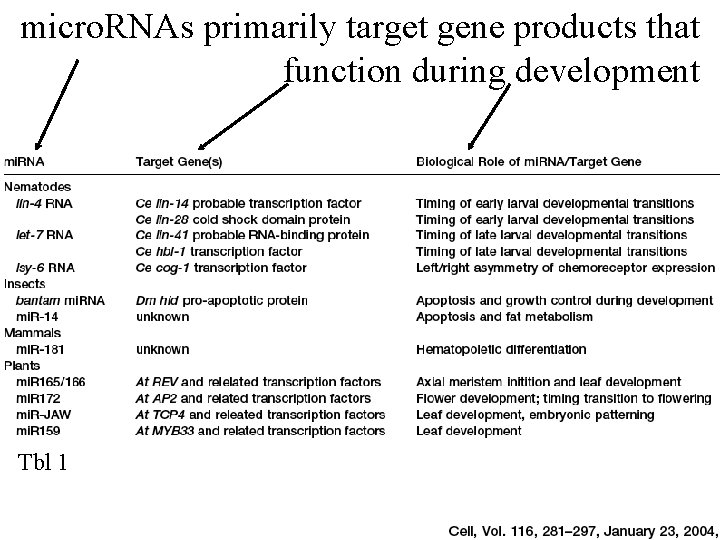
micro. RNAs primarily target gene products that function during development Tbl 1

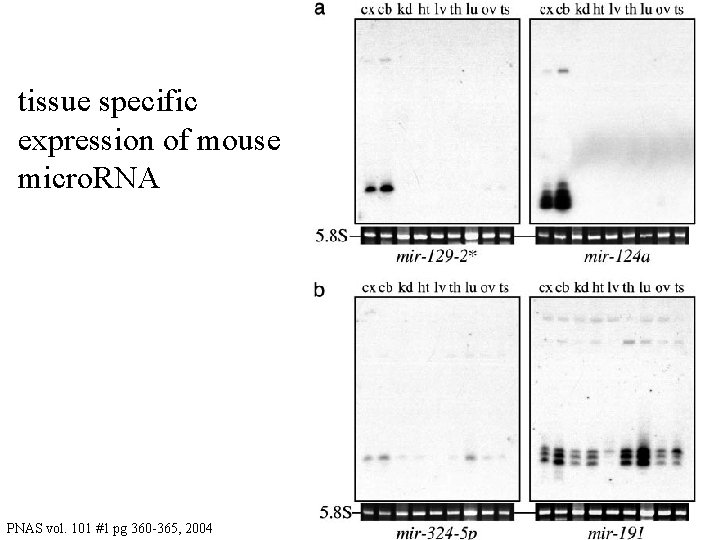
tissue specific expression of mouse micro. RNA PNAS vol. 101 #1 pg 360 -365, 2004

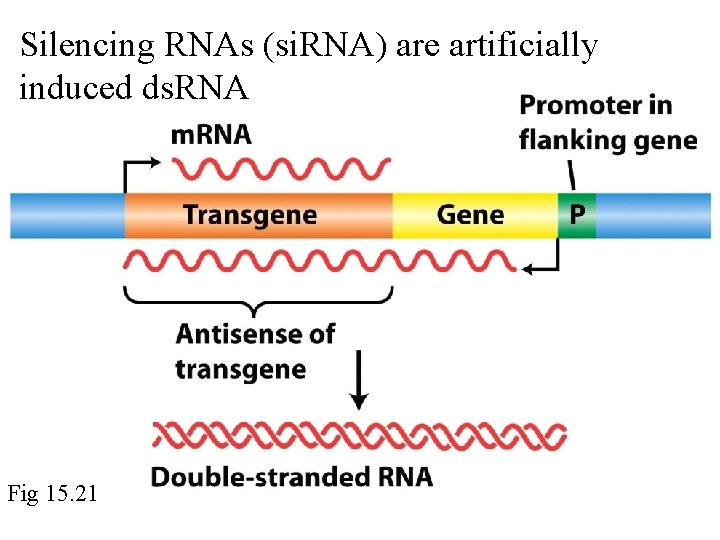
Silencing RNAs (si. RNA) are artificially induced ds. RNA Fig 15. 21

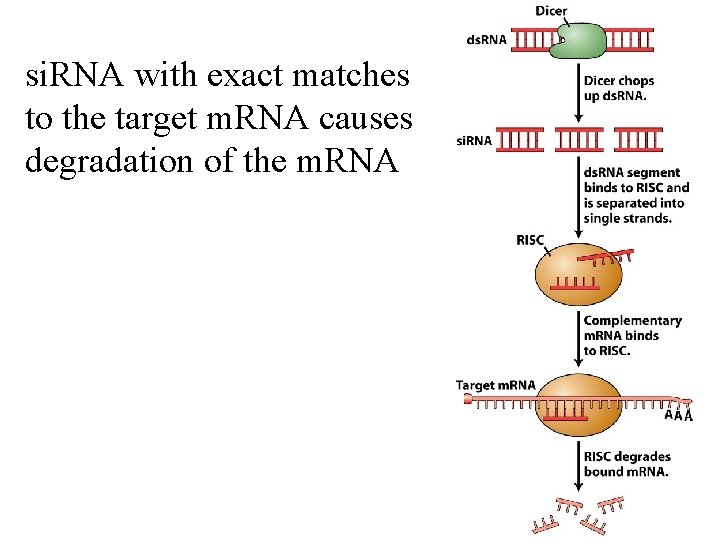
si. RNA with exact matches to the target m. RNA causes degradation of the m. RNA

micro. RNA Translation inhibited si. RNA m. RNA degraded

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 16. 1

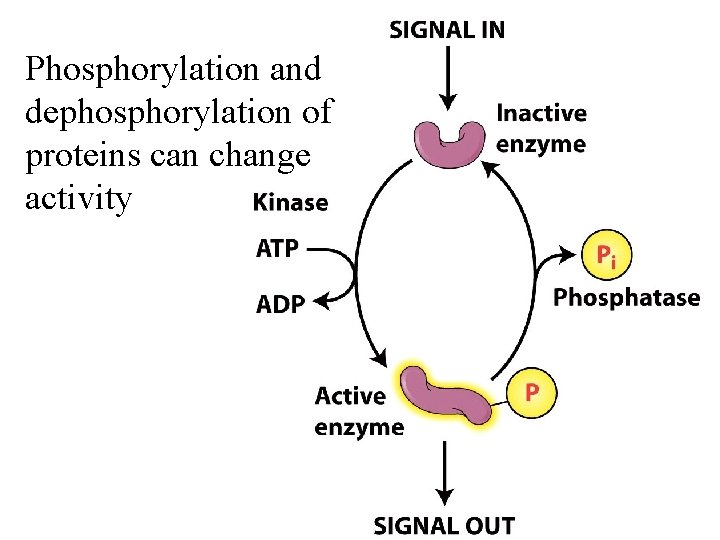
Phosphorylation and dephosphorylation of proteins can change activity

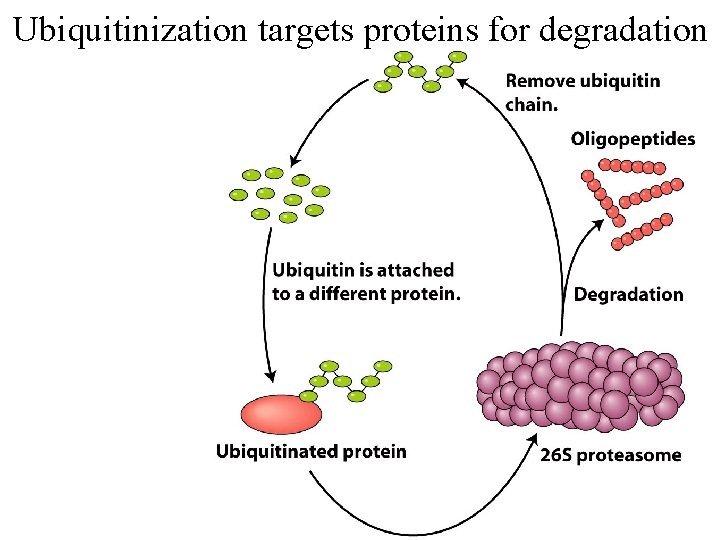
Ubiquitinization targets proteins for degradation

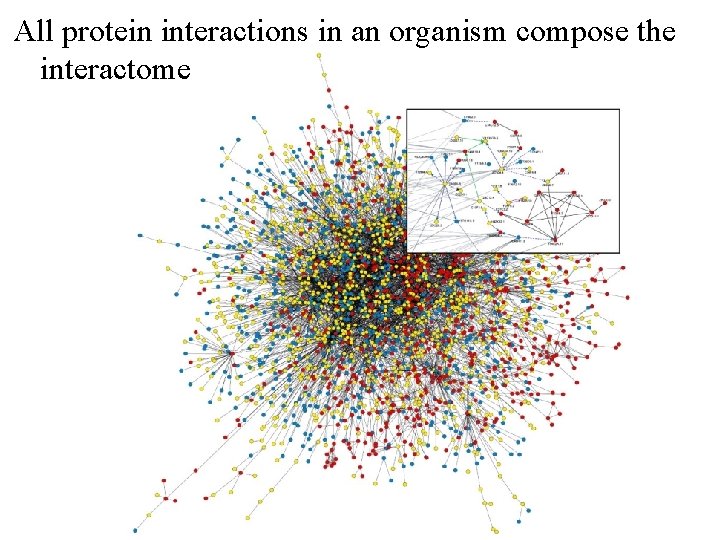
All protein interactions in an organism compose the interactome

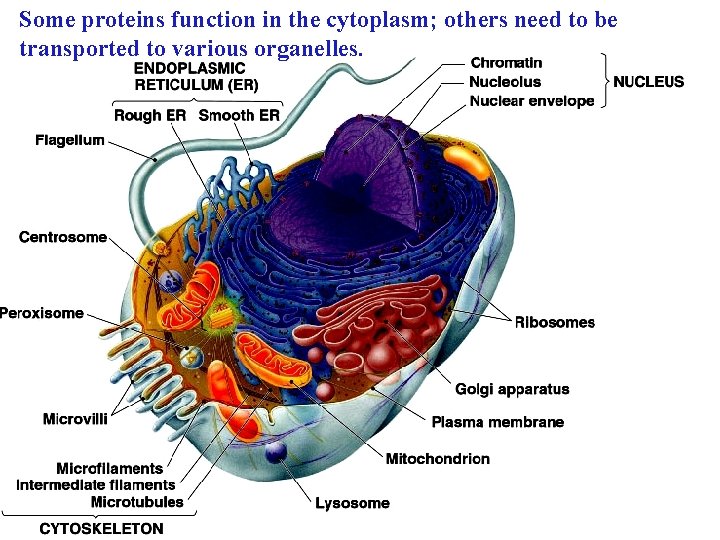
Some proteins function in the cytoplasm; others need to be transported to various organelles.

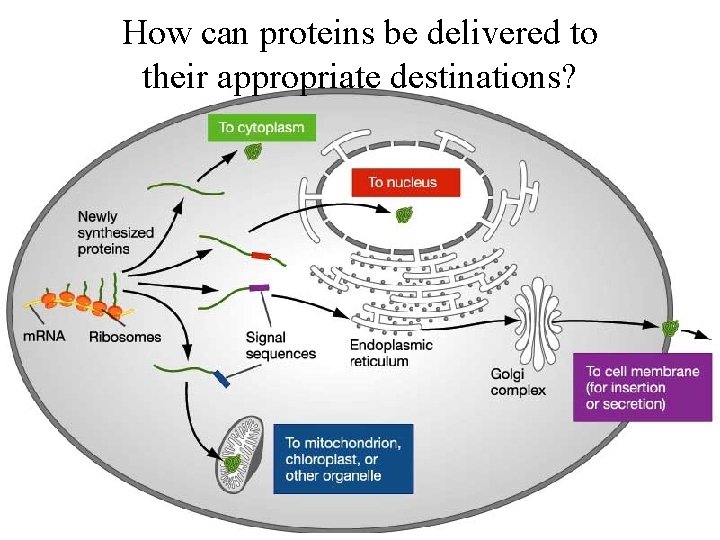
How can proteins be delivered to their appropriate destinations?

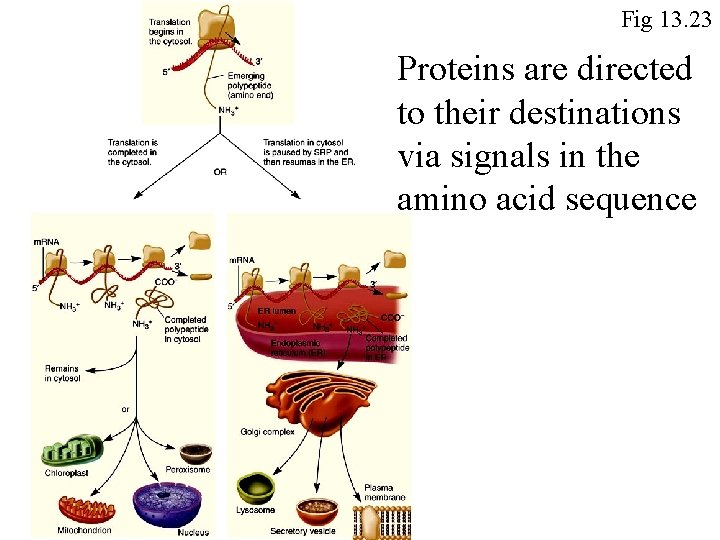
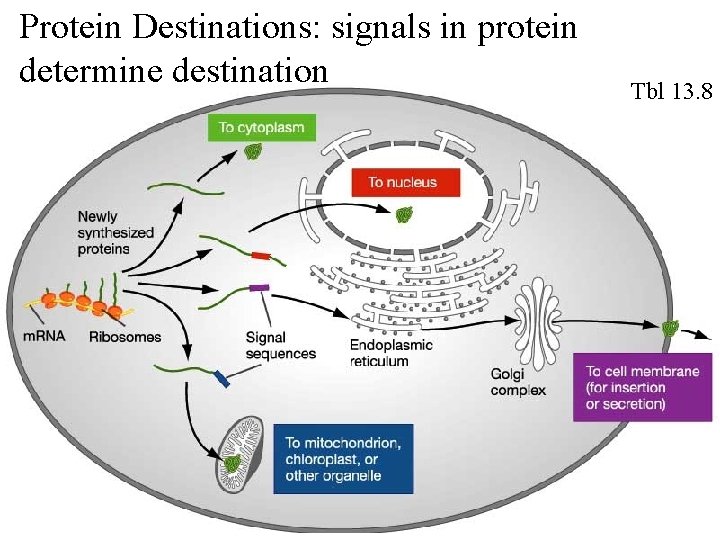
Fig 13. 23 Proteins are directed to their destinations via signals in the amino acid sequence

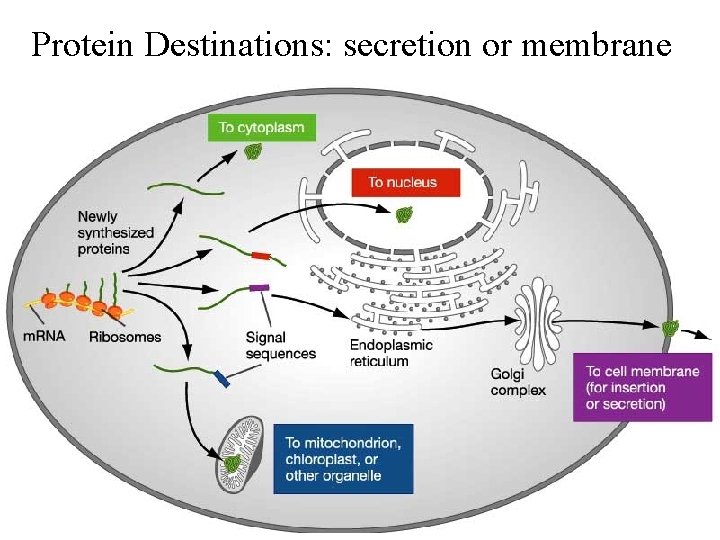
Protein Destinations: secretion or membrane

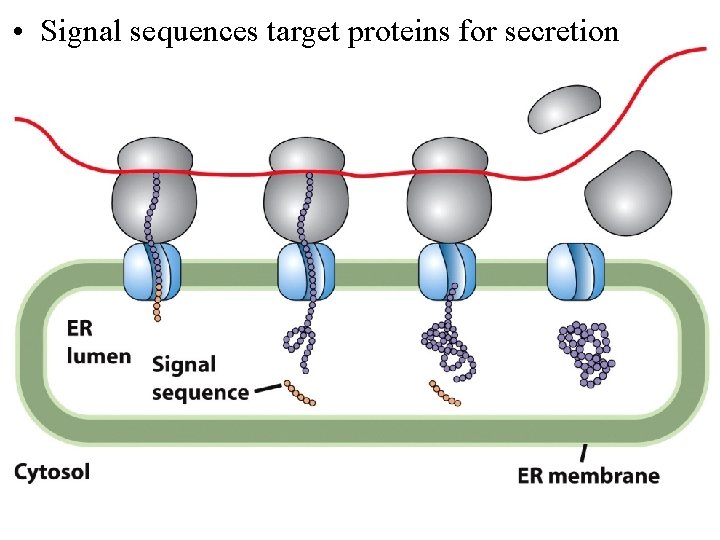
• Signal sequences target proteins for secretion

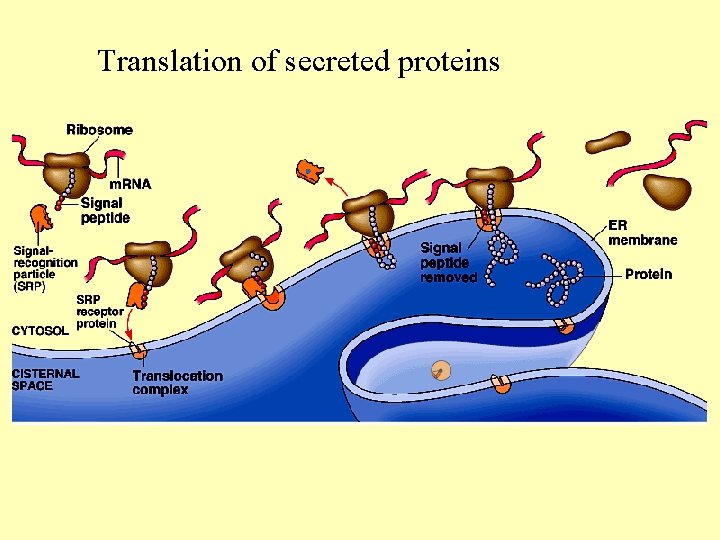
Translation of secreted proteins

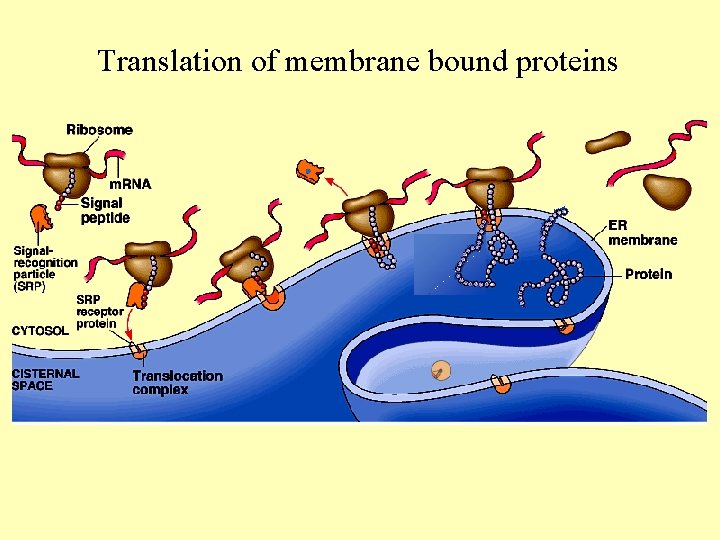
Translation of membrane bound proteins

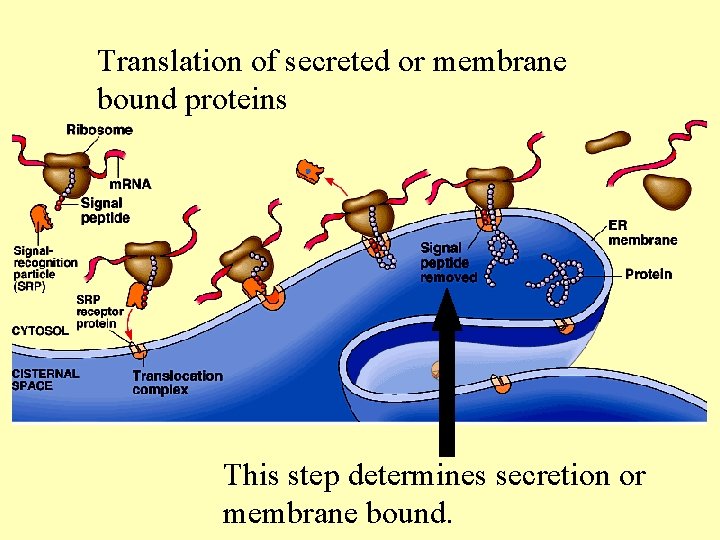
Translation of secreted or membrane bound proteins This step determines secretion or membrane bound.

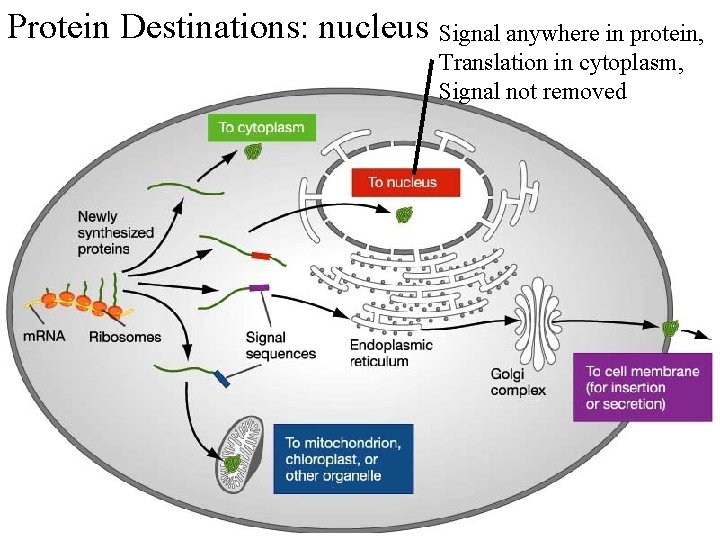
Protein Destinations: nucleus Signal anywhere in protein, Translation in cytoplasm, Signal not removed

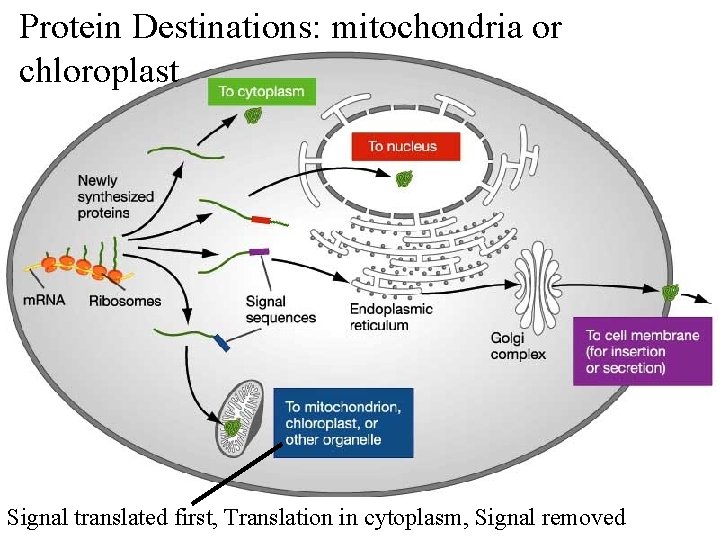
Protein Destinations: mitochondria or chloroplast Signal translated first, Translation in cytoplasm, Signal removed

Protein Destinations: signals in protein determine destination Tbl 13. 8

Development: differentiating cells to become an organism
