more regulating gene expression We looked at the








- Slides: 29

more regulating gene expression

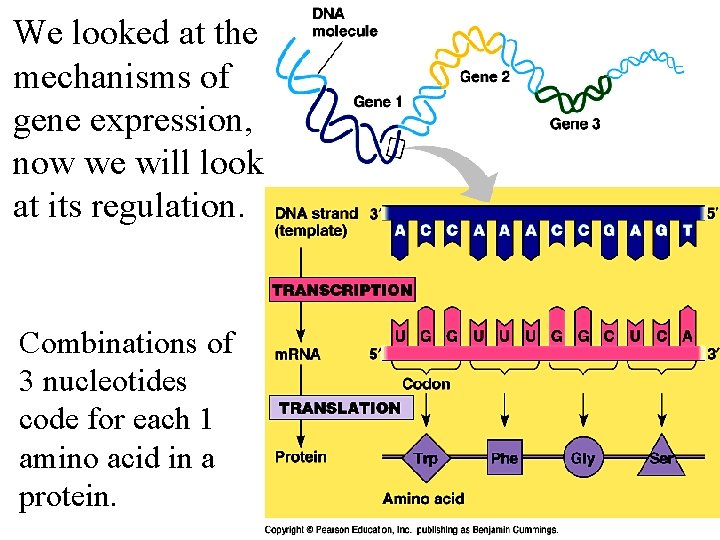
We looked at the mechanisms of gene expression, now we will look at its regulation. Combinations of 3 nucleotides code for each 1 amino acid in a protein.

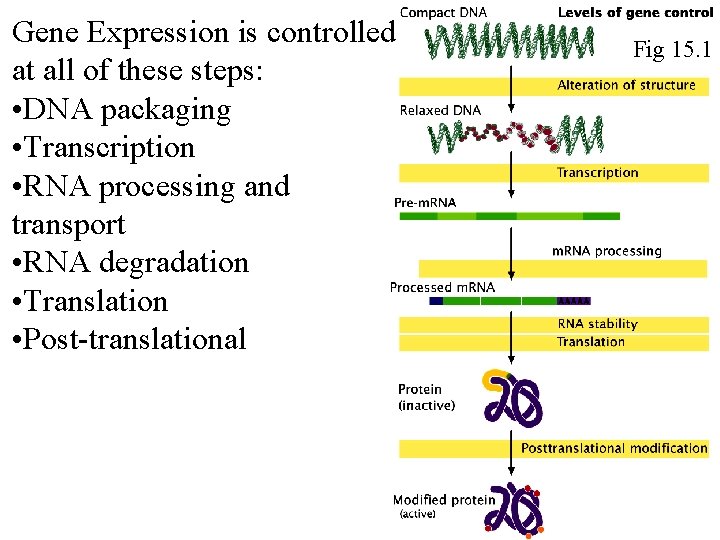
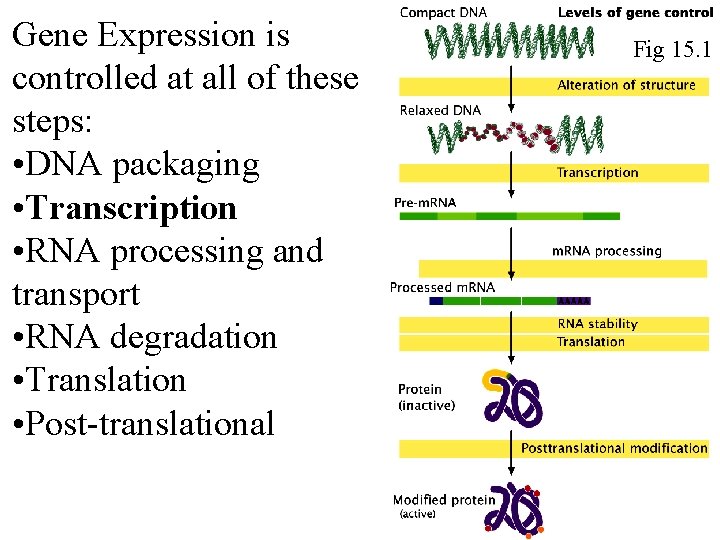
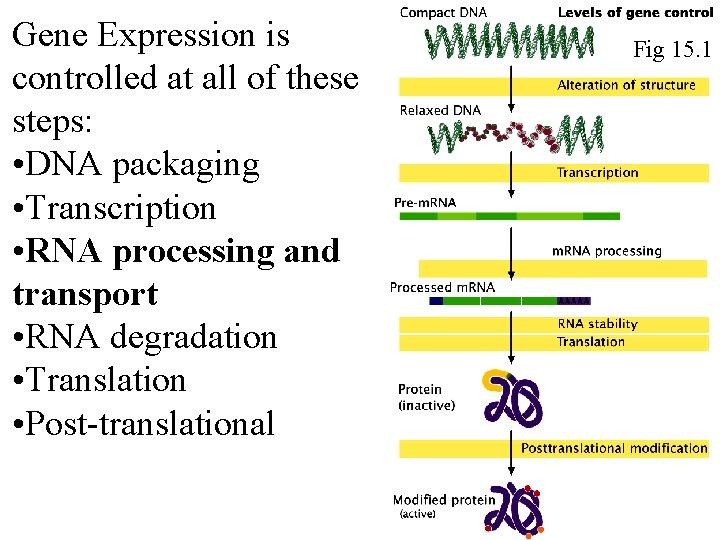
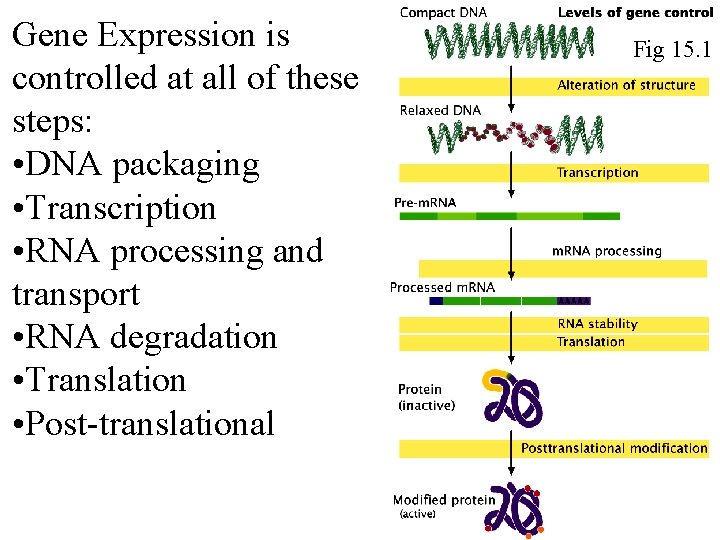
Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

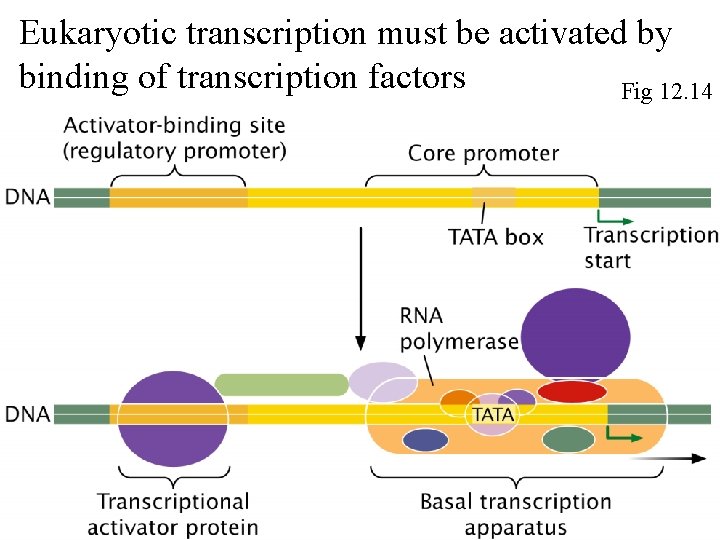
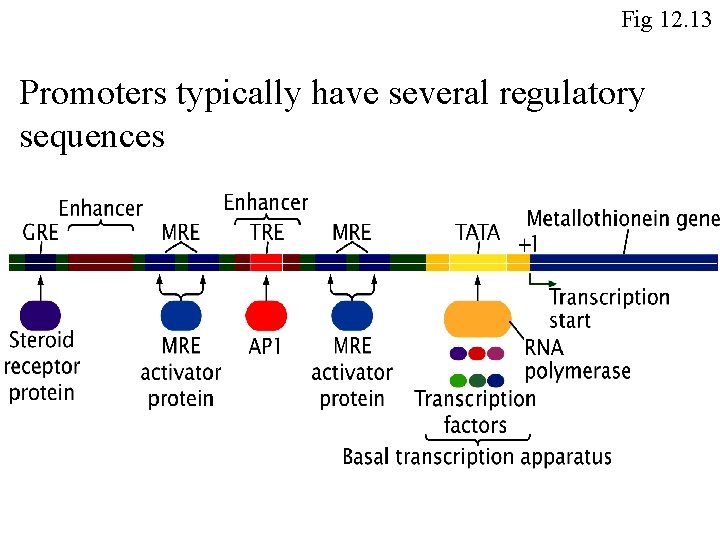
Eukaryotic transcription must be activated by binding of transcription factors Fig 12. 14

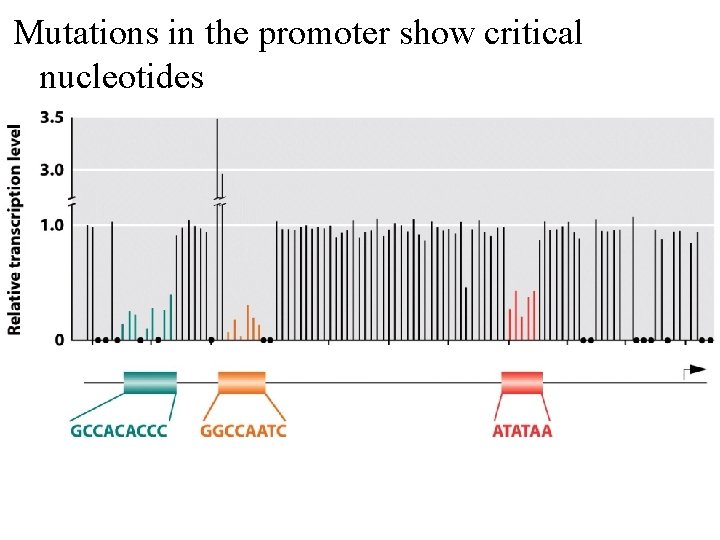
Mutations in the promoter show critical nucleotides

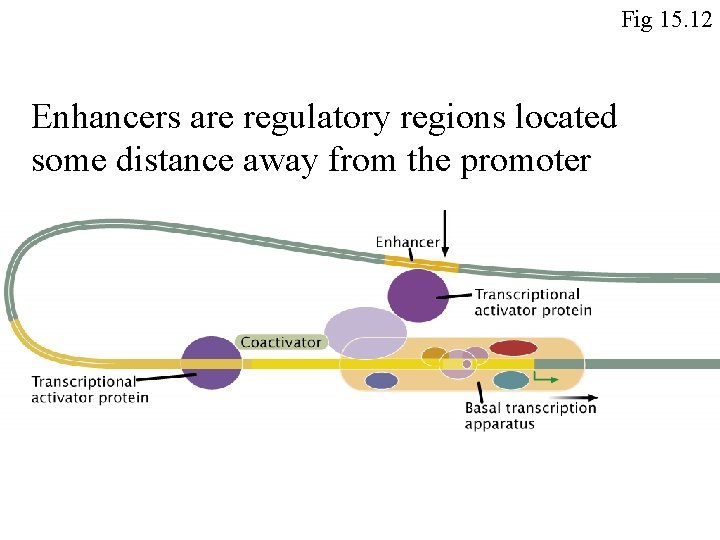
Fig 15. 12 Enhancers are regulatory regions located some distance away from the promoter

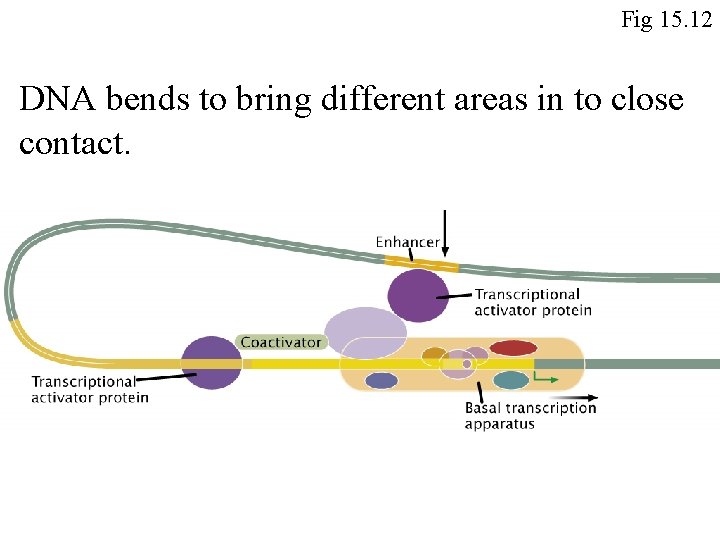
Proteins that help bend DNA can play an important role in transcription Fig 15. 12

Fig 15. 12 DNA bends to bring different areas in to close contact.

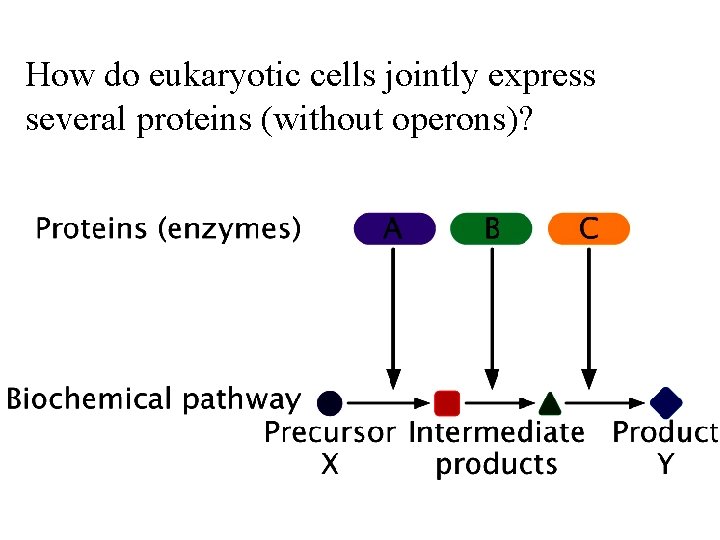
How do eukaryotic cells jointly express several proteins (without operons)?

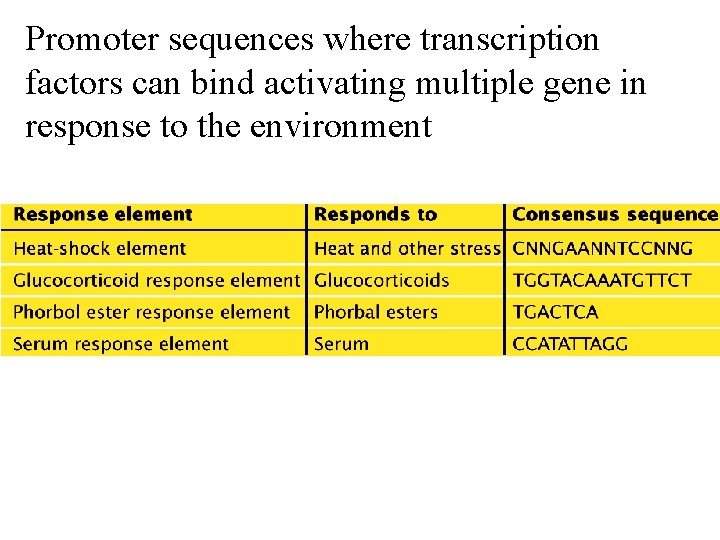
Promoter sequences where transcription factors can bind activating multiple gene in response to the environment

Fig 12. 13 Promoters typically have several regulatory sequences

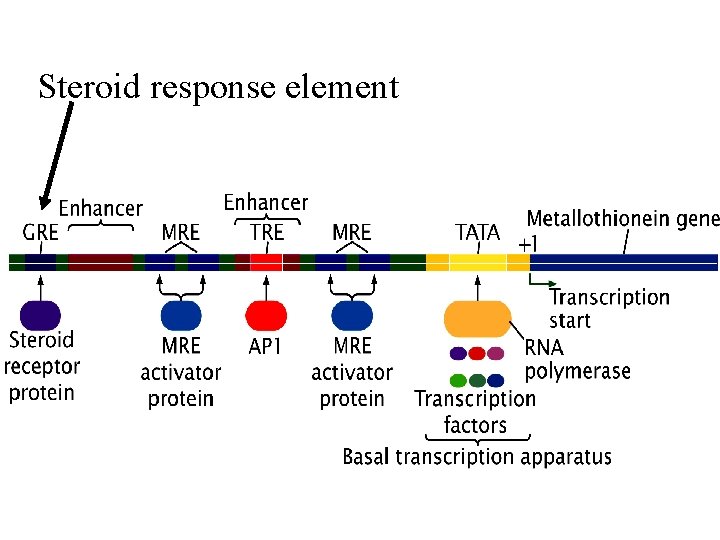
Steroid response element

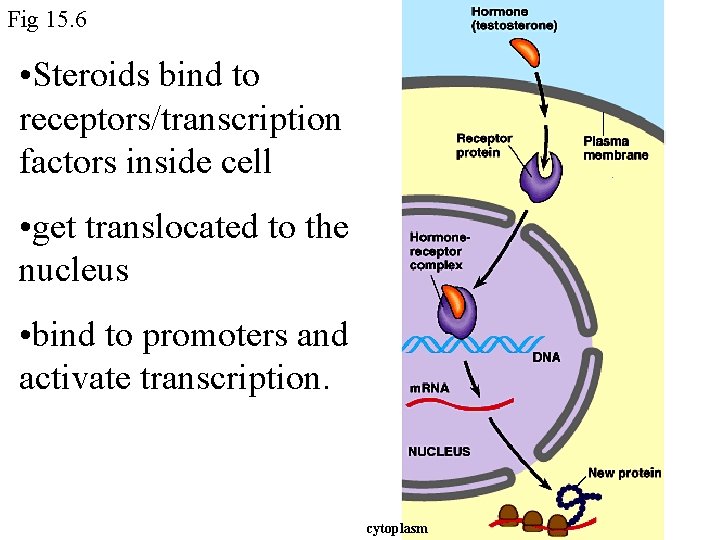
Fig 15. 6 • Steroids bind to receptors/transcription factors inside cell • get translocated to the nucleus • bind to promoters and activate transcription. cytoplasm

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1

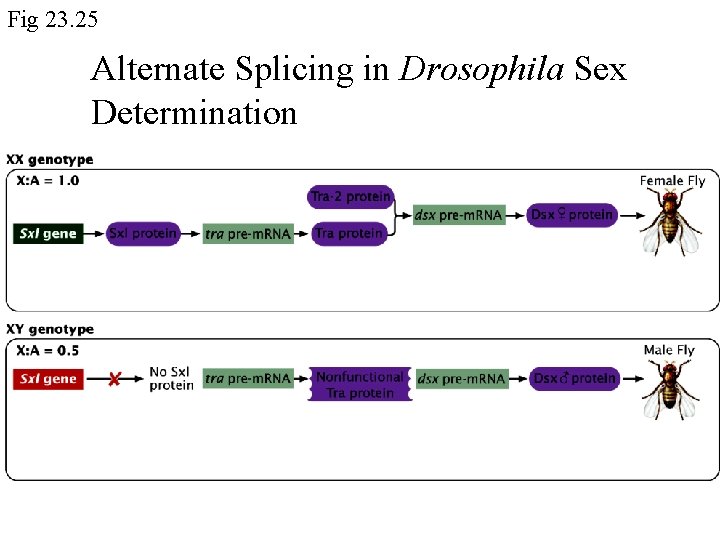
Fig 23. 25 Alternate Splicing in Drosophila Sex Determination

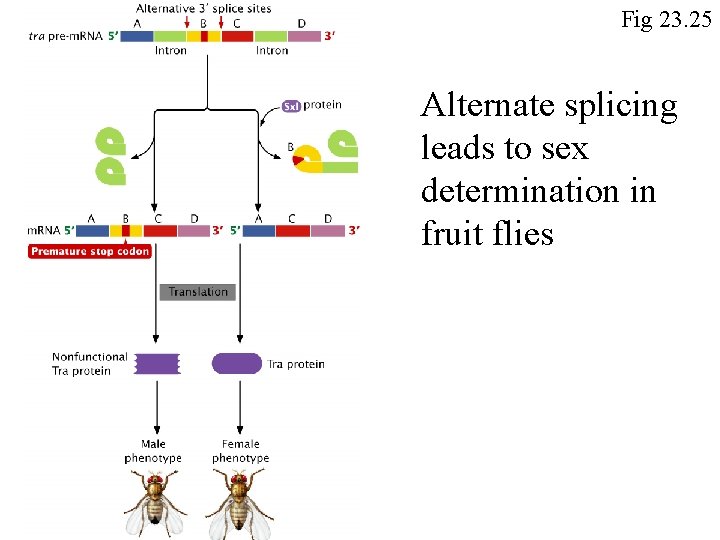
Fig 23. 25 Alternate splicing leads to sex determination in fruit flies

Mammalian m. RNA Splice-Isoform Selection Is Tightly Controlled Jennifer L. Chisa and David T. Burke Genetics, Vol. 175: 1079 -1087, March 2007 • Regulation of gene expression is often in response to a changing environment. • But how stable can alternative splicing be, and does it play a role in maintaining homeostasis?

• Alternative splicing modifies at least half of all primary m. RNA transcripts in mammals. • More than one alternative splice isoform can be maintained concurrently in the steady state m. RNA pool of a single tissue or cell type, and changes in the ratios of isoforms have been associated with physiological variation and susceptibility to disease. • Splice isoforms with opposing functions can be generated; for example, different isoforms of Bcl-x have pro-apoptotic and anti-apoptotic function. Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 1

Alternatively spliced versions of different genes were identified Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 1

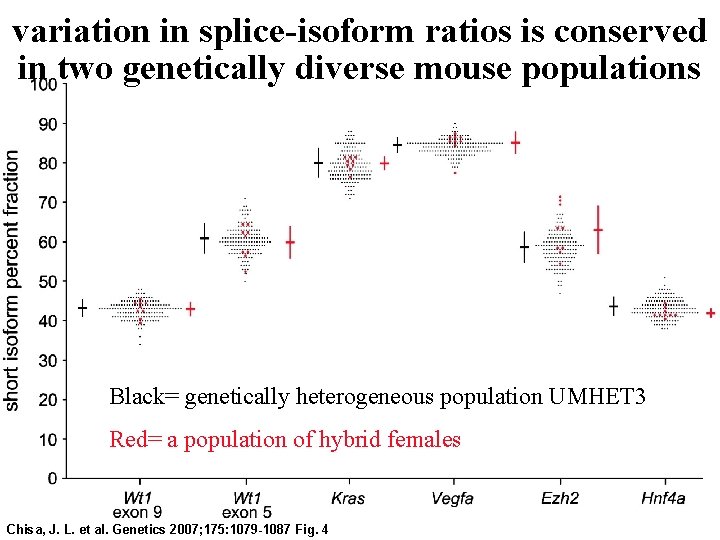
variation in splice-isoform ratios is conserved in two genetically diverse mouse populations Black= genetically heterogeneous population UMHET 3 Red= a population of hybrid females Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 4

In different individuals splice isoforms in different tissues are conserved Chisa, J. L. et al. Genetics 2007; 175: 1079 -1087 Fig. 5

Conclusions: • Alternate splicing for some genes is tightly regulated between different individuals. • Slight differences in alternative splicing may be indicative of abnormalities (disease).

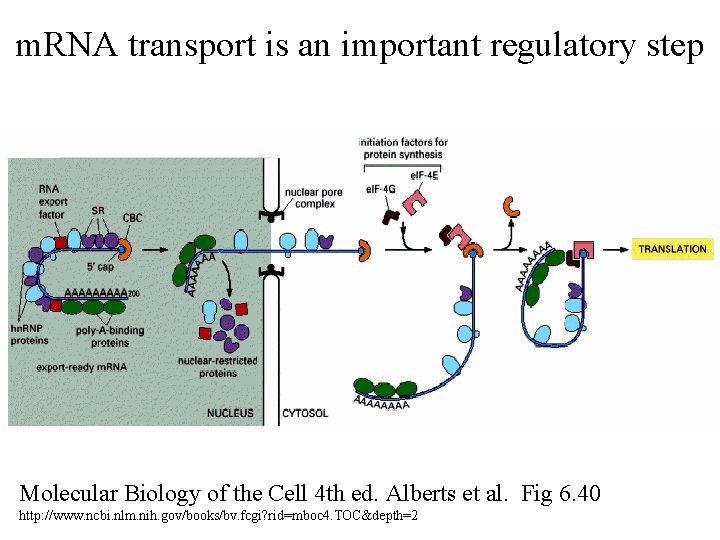
m. RNA transport is an important regulatory step Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 6. 40 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

m. RNA can be localized to a specific parts of a cell (from Drosophila embryo) Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 52 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

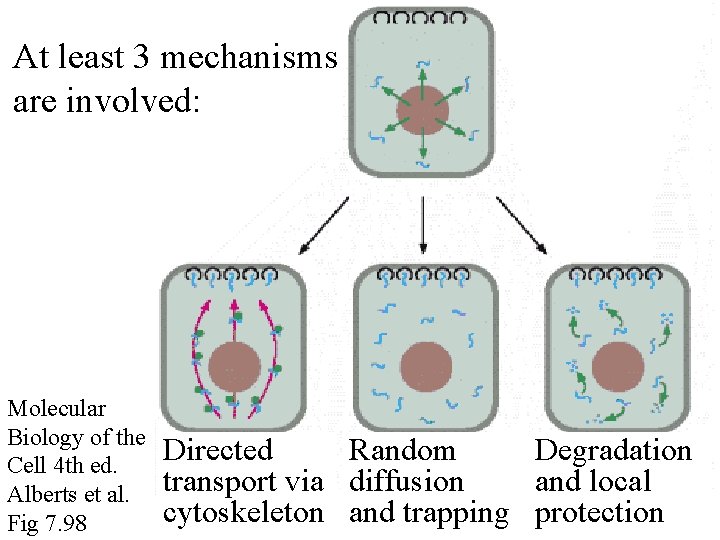
At least 3 mechanisms are involved: Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 98 Directed Random Degradation transport via diffusion and local cytoskeleton and trapping protection

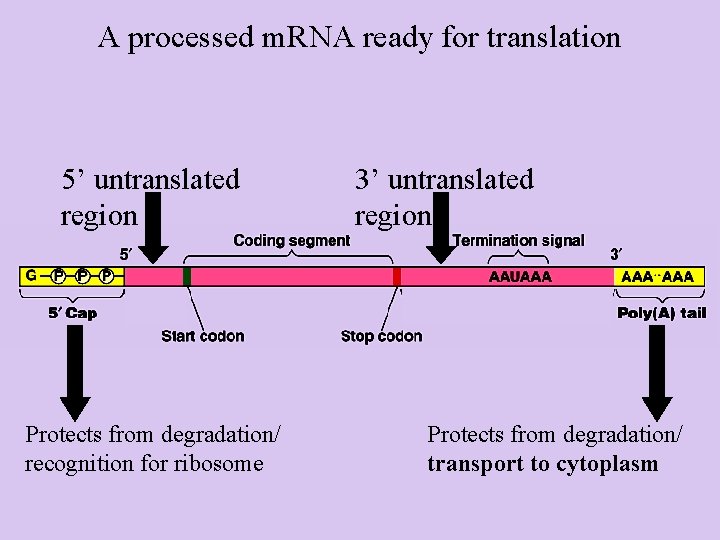
A processed m. RNA ready for translation 5’ untranslated region Protects from degradation/ recognition for ribosome 3’ untranslated region Protects from degradation/ transport to cytoplasm

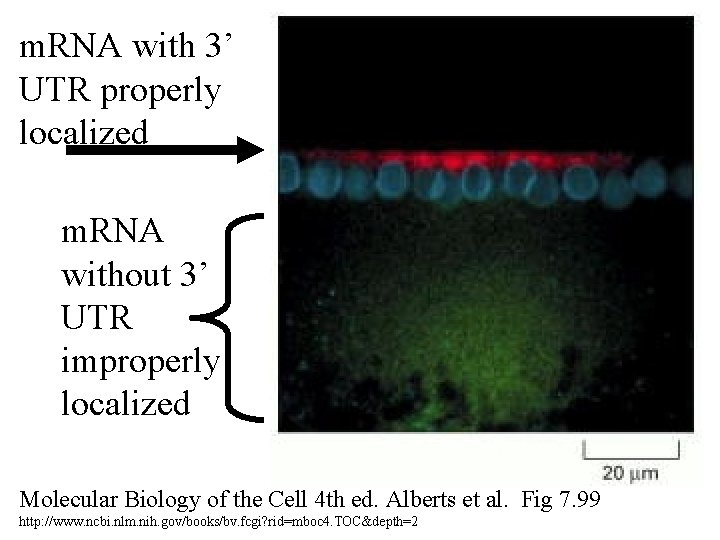
m. RNA with 3’ UTR properly localized m. RNA without 3’ UTR improperly localized Molecular Biology of the Cell 4 th ed. Alberts et al. Fig 7. 99 http: //www. ncbi. nlm. nih. gov/books/bv. fcgi? rid=mboc 4. TOC&depth=2

Gene Expression is controlled at all of these steps: • DNA packaging • Transcription • RNA processing and transport • RNA degradation • Translation • Post-translational Fig 15. 1 Fig 16. 1