Ch 18 Regulation of gene expression Gene expression























- Slides: 70

Ch 18 Regulation of gene expression

Gene expression must be altered in organisms in response to a changing environment

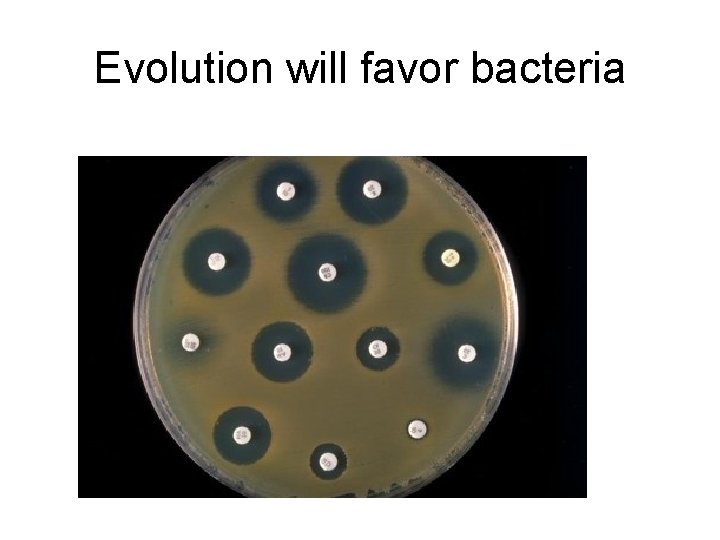
Evolution will favor bacteria

Regulation of anabolic metabolic pathway Abundance both inhibit feedback inhibition and Represses genes

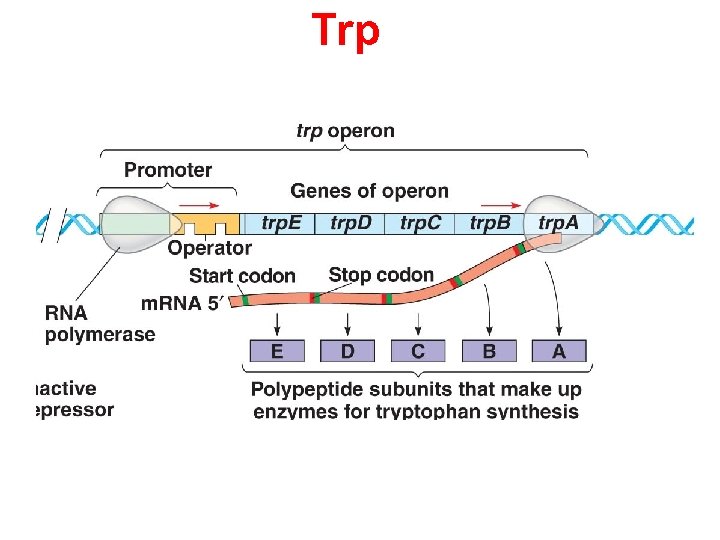
Operon Trp and lac Under coordinated control

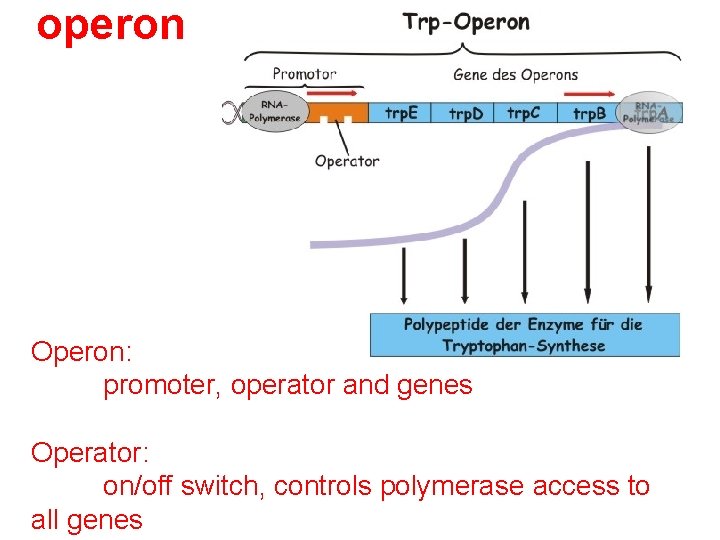
operon Operon: promoter, operator and genes Operator: on/off switch, controls polymerase access to all genes

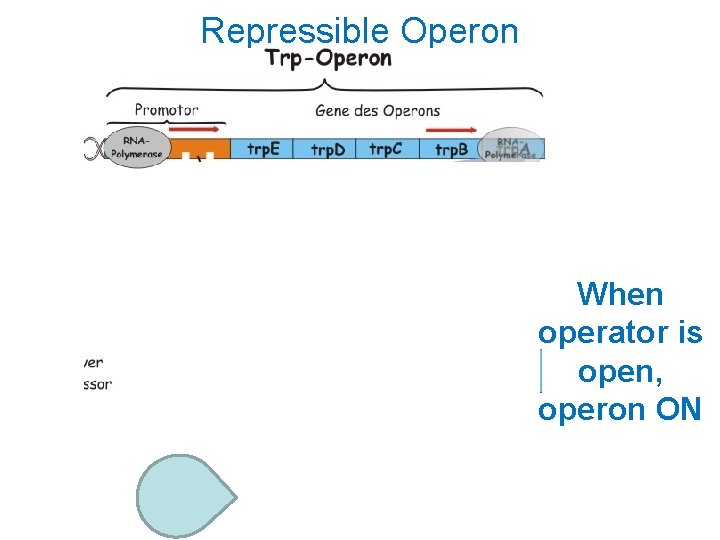
Repressible Operon When operator is open, operon ON

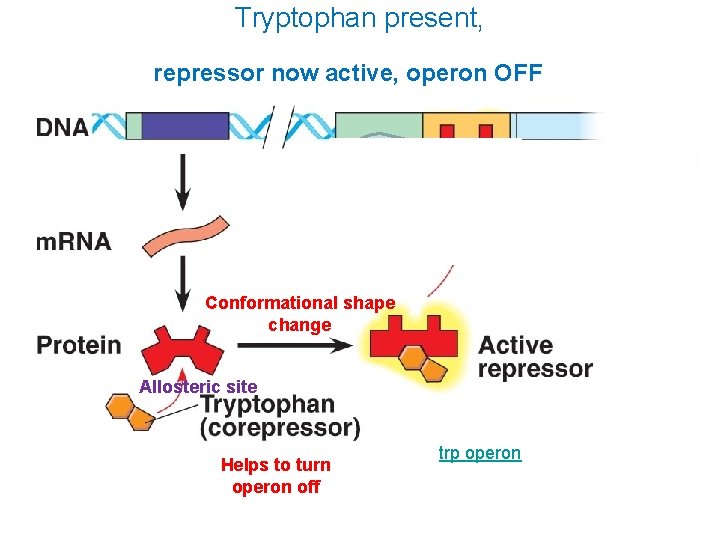
Trp

Regulatory gene Produces Inactive repressor

Tryptophan present, repressor now active, operon OFF Conformational shape change Allosteric site Helps to turn operon off trp operon

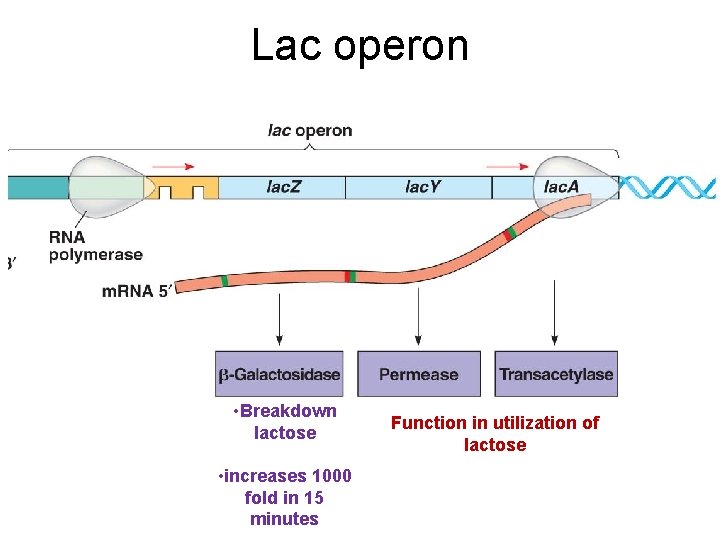
Lac operon • Breakdown lactose • increases 1000 fold in 15 minutes Function in utilization of lactose

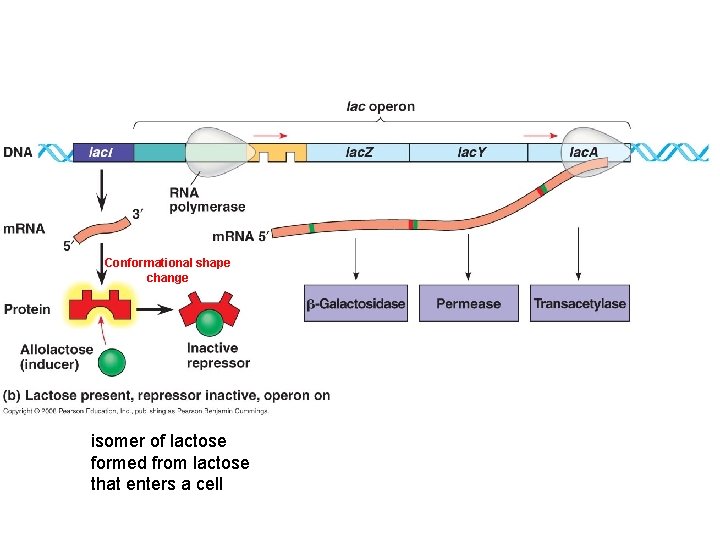
Lac: Inducible lac operon campbell

Conformational shape change isomer of lactose formed from lactose that enters a cell


Negative control of lac and trp operon • Operons are switched off by active repressor • Active Repressor blocks gene expression • Turns both operons off • (easier to “see” in trp operon)

Positive control • an active factor(regulatory protein) is required for gene expression • Regulatory protein turns transcription ON • CAP (Catabolic activator protein) binds to the DNA, transcription is increased.

• If Both glucose and lactose are present… • Glucose will be digested • It is the sugar of choice • It is easier to digest than lactose • Enzymes to digest glucose are always present

E. Coli prefer glucose… • But when Glucose is in short supply…. . • E. coli will use lactose… • Will need to synthesize enzymes to digest lactose SO…. How does it SENSE glucose concentration? • With the Regulatory protein(CAP) –allosteric protein

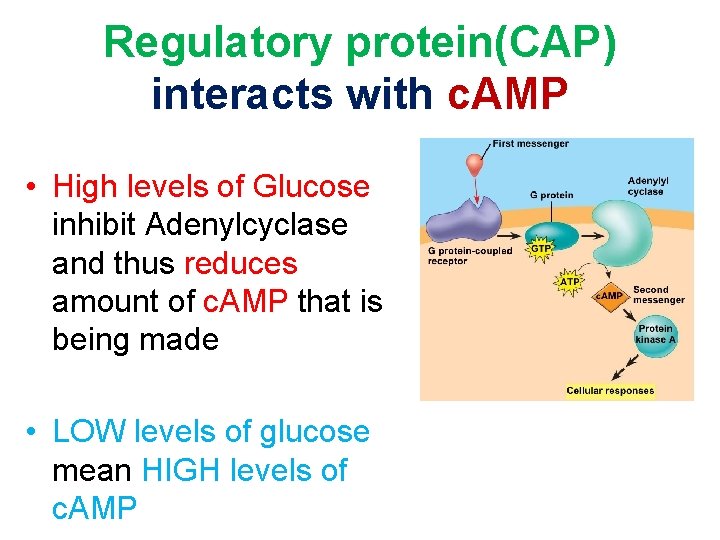
Regulatory protein(CAP) interacts with c. AMP • High levels of Glucose inhibit Adenylcyclase and thus reduces amount of c. AMP that is being made • LOW levels of glucose mean HIGH levels of c. AMP

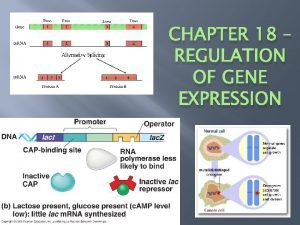
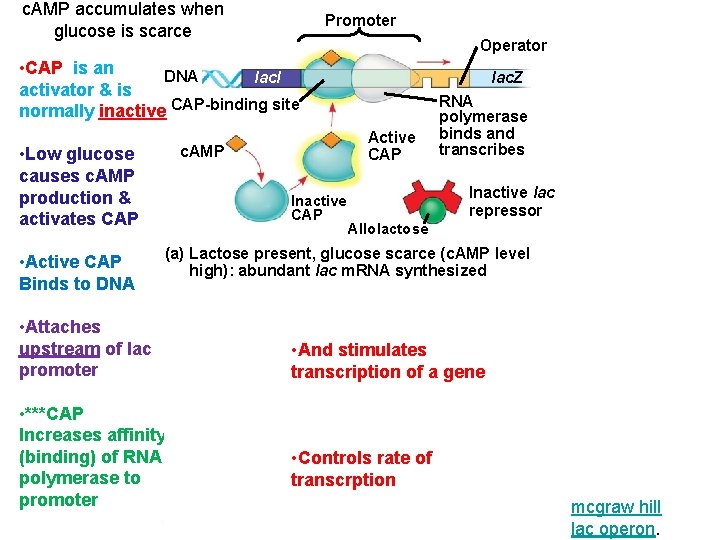
c. AMP accumulates when glucose is scarce Promoter Operator • CAP is an DNA lac. I activator & is normally inactive CAP-binding site • Low glucose causes c. AMP production & activates CAP • Active CAP Binds to DNA lac. Z RNA polymerase binds and transcribes Active CAP c. AMP Inactive CAP Inactive lac repressor Allolactose (a) Lactose present, glucose scarce (c. AMP level high): abundant lac m. RNA synthesized Promoter Operator • Attaches DNA lac. I lac. Z upstream of lac • And stimulates CAP-binding site promoter transcription of RNA a gene polymerase less likely to bind • ***CAP Inactive lac Increases affinity CAP repressor (binding) of RNA • Controls rate of polymerase to (b) Lactose present, transcrption glucose present (c. AMP level promoter low): little lac m. RNA synthesized mcgraw hill lac operon.

Recap… • c. AMP accumulates when glucose is scarce • CAP is an activator • Binds to DNA • Helps RNA polymerase bind to promoter • RNA polymerase has low affinity for the promoter, CAP helps it to bind • And stimulates transcription of a gene

• This is positive regulation

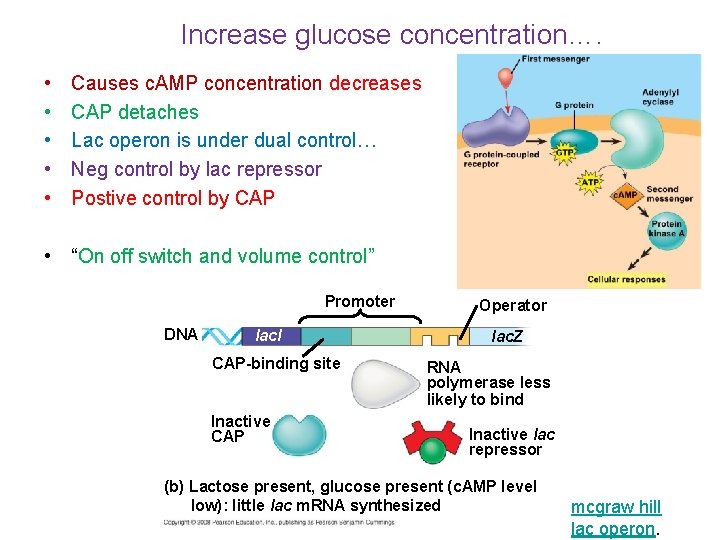
Promoter Increase glucose concentration…. Operator • • • DNA concentration lac. I Causes c. AMP decreases CAP-binding site CAP detaches Lac operon is under dual control…Active c. AMP CAP Neg control by lac repressor Postive control by CAP Inactive CAP lac. Z RNA polymerase binds and transcribes Inactive lac repressor Allolactose (a) Lactose present, glucose scarce (c. AMP level • “On off switch and volume control” high): abundant lac m. RNA synthesized Promoter DNA lac. I CAP-binding site Inactive CAP Operator lac. Z RNA polymerase less likely to bind Inactive lac repressor (b) Lactose present, glucose present (c. AMP level low): little lac m. RNA synthesized mcgraw hill lac operon.

Positive control • When regulatory protein interacts directly with the genome to switch transcription on • referred to as positive control because when the activator protein (CAP) binds to the DNA, transcription is increased. • Chemical present…gene is on

Gene regulation bozeman gene regulation Watch this! Paul Anderson will start with prokaryotic regulation with operons and then discuss eukaryotic regulation

Eukaryotic gene expression is regulated at many stages • All organisms whether prokaryotic or eukaryotic must regulate gene expression. • All turn genes on and off in response to environmental conditions • Signals can come from the external or internal environment • Regulation of gene expression is essential for cell specialization. • Different cells have different jobs and use different genes

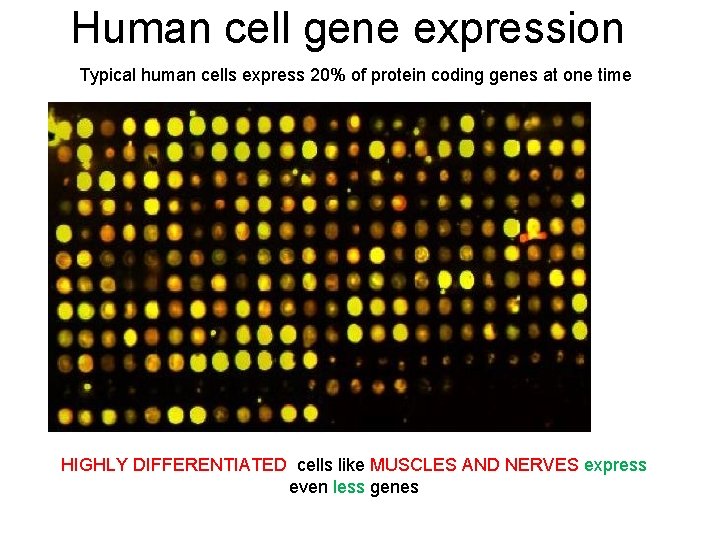
Human cell gene expression Typical human cells express 20% of protein coding genes at one time HIGHLY DIFFERENTIATED cells like MUSCLES AND NERVES express even less genes

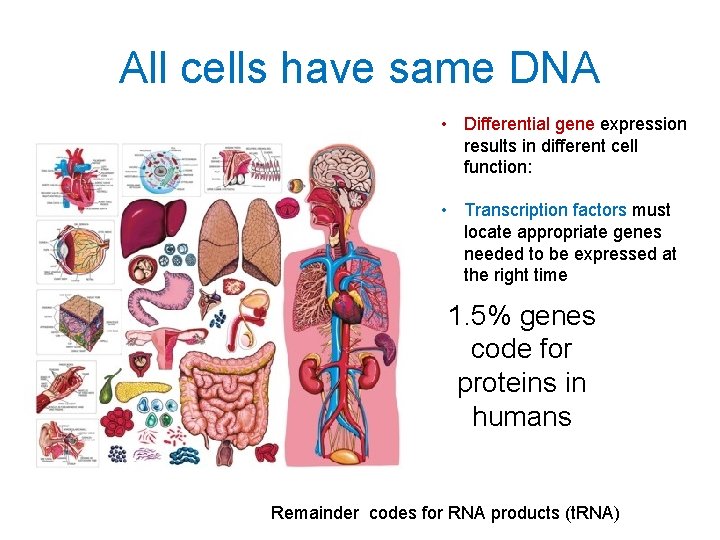
All cells have same DNA • Differential gene expression results in different cell function: • Transcription factors must locate appropriate genes needed to be expressed at the right time 1. 5% genes code for proteins in humans Remainder codes for RNA products (t. RNA)

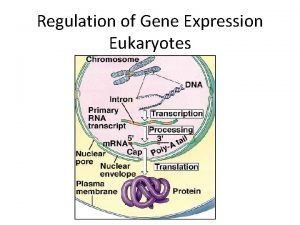
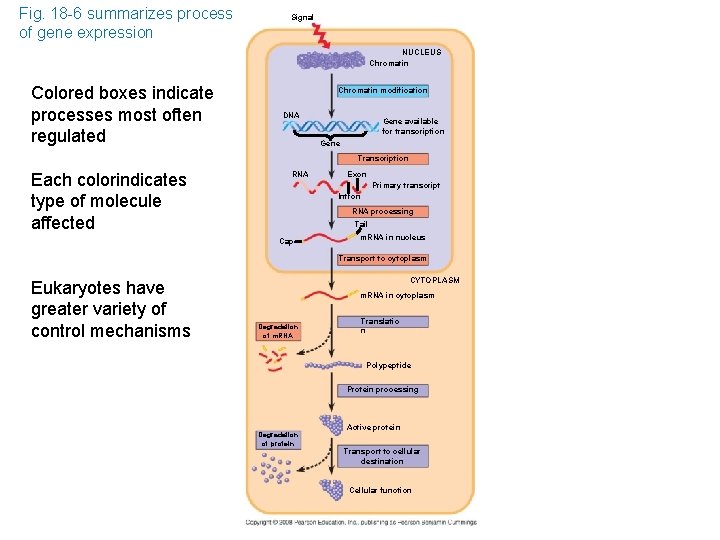
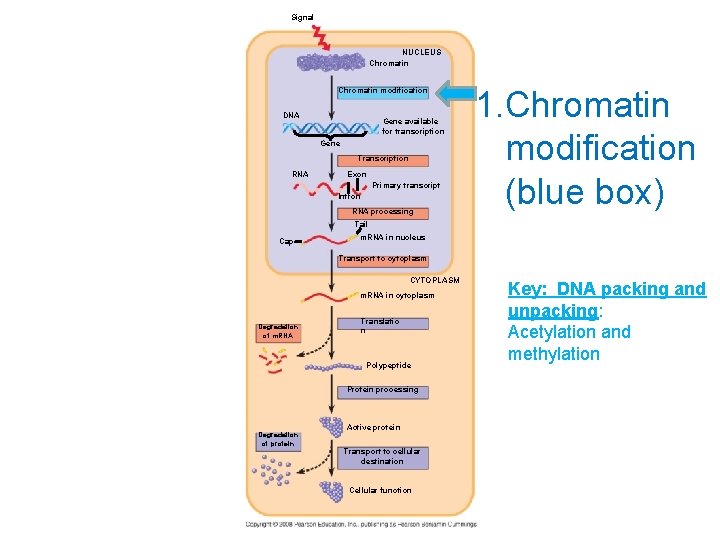
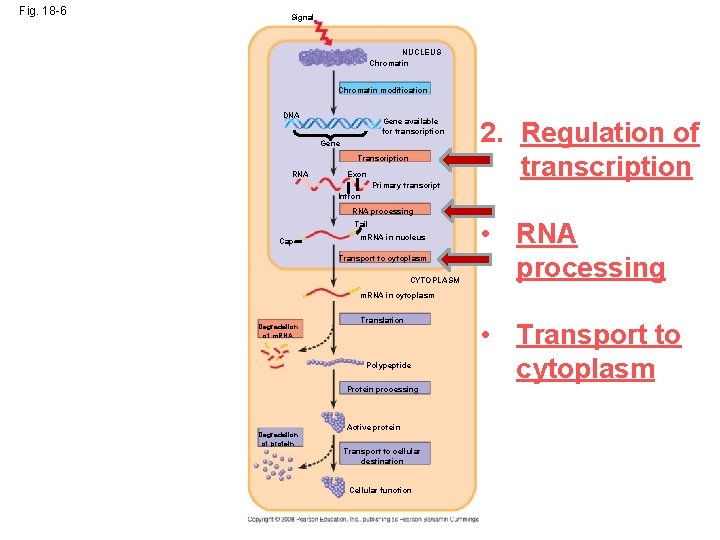
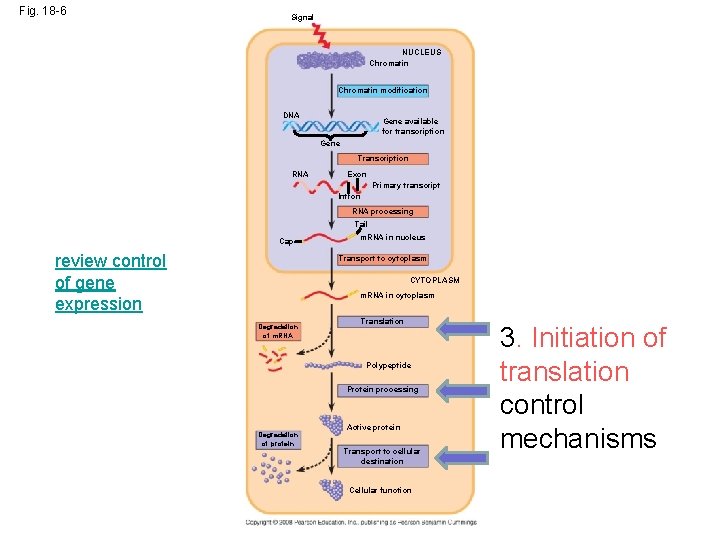
Fig. 18 -6 summarizes process of gene expression Signal NUCLEUS Chromatin Colored boxes indicate processes most often regulated Chromatin modification DNA Gene available for transcription Gene Transcription Each colorindicates type of molecule affected RNA Exon Primary transcript Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm Eukaryotes have greater variety of control mechanisms CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translatio n Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function

Signal NUCLEUS Chromatin modification DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing 1. Chromatin modification (blue box) Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translatio n Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function Key: DNA packing and unpacking: Acetylation and methylation

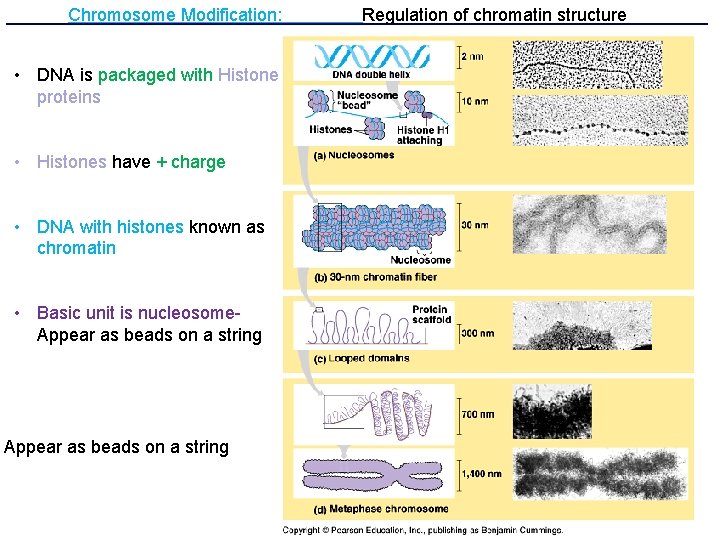
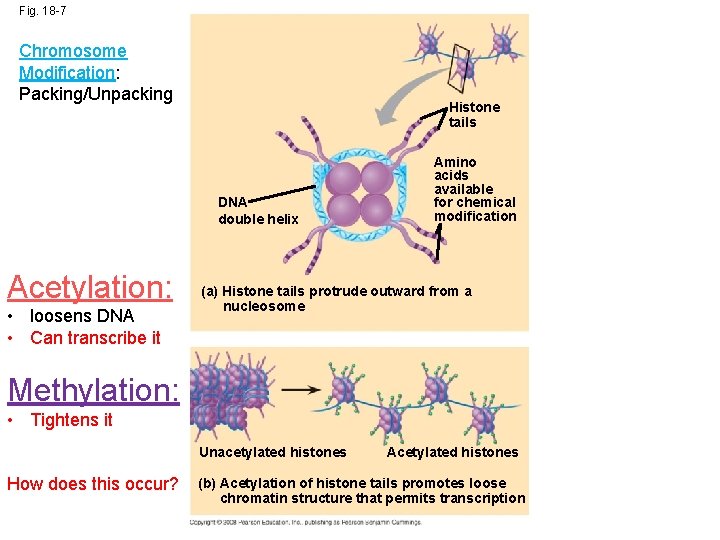
Chromosome Modification: • DNA is packaged with Histone proteins • Histones have + charge • DNA with histones known as chromatin • Basic unit is nucleosome. Appear as beads on a string Regulation of chromatin structure

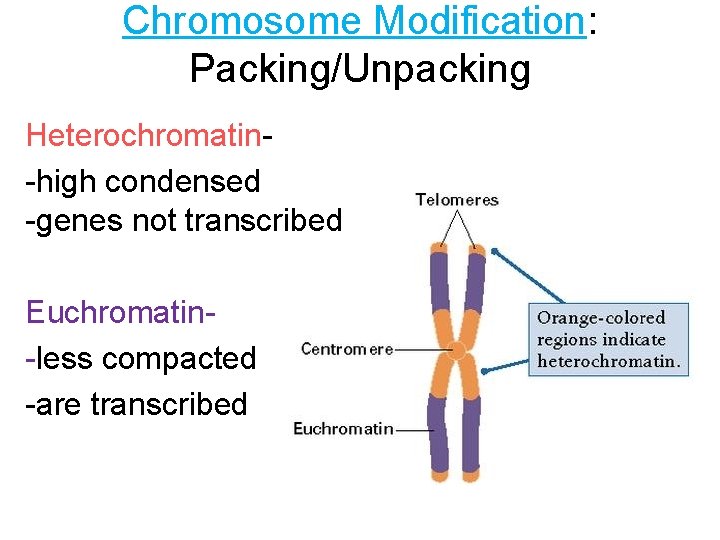
Chromosome Modification: Packing/Unpacking Heterochromatin-high condensed -genes not transcribed Euchromatin-less compacted -are transcribed

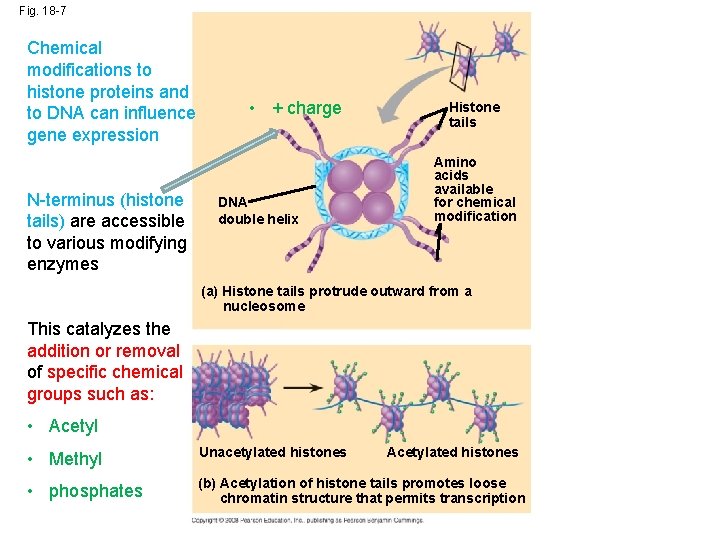
Fig. 18 -7 Chemical modifications to histone proteins and to DNA can influence gene expression N-terminus (histone tails) are accessible to various modifying enzymes • + charge DNA double helix Histone tails Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome This catalyzes the addition or removal of specific chemical groups such as: • Acetyl • Methyl Unacetylated histones • phosphates (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription Acetylated histones

Fig. 18 -7 Chromosome Modification: Packing/Unpacking Histone tails DNA double helix Acetylation: • loosens DNA • Can transcribe it Amino acids available for chemical modification (a) Histone tails protrude outward from a nucleosome Methylation: • Tightens it Unacetylated histones How does this occur? Acetylated histones (b) Acetylation of histone tails promotes loose chromatin structure that permits transcription

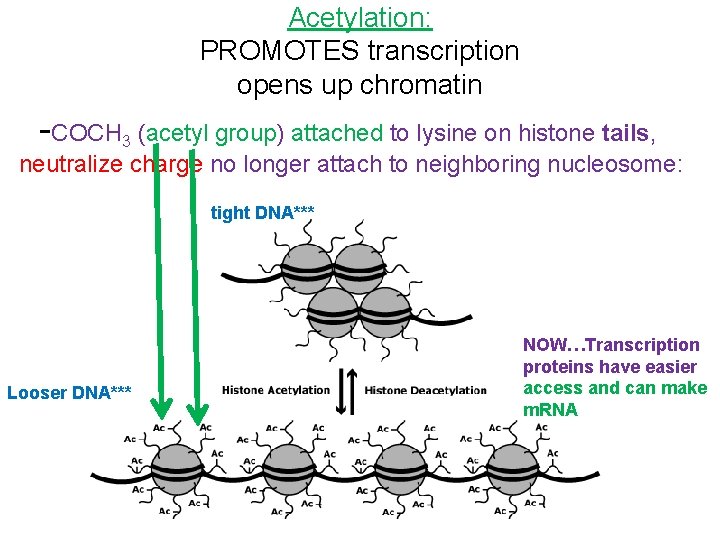
Acetylation: PROMOTES transcription opens up chromatin -COCH 3 (acetyl group) attached to lysine on histone tails, neutralize charge no longer attach to neighboring nucleosome: tight DNA*** Looser DNA*** NOW…Transcription proteins have easier access and can make m. RNA

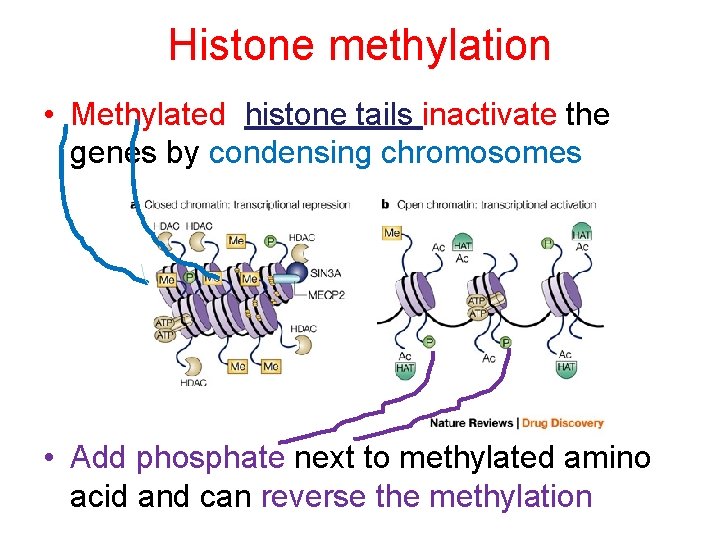
Histone methylation • Methylated histone tails inactivate the genes by condensing chromosomes • Add phosphate next to methylated amino acid and can reverse the methylation

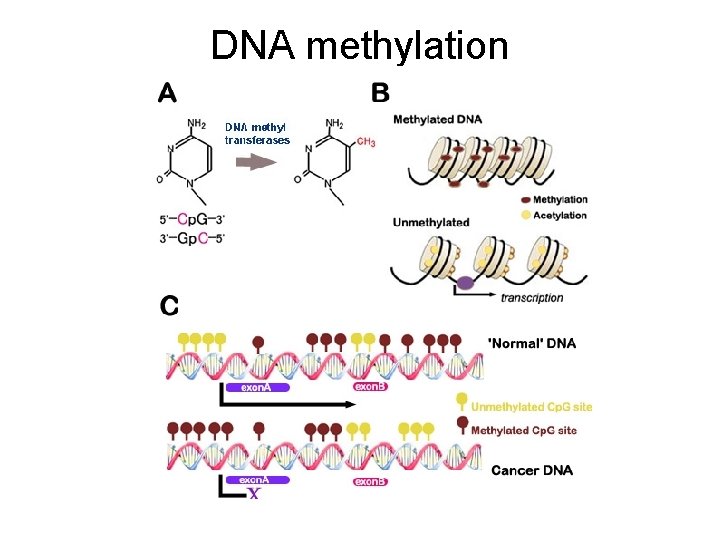
DNA methylation – Different enzymes methylate bases • (occurs at cytosine) – ***Examples: • Barr bodies (methylated DNA) X inactivation Bozeman • genomic imprinting permanent regulation of maternal and paternal allele genomic imprinting 6 min. should watch this one! Explains methylation and acetylation

Compare euchromatin to heterochromatin Lots of acetyl groups- loose DNA Less acetyl groups more methyl- loose DNA

DNA methylation

Summarize Regulation of chromatin structure DNA and chromatin regulation Khan academy

Epigenetics epigenetics Hank will discuss epigenome. I think you will enjoy him! He energetically describes how environment can effect epigenome. Also talks about how diet, stress levels of can possibly cause increases in diseases. Watch him!

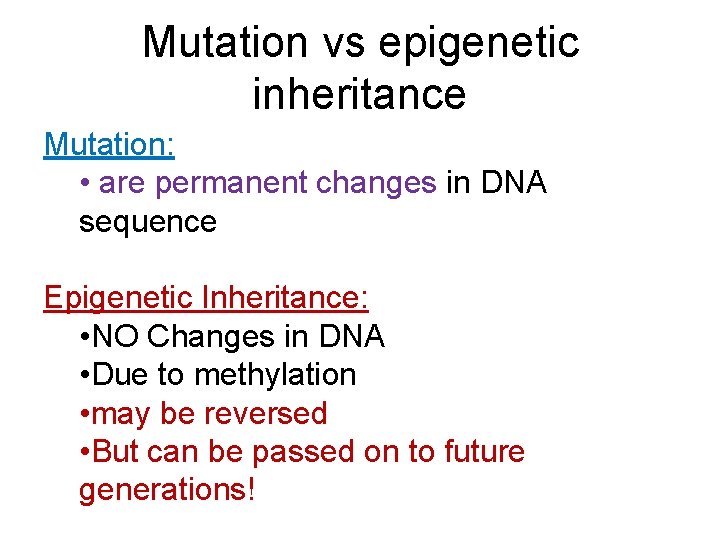
Mutation vs epigenetic inheritance Mutation: • are permanent changes in DNA sequence Epigenetic Inheritance: • NO Changes in DNA • Due to methylation • may be reversed • But can be passed on to future generations!

Epigenetic inheritance – BUT Modifications in chromatin can be reversed: methylation/acetylation – Ex: identical twins, one gets genetically based disease other doesn’t utah. edu epigenetics Refer to online activity!

Responsible for • 1 twin gets genetically based disease • Some cancers • Diabetes

Cloned mice with different DNA methylation

Now we that have loosened the DNA and it is now available for transcription

Fig. 18 -6 Signal NUCLEUS Chromatin modification DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript 2. Regulation of transcription Intron RNA processing Tail Cap m. RNA in nucleus Transport to cytoplasm CYTOPLASM • RNA processing m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function • Transport to cytoplasm

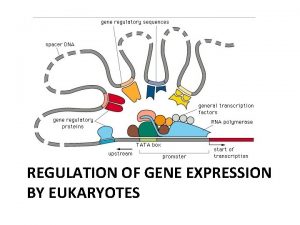
Lets review the structure of a typical eukaryotic gene… • Cluster of proteins called Transcription intiation complex assembles on promoter sequence upstream of the gene • One of these proteins: RNA polymerase transcribes it (pre m. RNA) • RNA processing occurs (exon, intron, 5’ cap, poly A tail) • = mature m. RNA

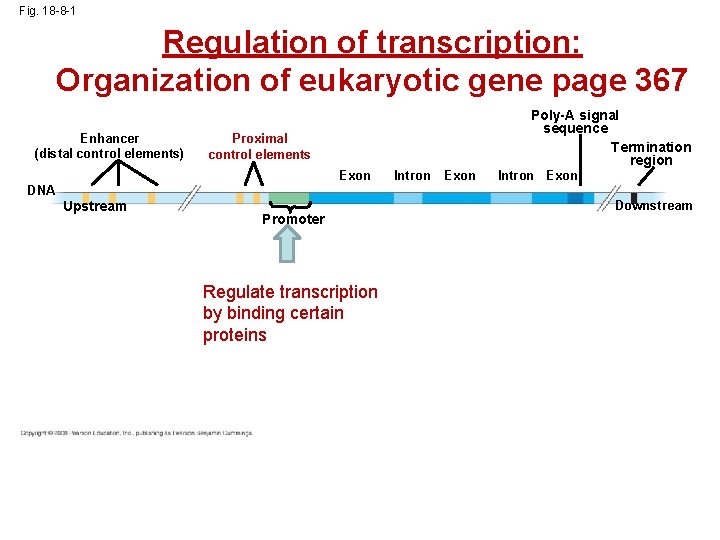
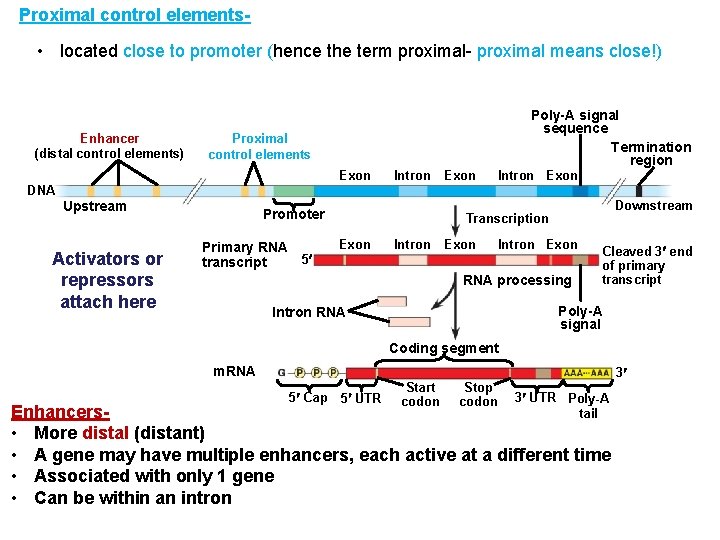
Fig. 18 -8 -1 Regulation of transcription: Organization of eukaryotic gene page 367 Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Promoter Regulate transcription by binding certain proteins Intron Exon Downstream

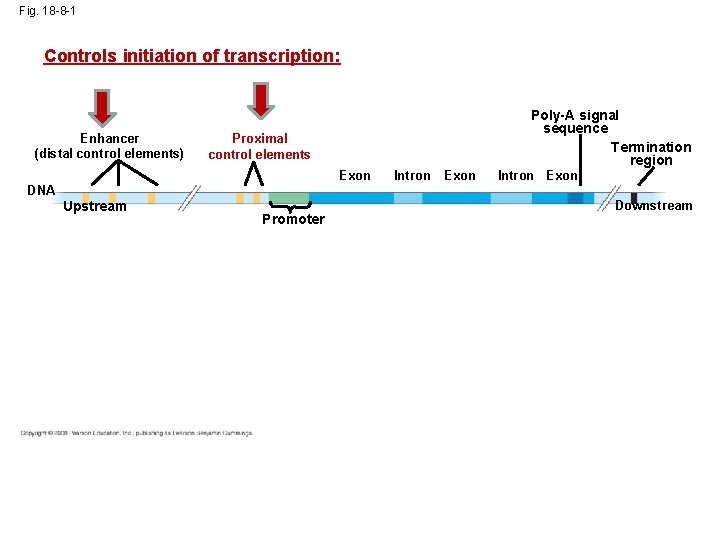
Fig. 18 -8 -1 Controls initiation of transcription: Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Promoter Intron Exon Downstream

Fig. 18 -8 -2 Controls initiation of transcription: Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Intron Promoter Primary RNA 5 transcript Exon Intron Exon Downstream Transcription Exon Intron Exon • Control elements are segments of NON CODING DNA that serve as binding sites for transcription factors • These REGULATE TRANSCRIPTION • These are critical to precise regulation of gene expression Cleaved 3 end of primary transcript Poly-A signal

Proximal control elements- • located close to promoter (hence the term proximal- proximal means close!) Enhancer (distal control elements) Poly-A signal sequence Termination region Proximal control elements Exon DNA Upstream Activators or repressors attach here Intron Promoter Primary RNA 5 transcript Exon Intron Exon Downstream Transcription Exon Intron Exon RNA processing Cleaved 3 end of primary transcript Poly-A signal Intron RNA Coding segment m. RNA 3 5 Cap 5 UTR Start codon Stop codon 3 UTR Poly-A tail Enhancers • More distal (distant) • A gene may have multiple enhancers, each active at a different time • Associated with only 1 gene • Can be within an intron

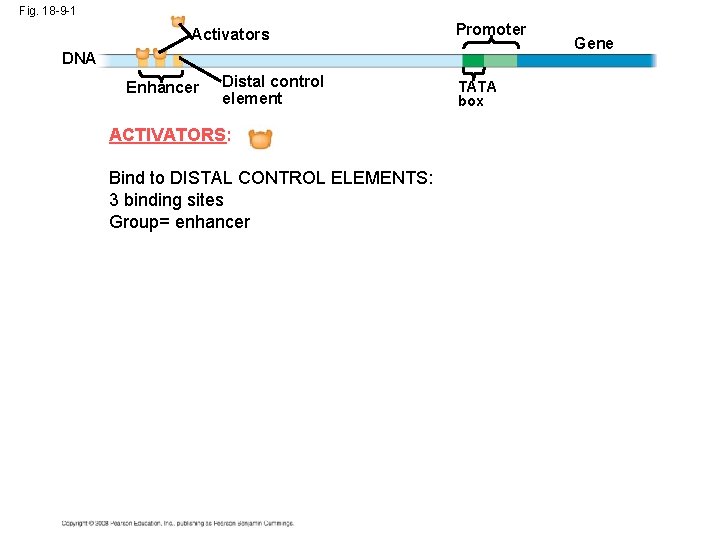
Fig. 18 -9 -1 Activators Promoter DNA Enhancer Distal control element ACTIVATORS: Bind to DISTAL CONTROL ELEMENTS: 3 binding sites Group= enhancer TATA box Gene

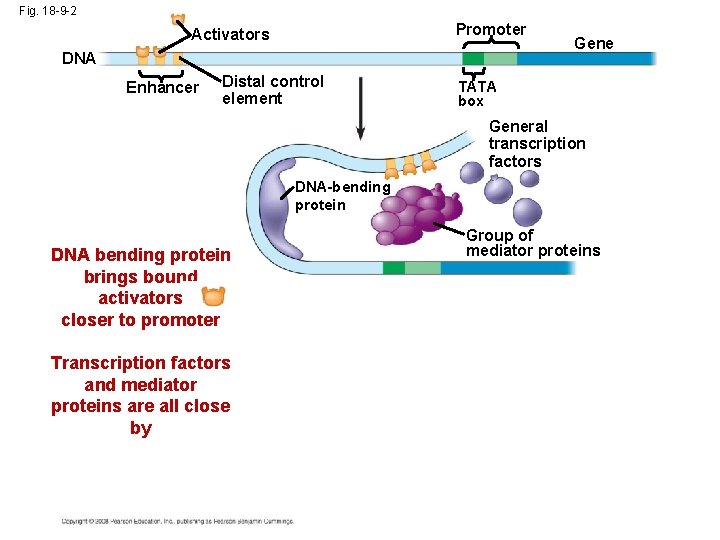
Fig. 18 -9 -2 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors DNA-bending protein DNA bending protein brings bound activators closer to promoter Transcription factors and mediator proteins are all close by Group of mediator proteins

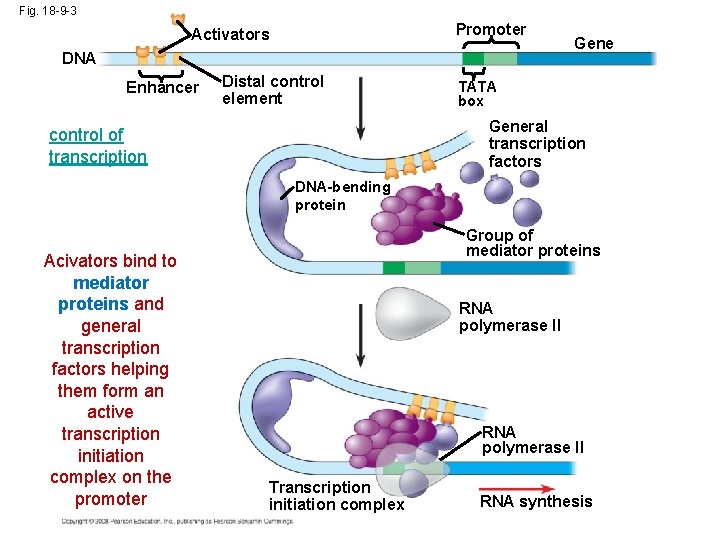
Fig. 18 -9 -3 Promoter Activators DNA Enhancer Distal control element Gene TATA box General transcription factors control of transcription DNA-bending protein Acivators bind to mediator proteins and general transcription factors helping them form an active transcription initiation complex on the promoter Group of mediator proteins RNA polymerase II Transcription initiation complex RNA synthesis

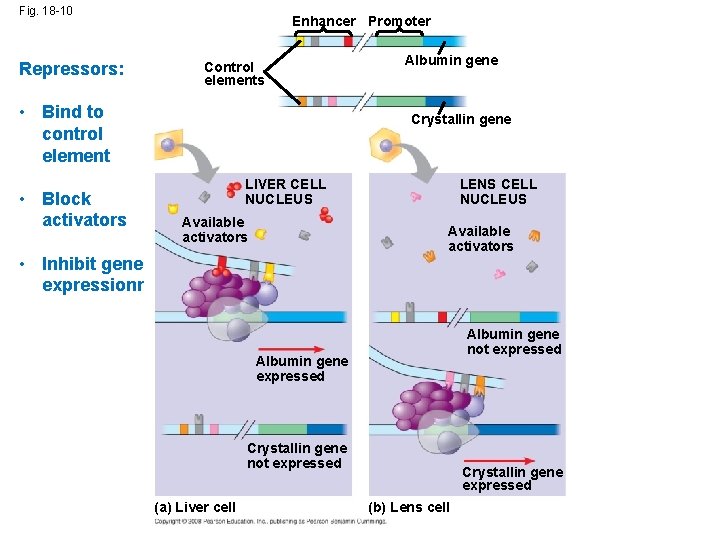
Fig. 18 -10 Repressors: Enhancer Promoter Control elements • Bind to control element • Block activators Albumin gene Crystallin gene LIVER CELL NUCLEUS Available activators LENS CELL NUCLEUS Available activators • Inhibit gene expressionr Albumin gene not expressed Albumin gene expressed Crystallin gene not expressed (a) Liver cell Crystallin gene expressed (b) Lens cell

• Transcription alone does not constitute gene expression. • Expression is ultimately measured by the AMOUNT of protein made by cell • Many regulatory mechanisms operate at various stages AFTER transcription • This allows for fine tuning gene expression in response to environmental change

How to regulate once gene has been transcribed

Post transcription control mechanisms Regulate gene expression Post transcription : 1. RNA processing – alternative splicing- make different m. RNA from same primary transcript– Determined by which segments are treated as exons and introns

alternative splicing Exons DNA Troponin T gene Primary RNA transcript RNA splicing m. RNA or

Post transcription control mechanisms • Once in cytoplasm…. Regulate gene expression Post transcription by: 2. m. RNA DEGRADATION • Longer m. RNA lasts the MORE protein is made • Sequence of UTR at 3’ end determine how long before degradation • Remove 5’ cap – Enzymes degrade post transcription control mechanisms

Fig. 18 -6 Signal NUCLEUS Chromatin modification DNA Gene available for transcription Gene Transcription RNA Exon Primary transcript Intron RNA processing Tail Cap review control of gene expression m. RNA in nucleus Transport to cytoplasm CYTOPLASM m. RNA in cytoplasm Degradation of m. RNA Translation Polypeptide Protein processing Degradation of protein Active protein Transport to cellular destination Cellular function 3. Initiation of translation control mechanisms

3. translationanother opportunity to regulate gene expression • Occurs in initiation stage • Can be BLOCKED by Regulatory protein attaching at UTR by the 5’ cap or 3’ end preventing attachment to ribosome post transcription control mechanisms Slide 6

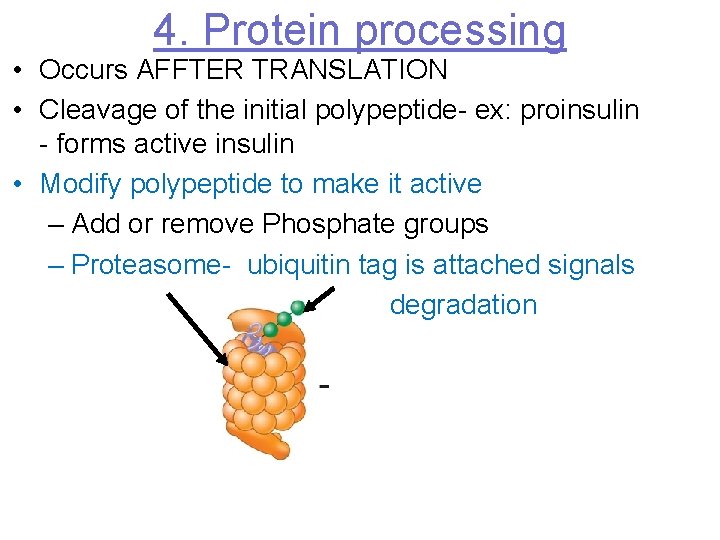
4. Protein processing • Occurs AFFTER TRANSLATION • Cleavage of the initial polypeptide- ex: proinsulin - forms active insulin • Modify polypeptide to make it active – Add or remove Phosphate groups – Proteasome- ubiquitin tag is attached signals degradation

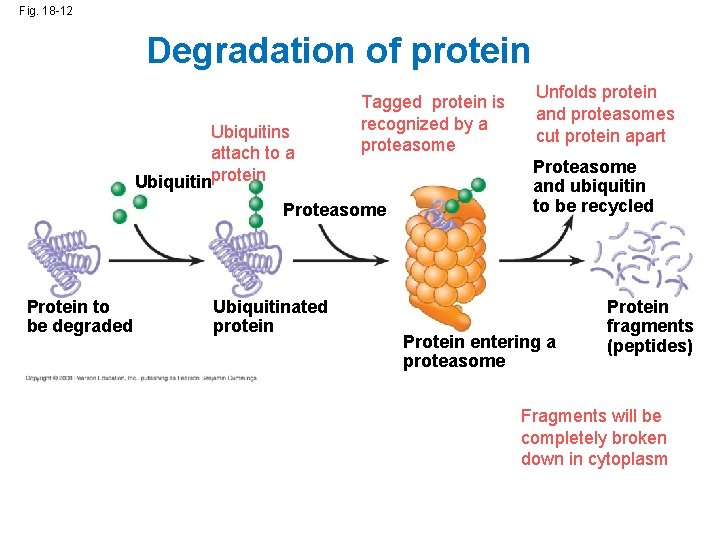
Fig. 18 -12 Degradation of protein Ubiquitins attach to a Ubiquitinprotein Tagged protein is recognized by a proteasome Protein to be degraded Ubiquitinated protein Unfolds protein and proteasomes cut protein apart Proteasome and ubiquitin to be recycled Protein entering a proteasome Protein fragments (peptides) Fragments will be completely broken down in cytoplasm

Micro. RNA micro RNA • Watch video clip • • gene silencing by micro RNA Key to switching genes on and off Micro. RNA sequence found in intron 22 nucleotides long Binds to dicer &argonaute proteins to help silence genes • Acts as a road block • Prevent from making too much protein

Medicinal value • Key treatment of diseases in the future • Such as cancers • May want to view videolink below and see how well you can follow what Dr. Valeri is saying • 12 min clip micro RNAs and colon cancer generation and action of si. RNA and micro RNA

Small interfering RNA- Si RNA • 20 nucleotides long what is small • Causes gene silencing interfering RNA • Regulate the stability and translation of m. RNA generation and action of si. RNA and micro RNA

• Only need to know that micro RNA and si. RNA are used to regulate translation of proteins

Next will be select portions of ch 19.
 Chapter 18
Chapter 18 Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Regulation of gene expression
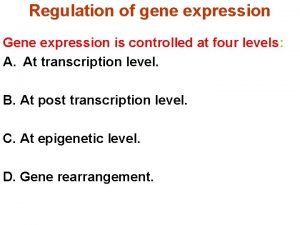
Regulation of gene expression Regulation of gene expression
Regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 18 regulation of gene expression
Chapter 18 regulation of gene expression Chapter 17 from gene to protein
Chapter 17 from gene to protein Section 12-1 dna answers
Section 12-1 dna answers Differential gene regulation
Differential gene regulation Section 12-5 gene regulation
Section 12-5 gene regulation What is gene regulation
What is gene regulation Positive vs negative controls
Positive vs negative controls Prokaryotes vs eukaryotes gene regulation
Prokaryotes vs eukaryotes gene regulation Gene regulation
Gene regulation Section 4 gene regulation and mutation
Section 4 gene regulation and mutation Section 4 gene regulation and mutation
Section 4 gene regulation and mutation Gene regulation
Gene regulation Gene by gene test results
Gene by gene test results Lyonization of gene expression
Lyonization of gene expression Corbett maths
Corbett maths Gene expression
Gene expression Prokaryotic
Prokaryotic Gene expression omnibus tutorial
Gene expression omnibus tutorial Gene expression
Gene expression טרנסלציה
טרנסלציה Gene expression
Gene expression Cells must control gene expression so that __________.
Cells must control gene expression so that __________. Genetic effects on gene expression across human tissues
Genetic effects on gene expression across human tissues Gene expression
Gene expression Trp operon
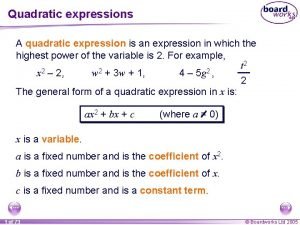
Trp operon Quadratic formula examples
Quadratic formula examples Homometric regulation
Homometric regulation What is emergency bilge suction valve in engine room
What is emergency bilge suction valve in engine room Board regulation 1 series of 2014
Board regulation 1 series of 2014 Social media yoga
Social media yoga Cimah regulation
Cimah regulation Efficiency of transformer formula
Efficiency of transformer formula Transcetolisation
Transcetolisation George stigler the theory of economic regulation
George stigler the theory of economic regulation Phosphoenolpyruvate
Phosphoenolpyruvate Ihr 1969
Ihr 1969 Principal cells
Principal cells Human regulation
Human regulation Difficulty in emotion regulation scale
Difficulty in emotion regulation scale Wa department of water and environmental regulation
Wa department of water and environmental regulation Thermoregulation
Thermoregulation Malaysia elv regulation
Malaysia elv regulation Commercial banking regulation
Commercial banking regulation Expanded withholding tax revenue regulation
Expanded withholding tax revenue regulation Cushing's reflex
Cushing's reflex What is the biomedical model of health
What is the biomedical model of health Unarmed regulation drill sequence
Unarmed regulation drill sequence Fgda color wheel
Fgda color wheel Line regulation formula
Line regulation formula Ots energy
Ots energy Better regulation cos'è
Better regulation cos'è What animals are cold blooded
What animals are cold blooded Regulation of recruitment and placement activities
Regulation of recruitment and placement activities Regulation asservissement
Regulation asservissement Vertical zones of the ocean
Vertical zones of the ocean Vietnam elv regulation
Vietnam elv regulation Regulation of breathing
Regulation of breathing Deviance regulation theory
Deviance regulation theory Fsa allergy chef cards
Fsa allergy chef cards Ar 600-8-10
Ar 600-8-10 Ncoer non rated codes
Ncoer non rated codes What is emotional regulation
What is emotional regulation Department of licensing and regulatory affairs
Department of licensing and regulatory affairs Nwkraft
Nwkraft Régulation
Régulation Regulation of kidney function class 11
Regulation of kidney function class 11