Gene Expression M Tevfik DORAK http www dorak









- Slides: 84

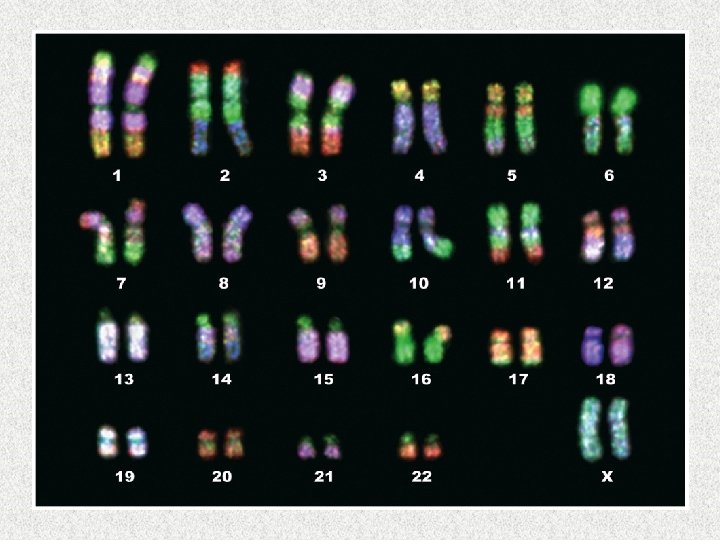
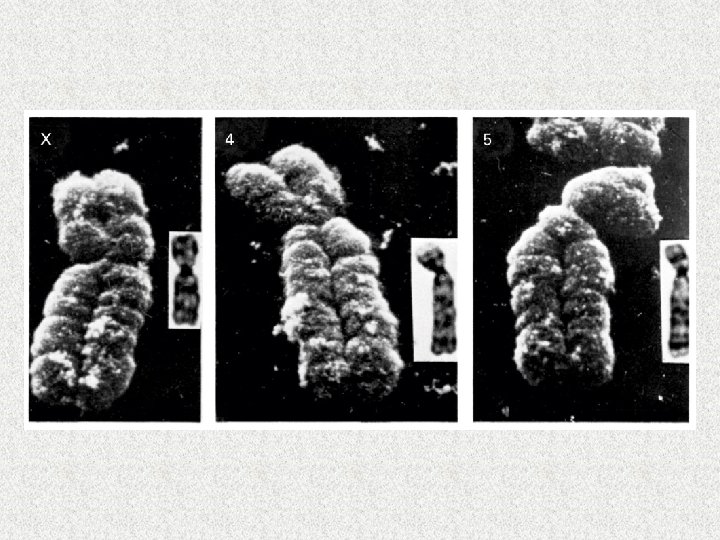
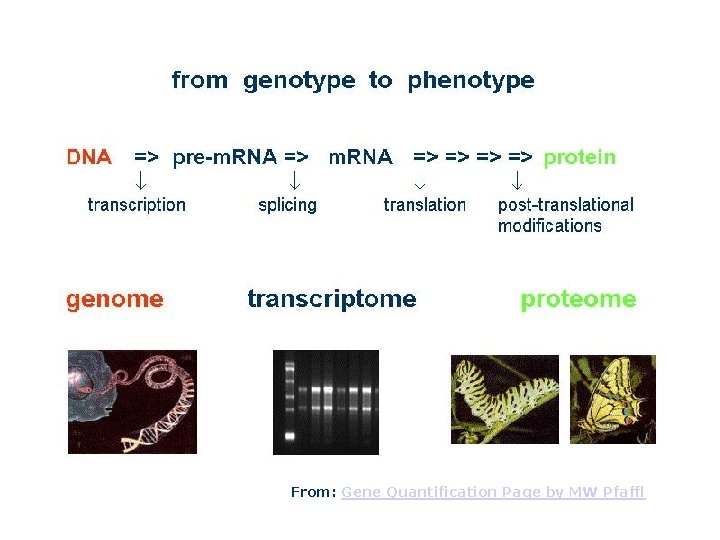
Gene Expression M. Tevfik DORAK http: //www. dorak. info



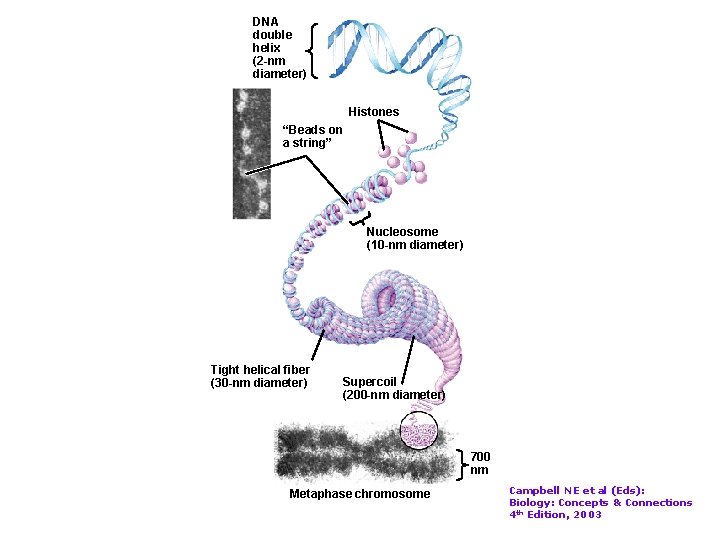
DNA double helix (2 -nm diameter) Histones “Beads on a string” Nucleosome (10 -nm diameter) Tight helical fiber (30 -nm diameter) Supercoil (200 -nm diameter) 700 nm Metaphase chromosome Campbell NE et al (Eds): Biology: Concepts & Connections 4 th Edition, 2003


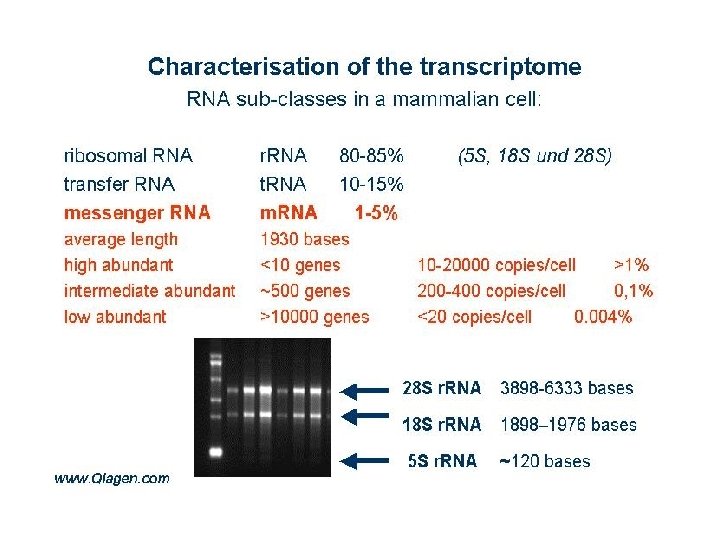
From: Gene Quantification Page by MW Pfaffl

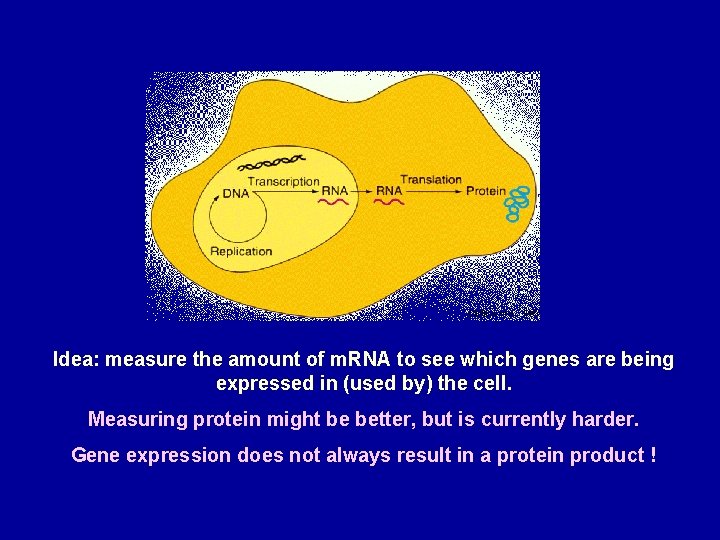
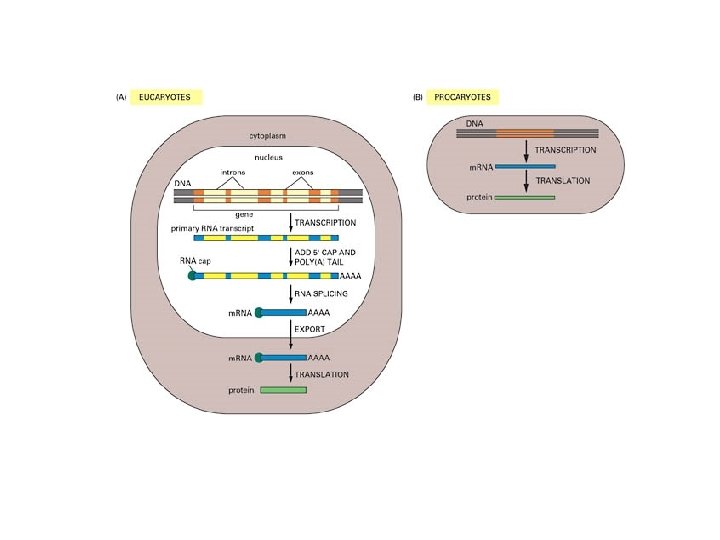
Idea: measure the amount of m. RNA to see which genes are being expressed in (used by) the cell. Measuring protein might be better, but is currently harder. Gene expression does not always result in a protein product !

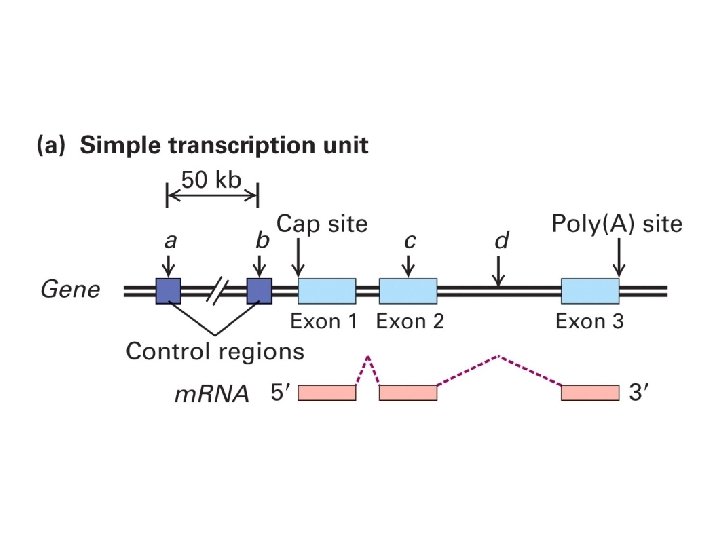
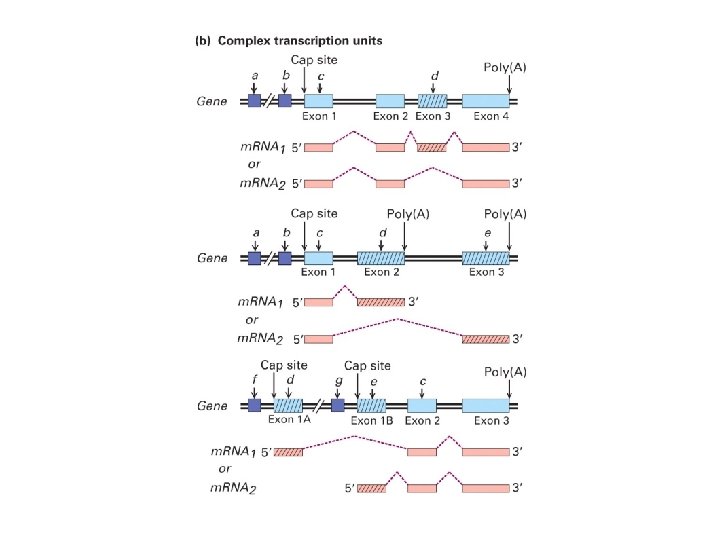
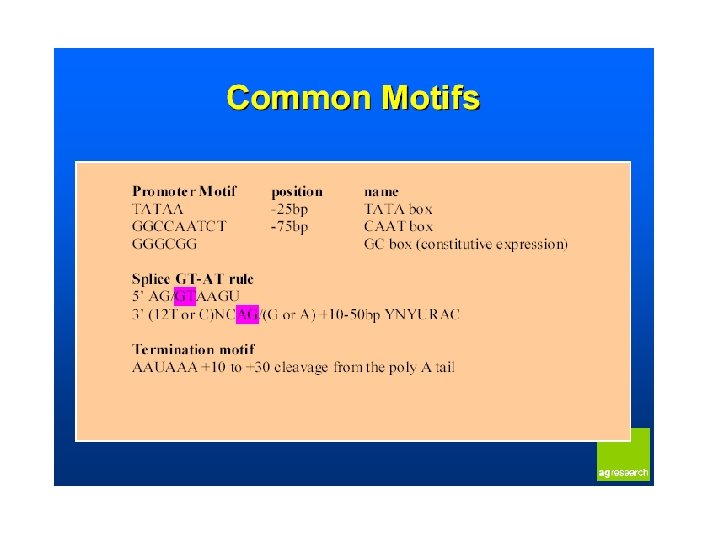
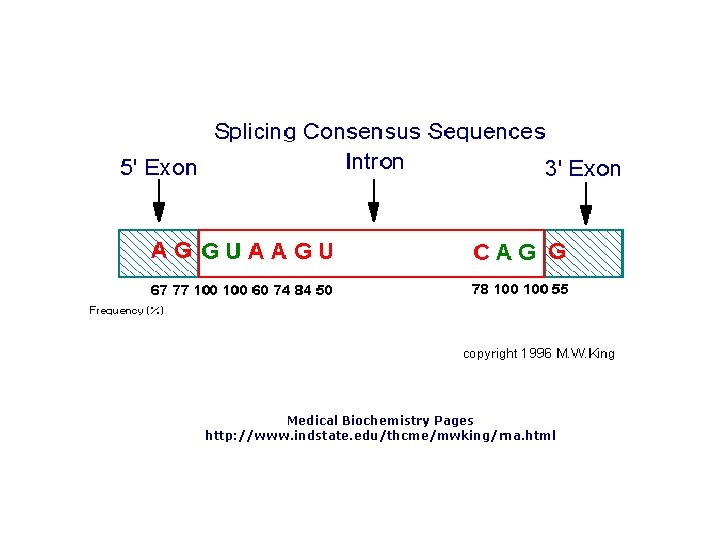
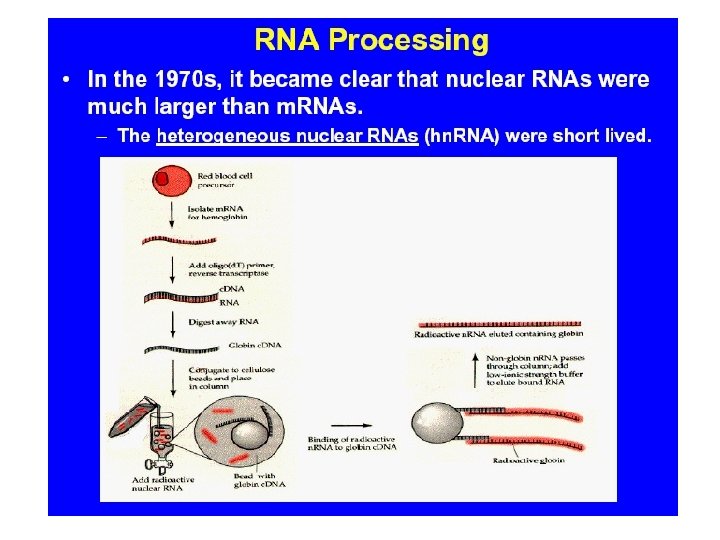
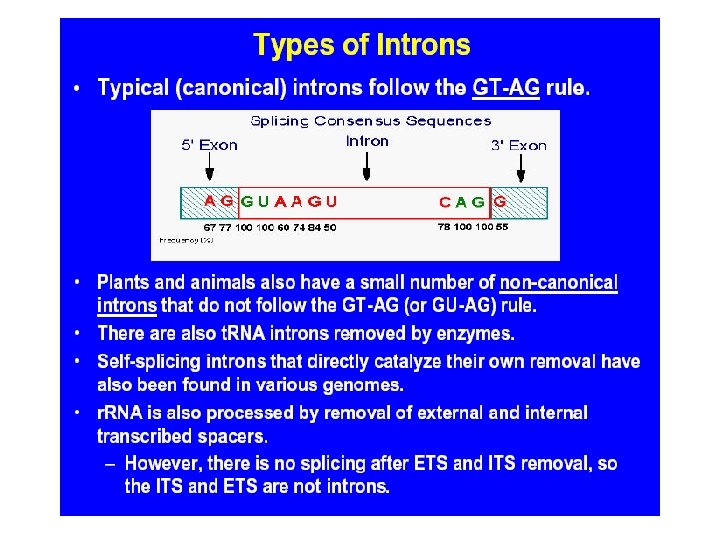
Transcribed and Nontranscribed Strands

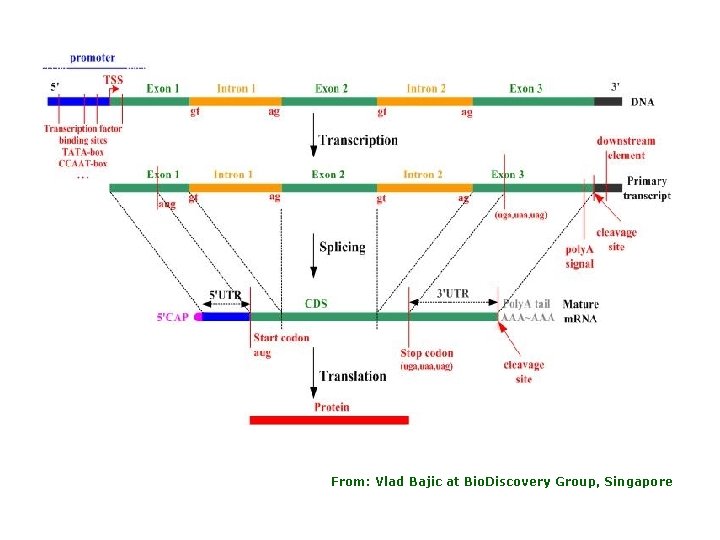
From: Vlad Bajic at Bio. Discovery Group, Singapore



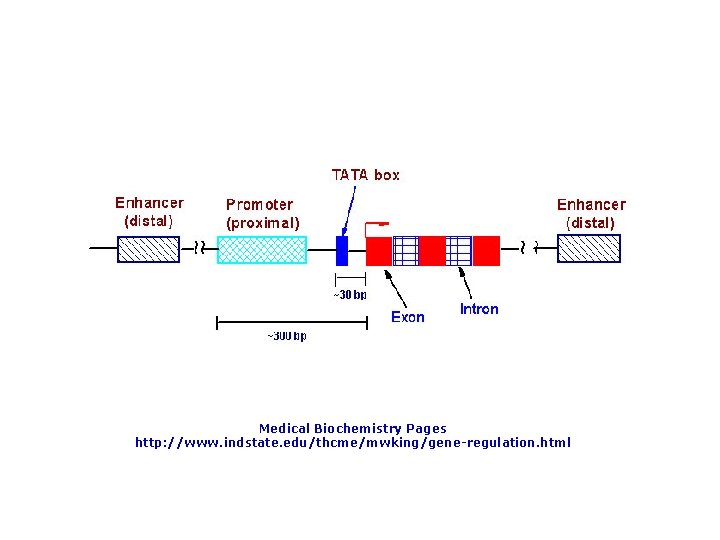
Medical Biochemistry Pages http: //www. indstate. edu/thcme/mwking/gene-regulation. html


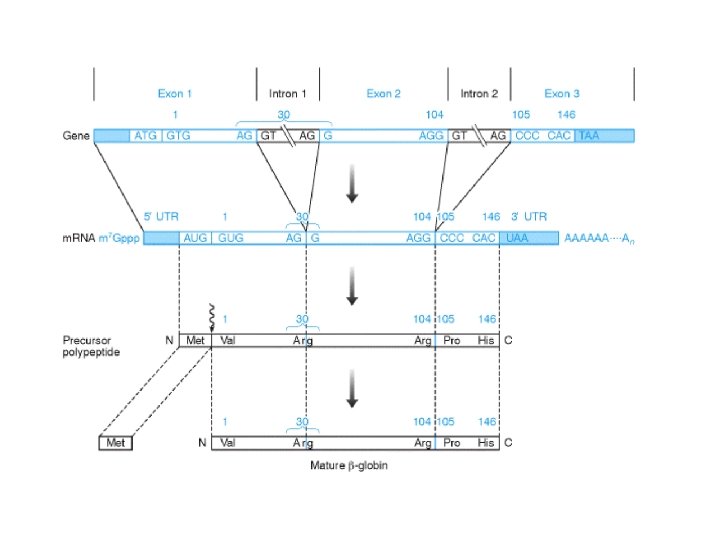
Expression of the human b-globin gene. Exons 1 and 3 each contain noncoding sequences (shaded bars) at their extremities, which are transcribed and are present at the 5’ and 3’ ends of the b-globin m. RNA, but are not translated to specify polypeptide synthesis. Such 5’ and 3’ untranslated regions (5’ UTR and 3’ UTR), however, are thought to be important in ensuring high efficiency of translation. The stop codon UAA represents the first three nucleotides of the 3’ untranslated region. Note that the initial translation product has 147 amino acids, but that the N-terminal methionine is removed by post-translational processing to generate the mature bglobin polypeptide. From: Human Molecular Genetics by Strachan & Read. NCBI Books Online

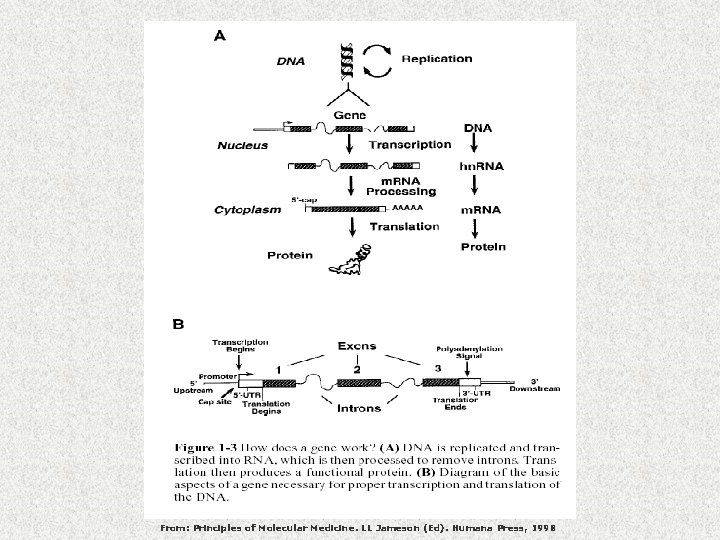
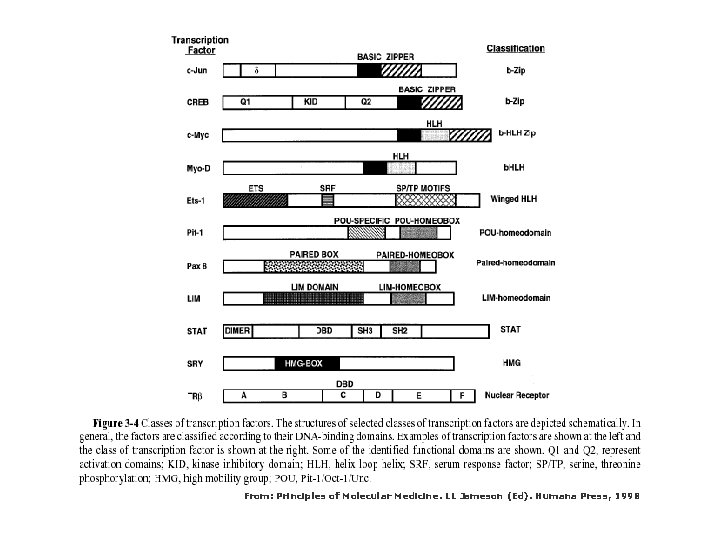
From: Principles of Molecular Medicine. LL Jameson (Ed). Humana Press, 1998

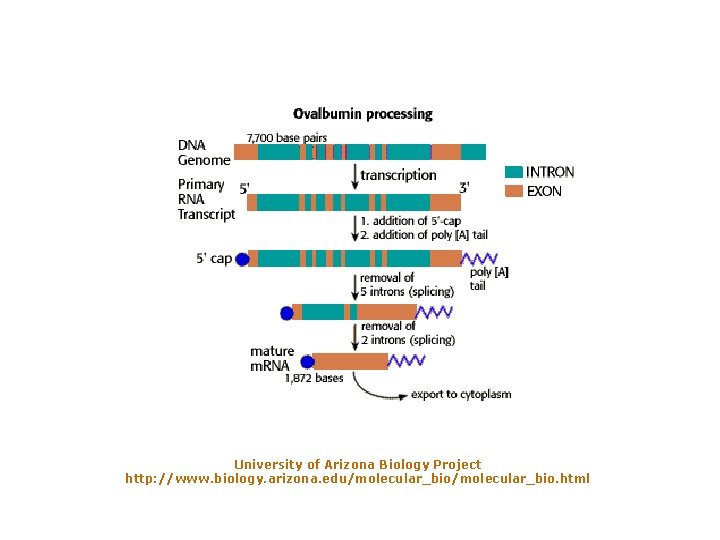
University of Arizona Biology Project http: //www. biology. arizona. edu/molecular_bio. html

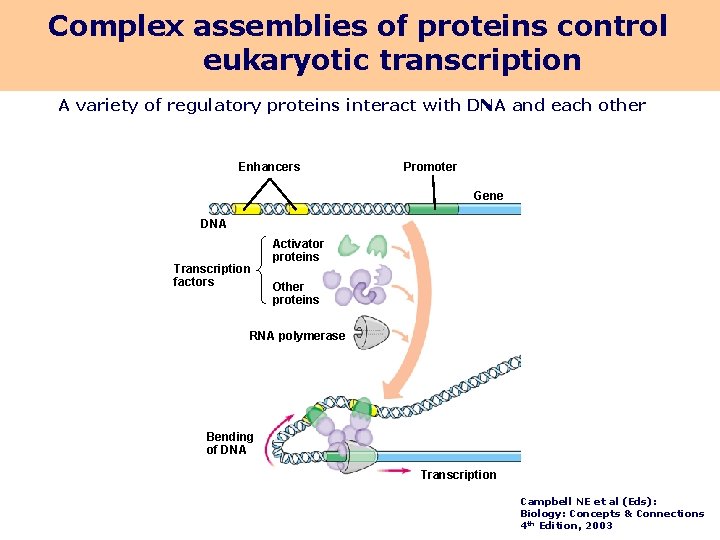
Complex assemblies of proteins control eukaryotic transcription A variety of regulatory proteins interact with DNA and each other Enhancers Promoter Gene DNA Transcription factors Activator proteins Other proteins RNA polymerase Bending of DNA Transcription Campbell NE et al (Eds): Biology: Concepts & Connections 4 th Edition, 2003

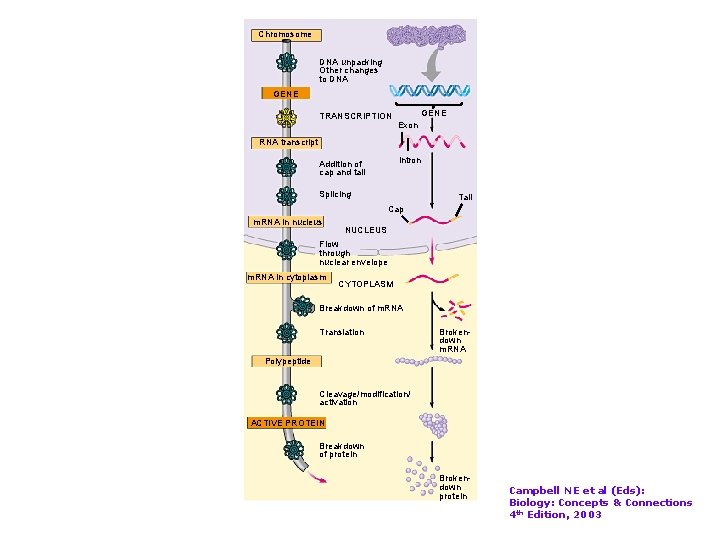
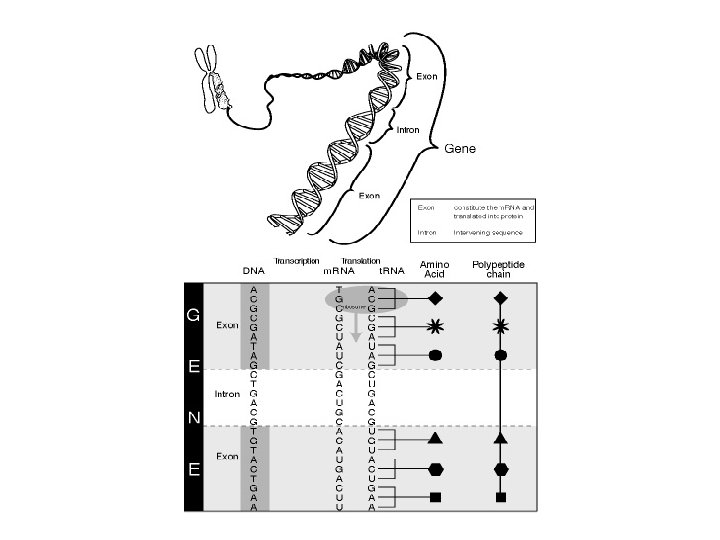
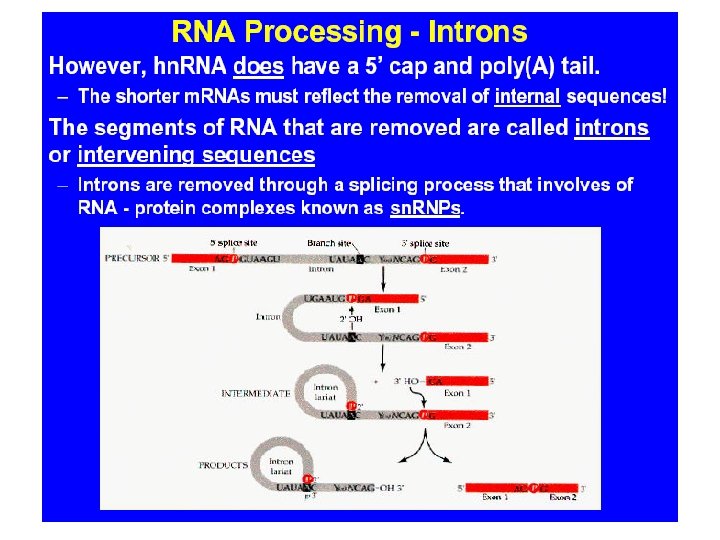
Chromosome DNA unpacking Other changes to DNA GENE TRANSCRIPTION GENE Exon RNA transcript Intron Addition of cap and tail Splicing Tail Cap m. RNA in nucleus NUCLEUS Flow through nuclear envelope m. RNA in cytoplasm CYTOPLASM Breakdown of m. RNA Translation Brokendown m. RNA Polypeptide Cleavage/modification/ activation ACTIVE PROTEIN Breakdown of protein Brokendown protein Campbell NE et al (Eds): Biology: Concepts & Connections 4 th Edition, 2003


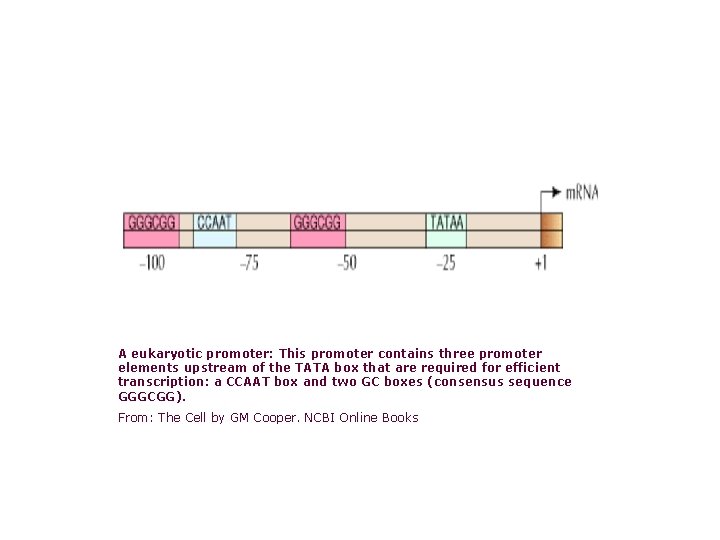
A eukaryotic promoter: This promoter contains three promoter elements upstream of the TATA box that are required for efficient transcription: a CCAAT box and two GC boxes (consensus sequence GGGCGG). From: The Cell by GM Cooper. NCBI Online Books



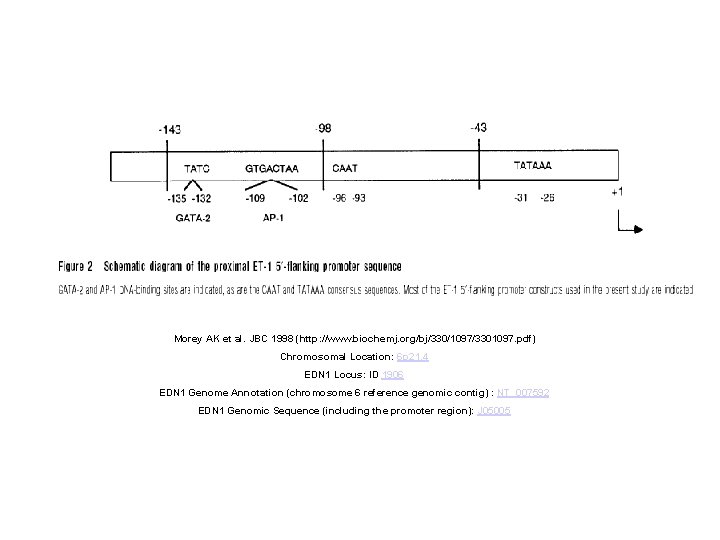
Morey AK et al. JBC 1998 (http: //www. biochemj. org/bj/330/1097/3301097. pdf) Chromosomal Location: 6 p 21. 4 EDN 1 Locus: ID 1906 EDN 1 Genome Annotation (chromosome 6 reference genomic contig) : NT_007592 EDN 1 Genomic Sequence (including the promoter region): J 05005

EDN 1 (Gene. ID 1906) GI: 340555 repeat_region 98. . . 383 /rpt_family=" Alu" protein_bind 739. . . 745 /bound_moiety="acute phase reactant regulatory element" misc_feature 979. . 1039 /note="Z-DNA region; putative" protein_bind 2183. . 2188 /bound_moiety="acute phase reactant regulatory element" protein_bind 2951. . 2958 /bound_moiety="TPA/JUN" protein_bind 3241. . 3248 /bound_moiety="TPA/JUN" protein_bind 3316. . 3328 /bound_moiety="NF-1" protein_bind 3499. . 3505 /bound_moiety="TPA/JUN" CAAT_signal 3510. . 3515 /gene="EDN 1" TATA_signal 3577. . 3582 /gene="EDN 1" Exon 1 3608. . 3939 /gene="EDN 1"

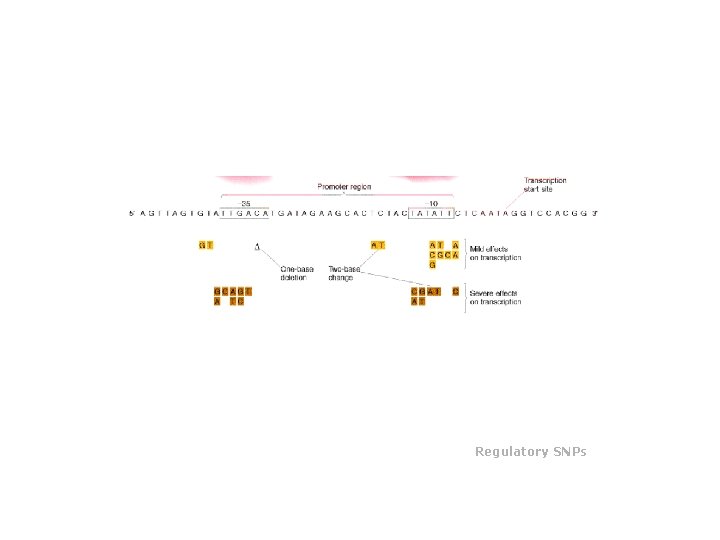
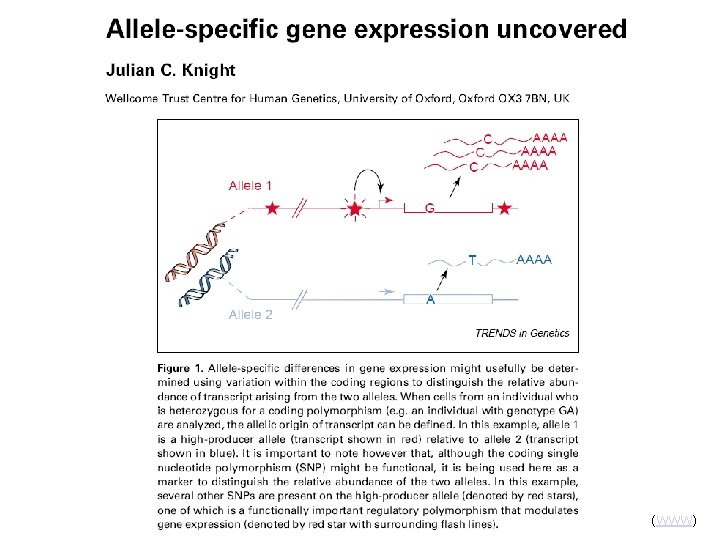
Regulatory SNPs

Medical Biochemistry Pages http: //www. indstate. edu/thcme/mwking/rna. html

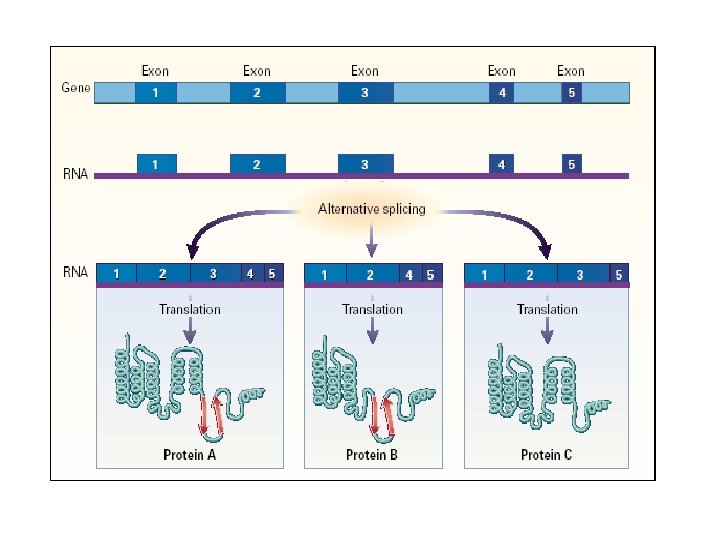
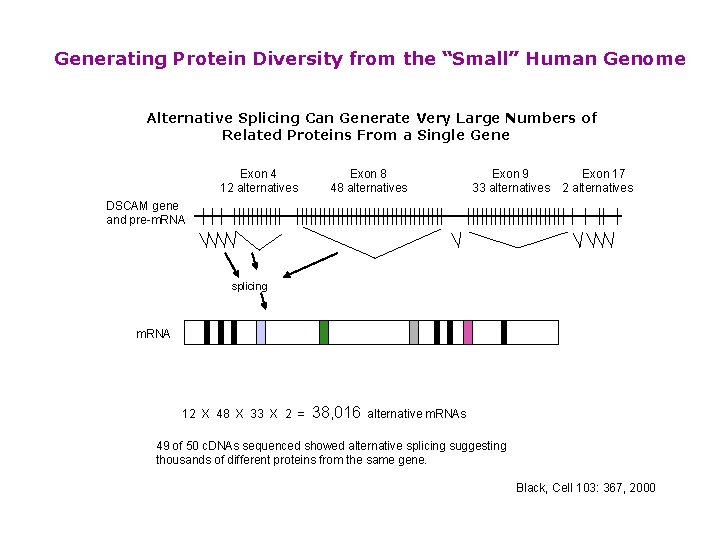
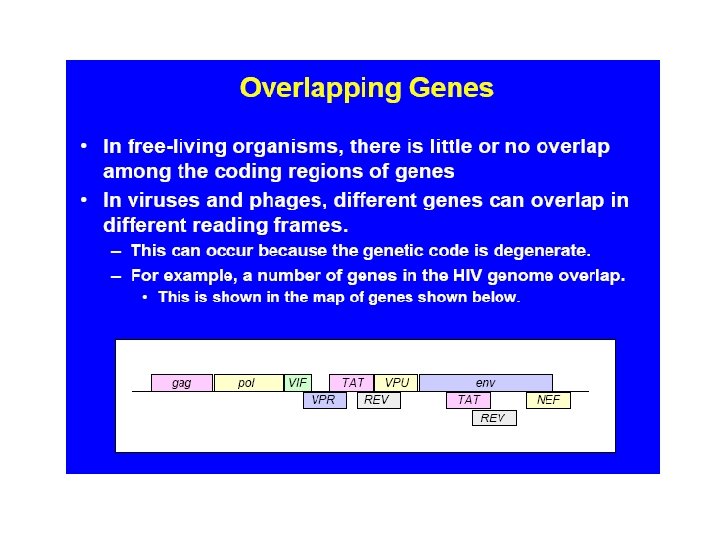
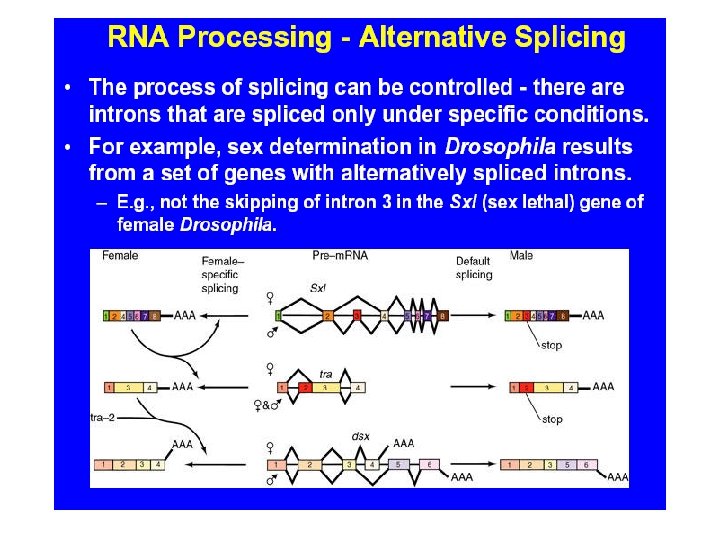
Generating Protein Diversity from the “Small” Human Genome Alternative Splicing Can Generate Very Large Numbers of Related Proteins From a Single Gene Exon 4 12 alternatives Exon 8 48 alternatives Exon 9 33 alternatives Exon 17 2 alternatives DSCAM gene and pre-m. RNA splicing m. RNA 12 X 48 X 33 X 2 = 38, 016 alternative m. RNAs 49 of 50 c. DNAs sequenced showed alternative splicing suggesting thousands of different proteins from the same gene. Black, Cell 103: 367, 2000

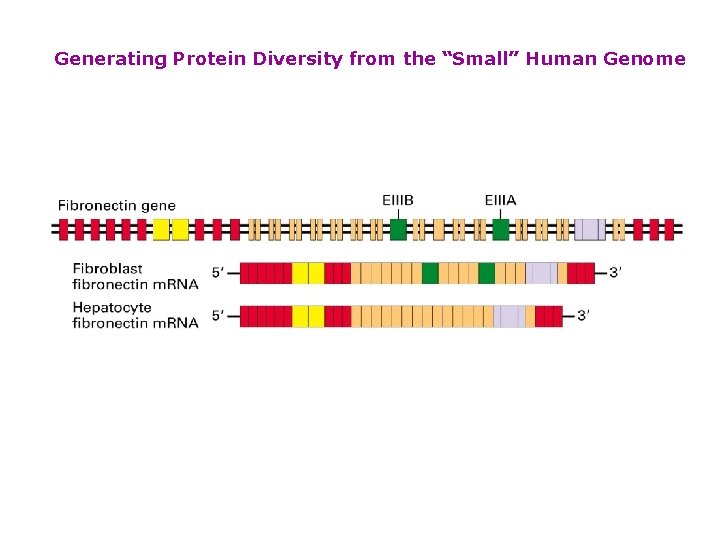
Generating Protein Diversity from the “Small” Human Genome

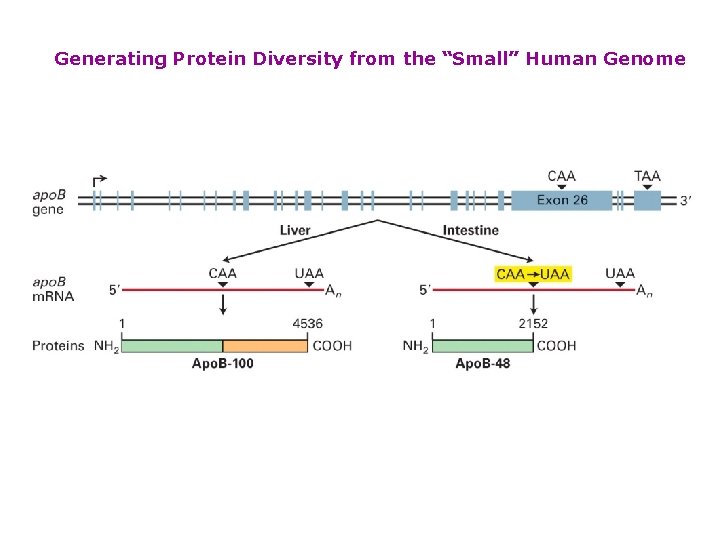
Generating Protein Diversity from the “Small” Human Genome




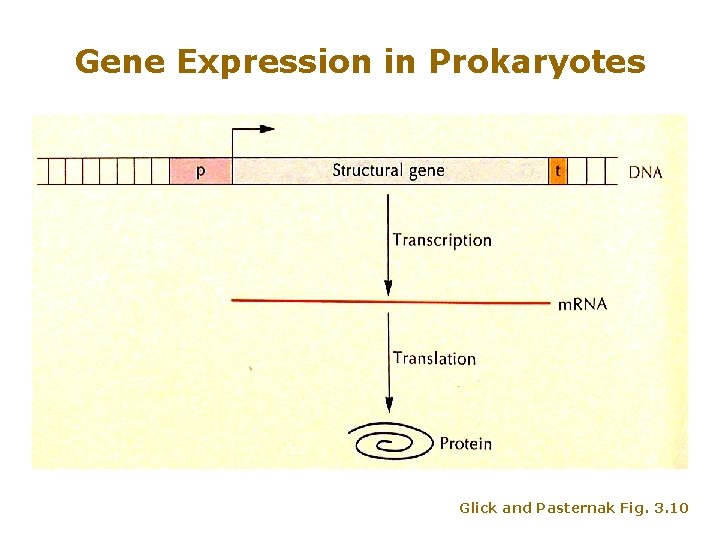
Gene Expression in Prokaryotes Glick and Pasternak Fig. 3. 10

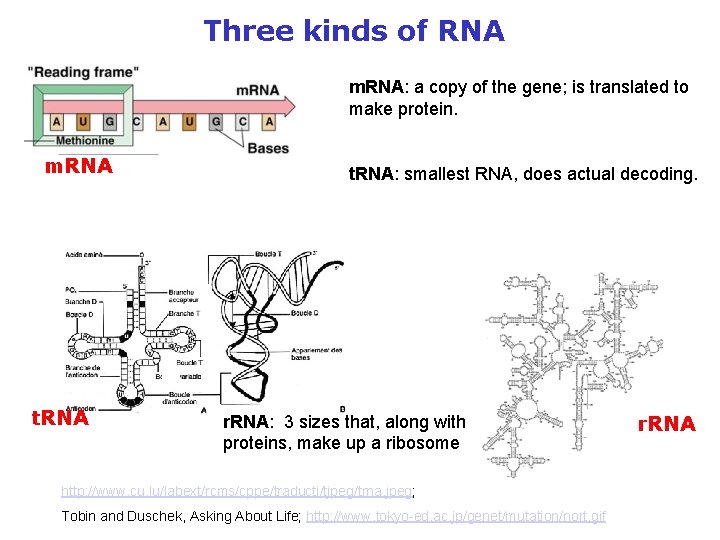
Three kinds of RNA m. RNA: a copy of the gene; is translated to make protein. m. RNA t. RNA: smallest RNA, does actual decoding. r. RNA: 3 sizes that, along with proteins, make up a ribosome http: //www. cu. lu/labext/rcms/cppe/traducti/tjpeg/trna. jpeg; Tobin and Duschek, Asking About Life; http: //www. tokyo-ed. ac. jp/genet/mutation/nort. gif r. RNA




From: Principles of Molecular Medicine. LL Jameson (Ed). Humana Press, 1998


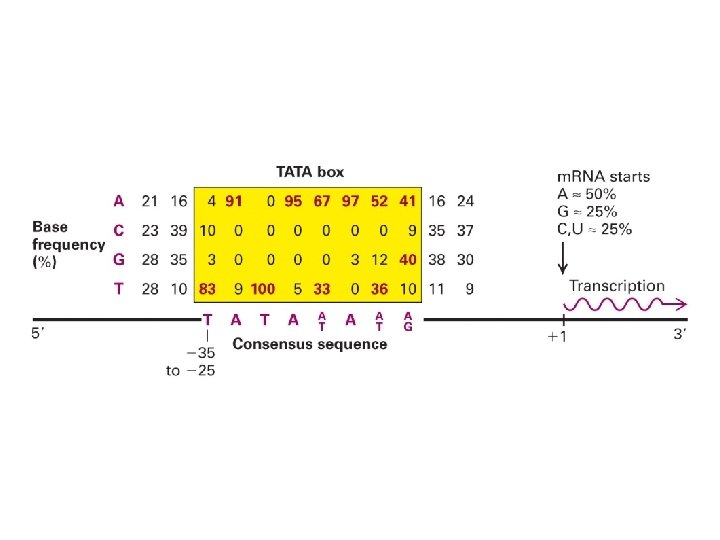
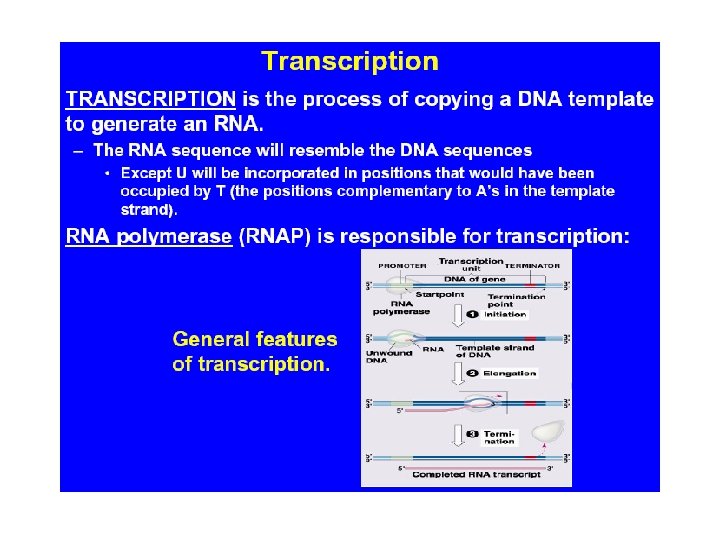
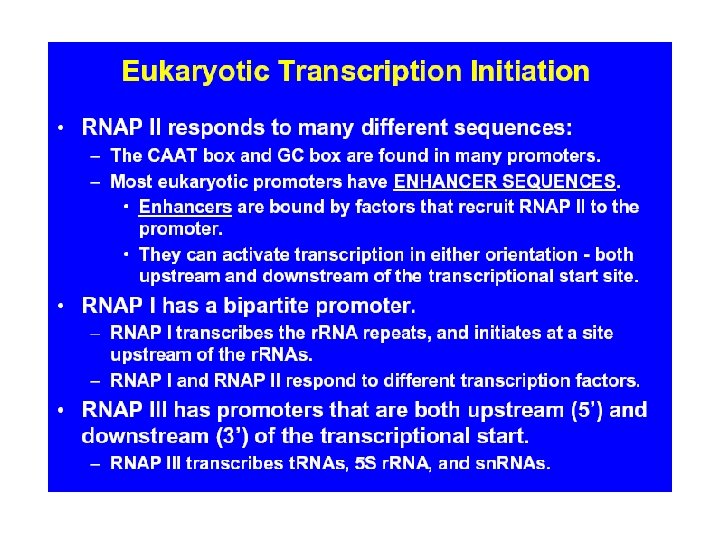
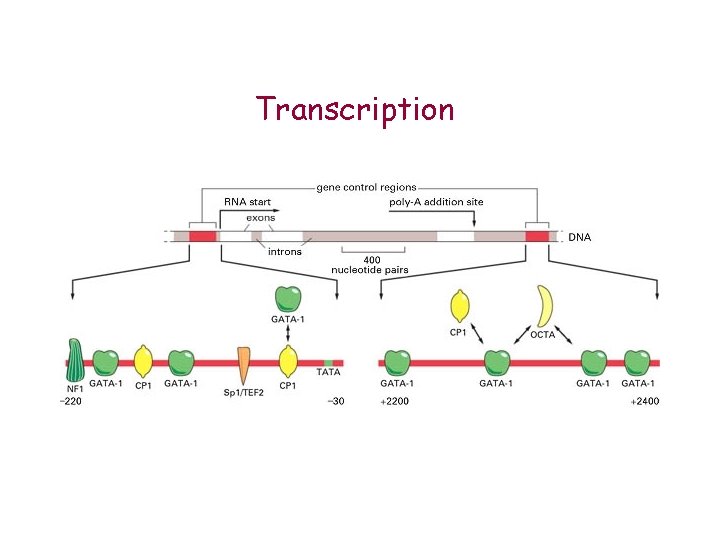
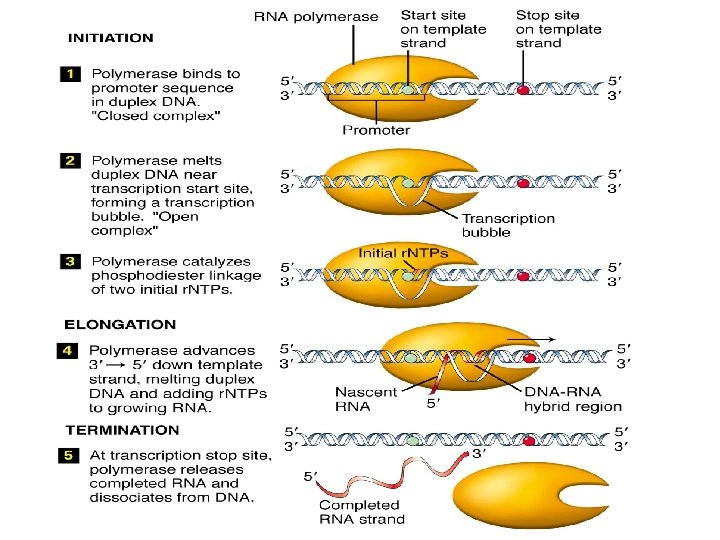
Transcription


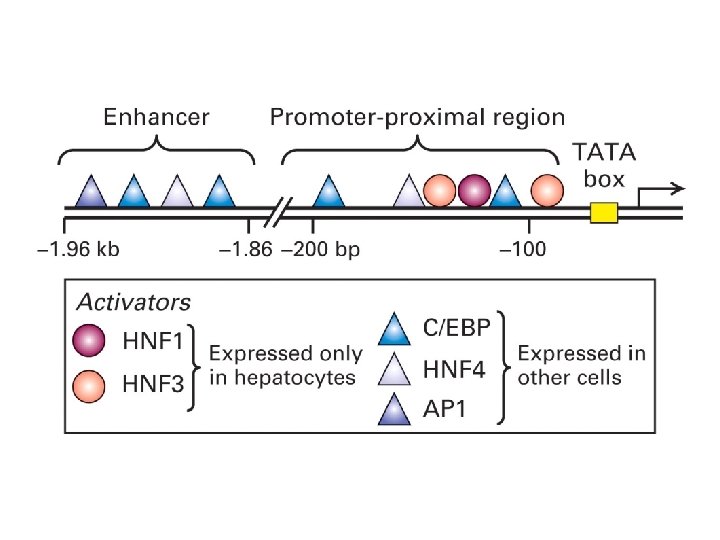
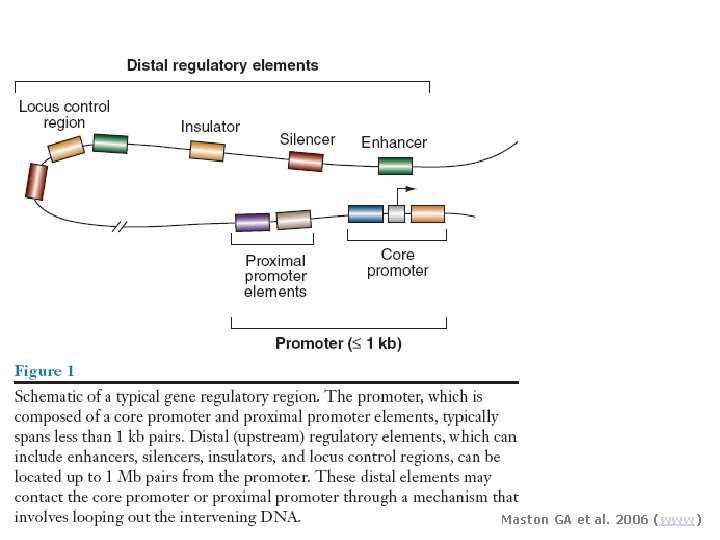
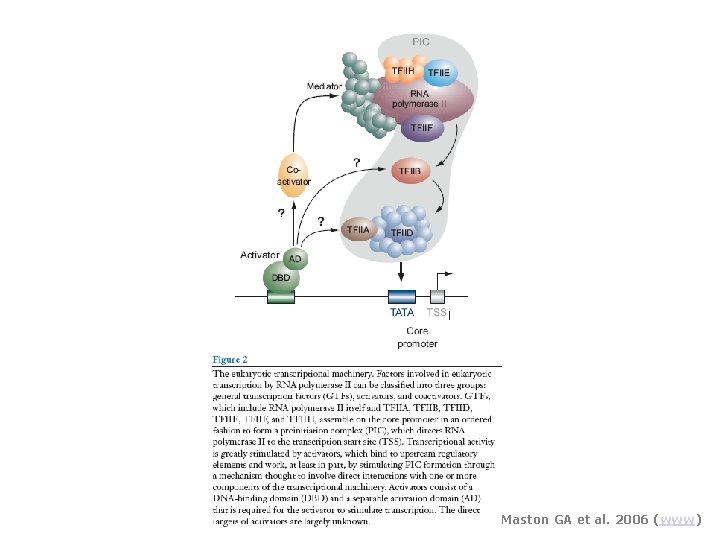
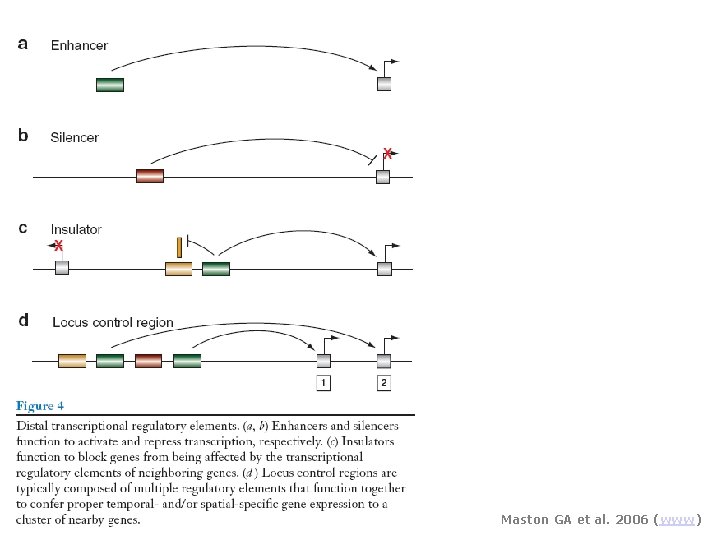
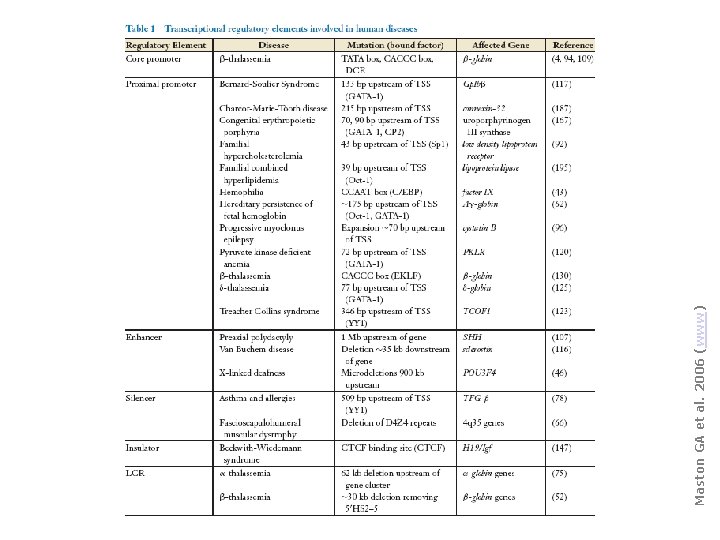
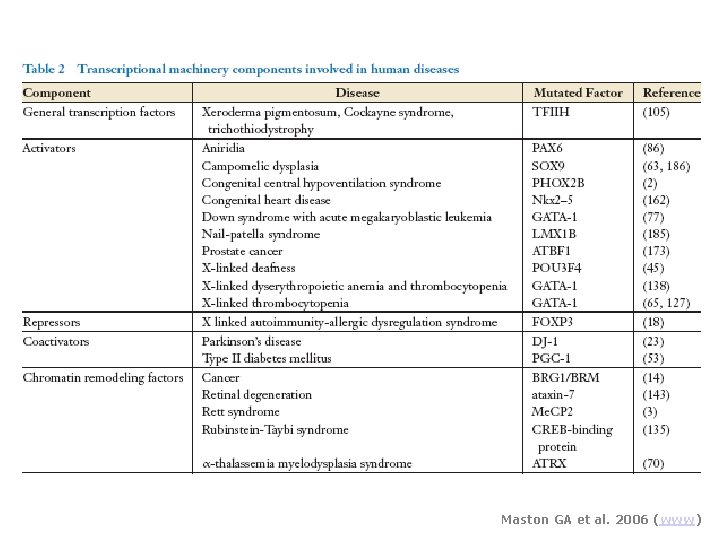
Maston GA et al. 2006 (www)

Maston GA et al. 2006 (www)

Maston GA et al. 2006 (www)

Maston GA et al. 2006 (www)

Maston GA et al. 2006 (www)

(WWW)












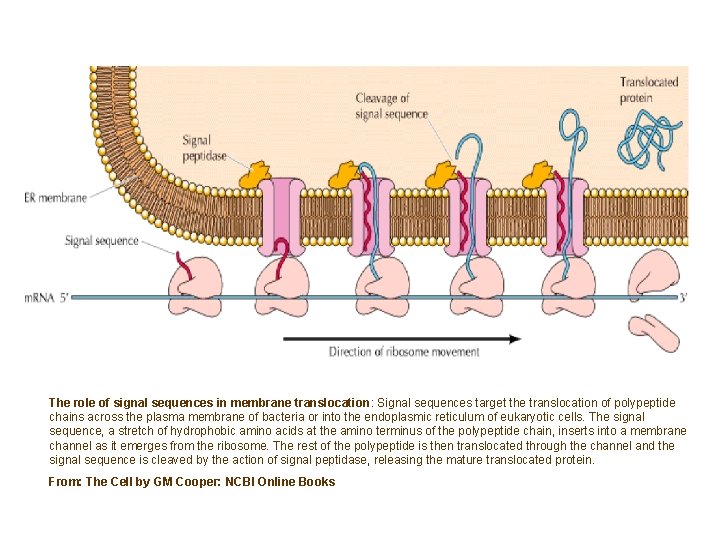
The role of signal sequences in membrane translocation: Signal sequences target the translocation of polypeptide chains across the plasma membrane of bacteria or into the endoplasmic reticulum of eukaryotic cells. The signal sequence, a stretch of hydrophobic amino acids at the amino terminus of the polypeptide chain, inserts into a membrane channel as it emerges from the ribosome. The rest of the polypeptide is then translocated through the channel and the signal sequence is cleaved by the action of signal peptidase, releasing the mature translocated protein. From: The Cell by GM Cooper: NCBI Online Books

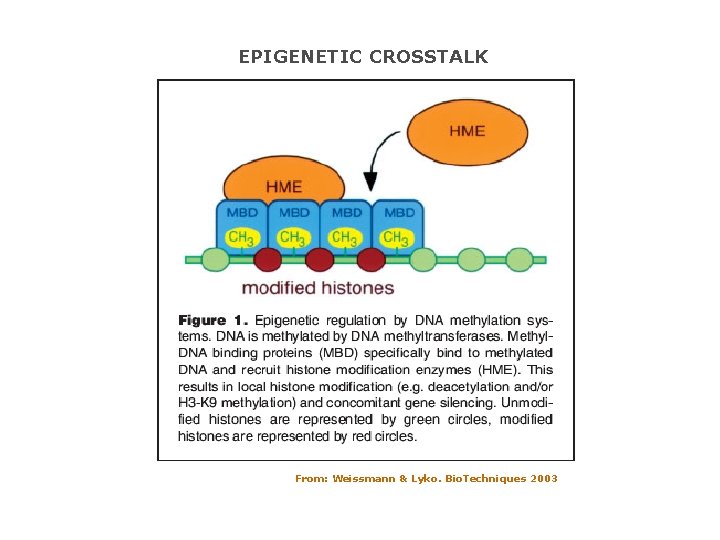
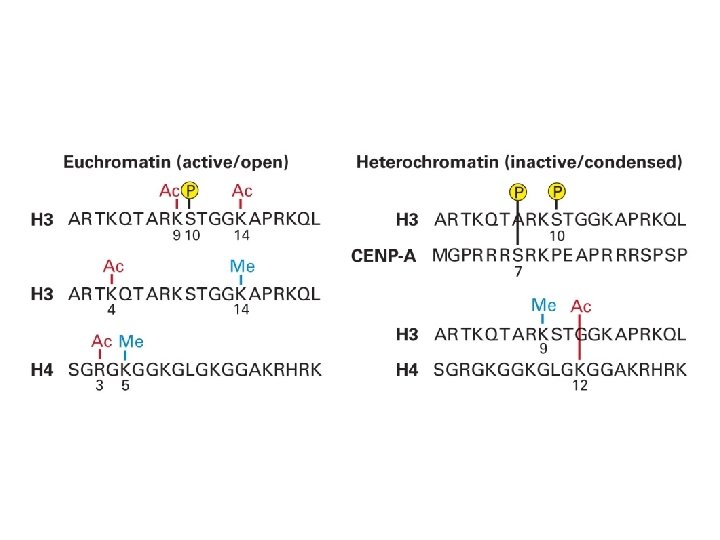
EPIGENETIC CROSSTALK From: Weissmann & Lyko. Bio. Techniques 2003

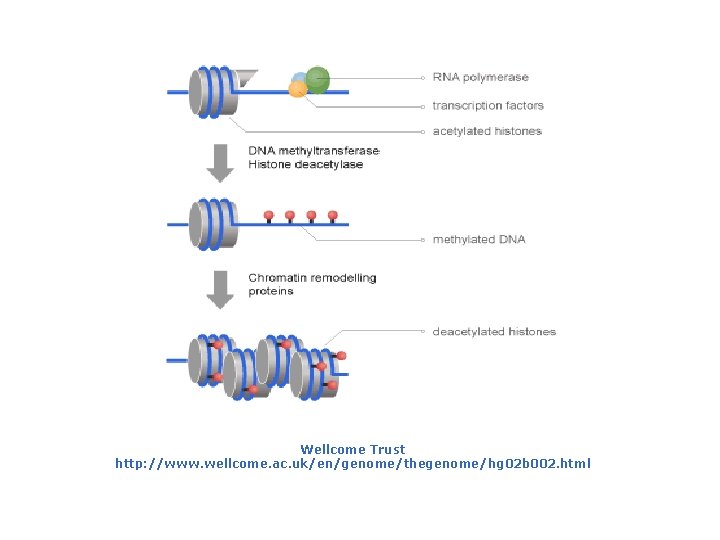
Wellcome Trust http: //www. wellcome. ac. uk/en/genome/thegenome/hg 02 b 002. html


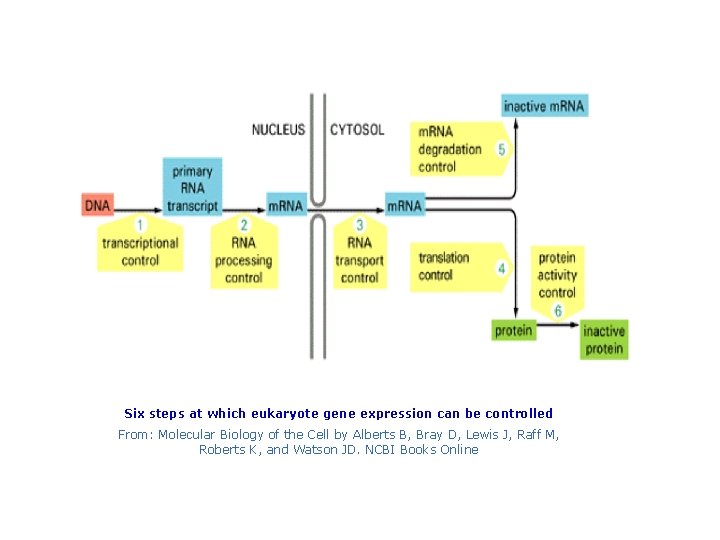
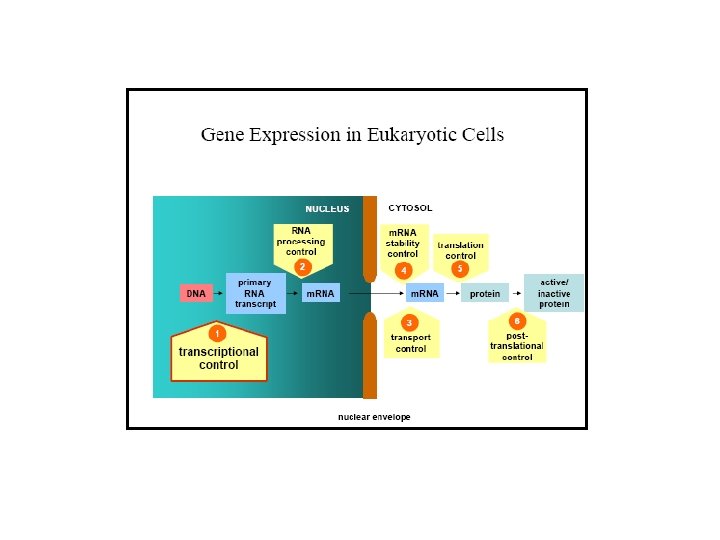
Six steps at which eukaryote gene expression can be controlled From: Molecular Biology of the Cell by Alberts B, Bray D, Lewis J, Raff M, Roberts K, and Watson JD. NCBI Books Online



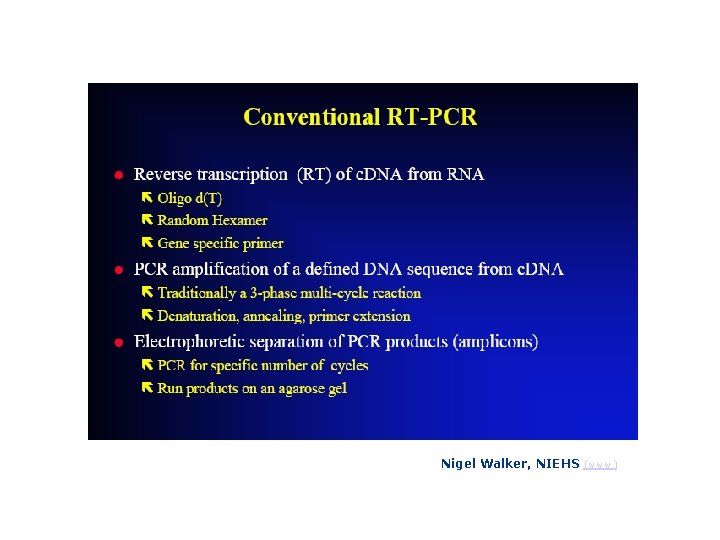
Nigel Walker, NIEHS (www)

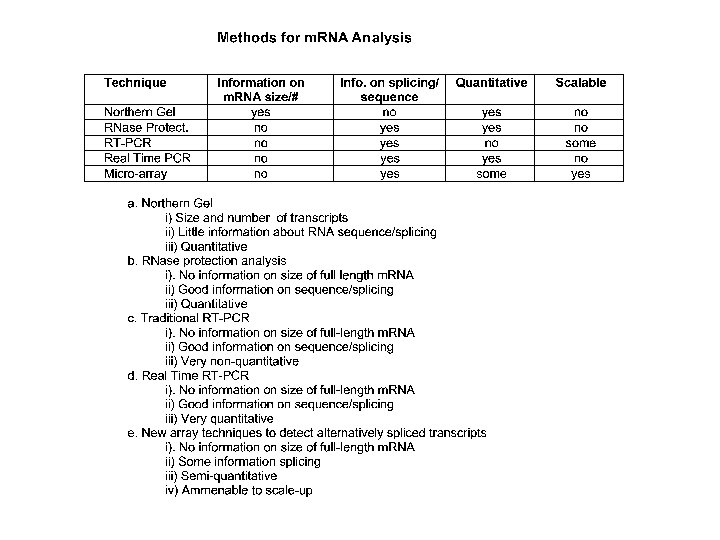
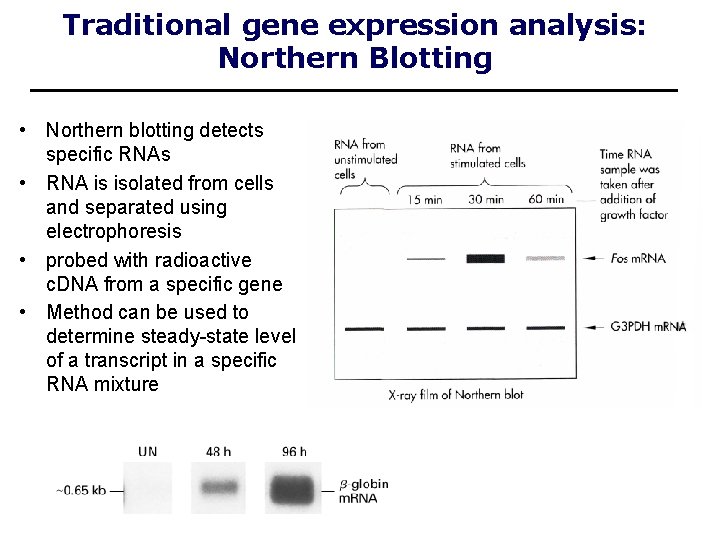
Traditional gene expression analysis: Northern Blotting • Northern blotting detects specific RNAs • RNA is isolated from cells and separated using electrophoresis • probed with radioactive c. DNA from a specific gene • Method can be used to determine steady-state level of a transcript in a specific RNA mixture

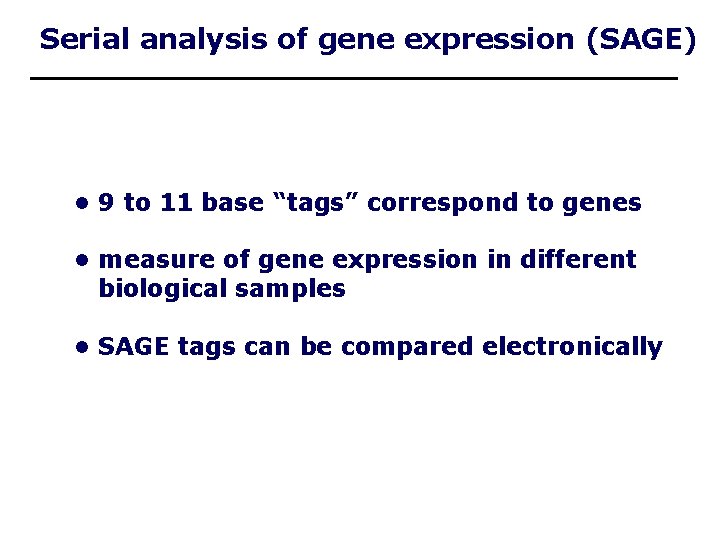
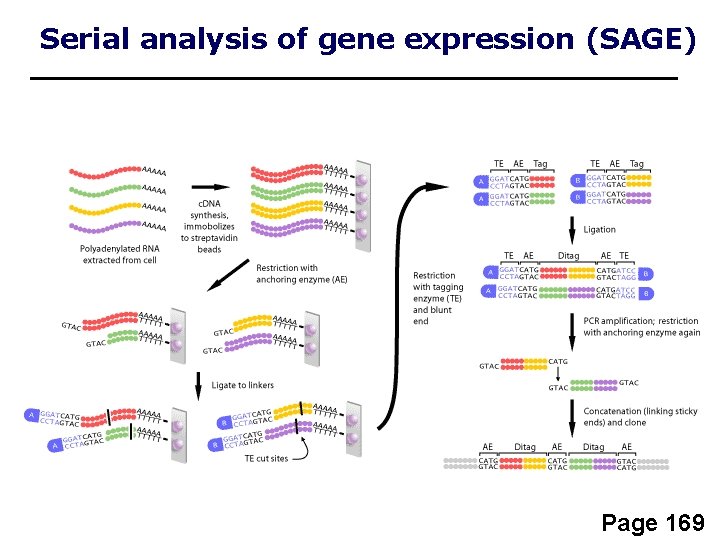
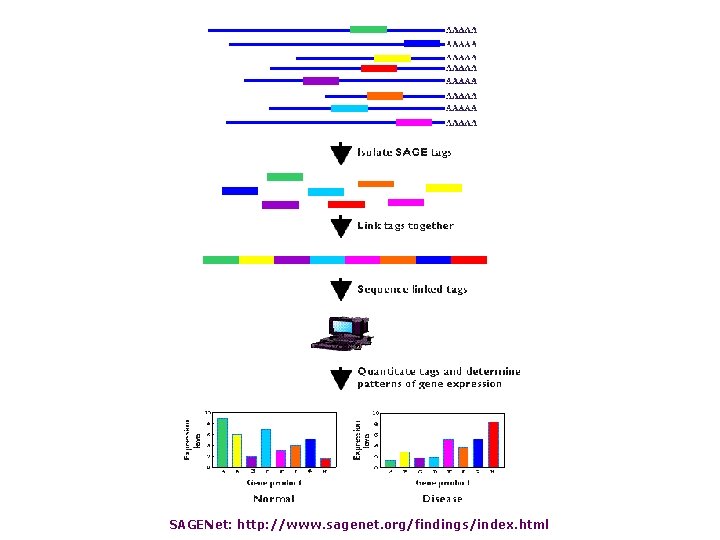
Serial analysis of gene expression (SAGE) • 9 to 11 base “tags” correspond to genes • measure of gene expression in different biological samples • SAGE tags can be compared electronically

Serial analysis of gene expression (SAGE) Page 169

SAGENet: http: //www. sagenet. org/findings/index. html


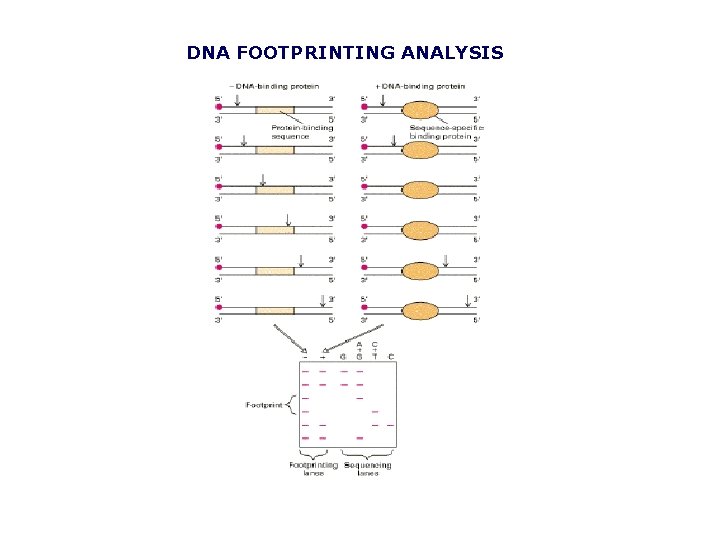
DNA FOOTPRINTING ANALYSIS

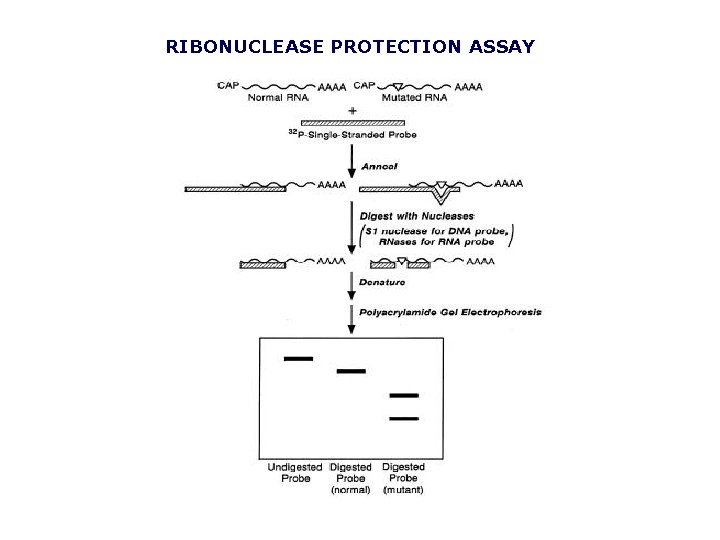
RIBONUCLEASE PROTECTION ASSAY

NUCLEASE PROTECTION ASSAY In this example, an m. RNA containing a point mutation (indicated by the inverted triangle in the m. RNA on the right) is distinguished from its normal, non-mutated counterpart (m. RNA on the left). The m. RNA is mixed with a single-stranded 32 Plabeled DNA or RNA probe that (1) has sequences perfectly complementary to the nonmutated region of interest in the m. RNA, and (2) extends for some length beyond the m. RNA. The mixture is heated then cooled to allow the probe to anneal to its complementary sequences in the m. RNA. The annealed mixture is then treated with single-strand specific nucleases (S 1 nuclease for a DNA probe, or RNAses for an RNA probe). This results in digestion of the probe at all singlestranded areas: the extension beyond the m. RNA sequences, and the single basepair mismatch overlying the mutation (right). The radioactive digestion products are then separated by electrophoresis through a urea-containing polyacrylamide gel. The probe that annealed to normal, nonmutated m. RNA is smaller than the undigested probe (by the length of the extended region not complementary to the m. RNA) and will therefore migrate farther than undigested probe. The probe that annealed to the mutated m. RNA will have been digested into two fragments whose summed length will equal that of the digested probe that annealed to nonmutated m. RNA.


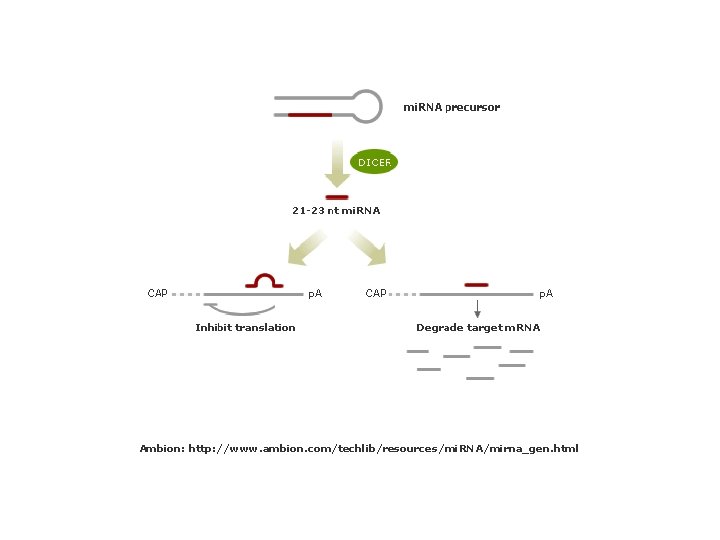
Ambion: http: //www. ambion. com/techlib/resources/mi. RNA/mirna_gen. html

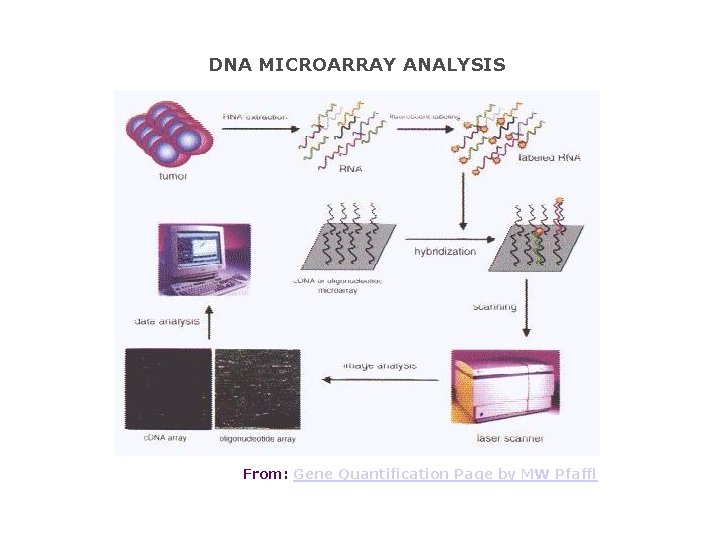
DNA MICROARRAY ANALYSIS From: Gene Quantification Page by MW Pfaffl

DNA MICROARRAY ANALYSIS RNA extracted from a tumour is end-labelled with a fluorescent marker, then allowed to hybridise to a chip consisting of c. DNAs or oligonucleotides. The precise location of RNA hybridisation to the chip can be determined using a laser scanner. Since the position of each unique c. DNA or oligonucleotide is known, the presence of a cognate RNA for any given unique sequence can be determined.


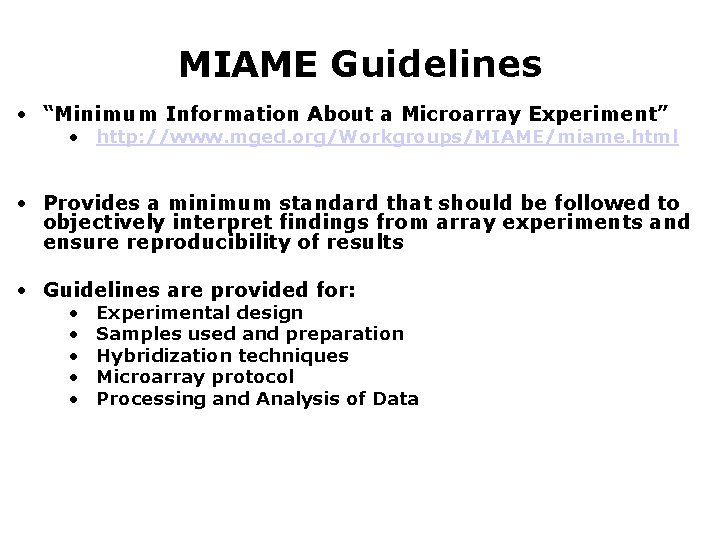
MIAME Guidelines • “Minimum Information About a Microarray Experiment” • http: //www. mged. org/Workgroups/MIAME/miame. html • Provides a minimum standard that should be followed to objectively interpret findings from array experiments and ensure reproducibility of results • Guidelines are provided for: • • • Experimental design Samples used and preparation Hybridization techniques Microarray protocol Processing and Analysis of Data

Davidson University (Microarray Animation) http: //www. bio. davidson. edu/courses/genomics/chip. html Imagecyte (Microarray Animation) http: //www. imagecyte. com/array 2. html Microarray Data Analysis (Microarray Bibliography) http: //www. nslij-genetics. org/microarray

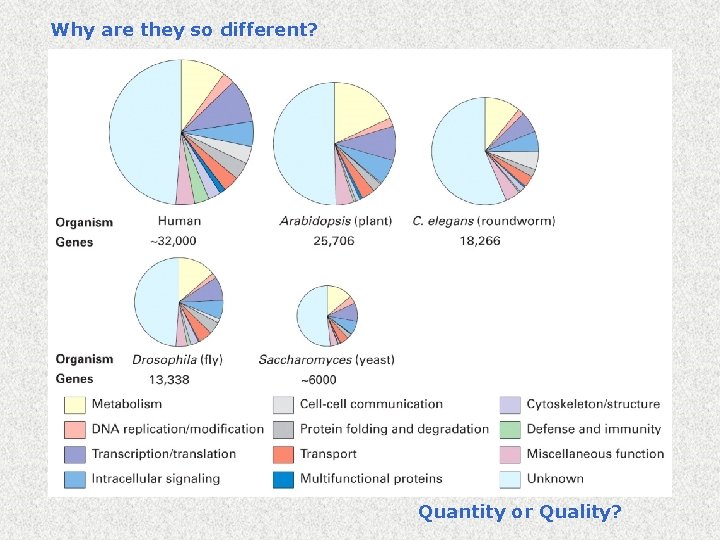
Why are they so different? Quantity or Quality?

(www)
