Applications of GO Goals of Gene Ontology Project











- Slides: 29

Applications of GO

Goals of Gene Ontology Project

Goals of Gene Ontology Project 1. Create controlled vocabularies – terms and definitions

Goals of Gene Ontology Project 1. Create controlled vocabularies – terms and definitions 2. Produce annotations to terms – gene product -> GO terms

Goals of Gene Ontology Project 1. Create controlled vocabularies – terms and definitions 2. Produce annotations to terms – gene product -> GO terms 3. Produce GO tools – browsing, searching and editing

Goals of Gene Ontology Project 1. Create controlled vocabularies – terms and definitions 2. Produce annotations to terms – gene product -> GO terms 3. Produce GO tools – browsing, searching and editing • Make everything publicly available

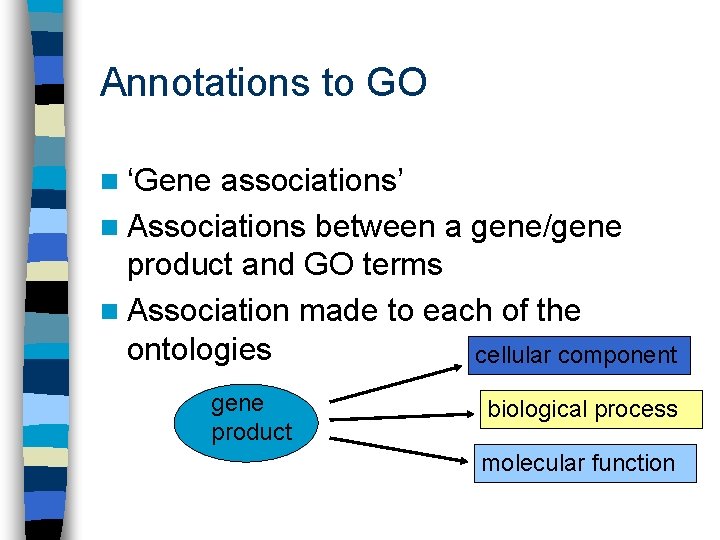
Annotations to GO n ‘Gene associations’ n Associations between a gene/gene product and GO terms n Association made to each of the ontologies cellular component gene product biological process molecular function

Annotations to GO n Three key parts: – gene name/id

Annotations to GO n Three key parts: – gene name/id – GO term

Annotations to GO n Three key parts: – gene name/id – GO term(s) – evidence for association

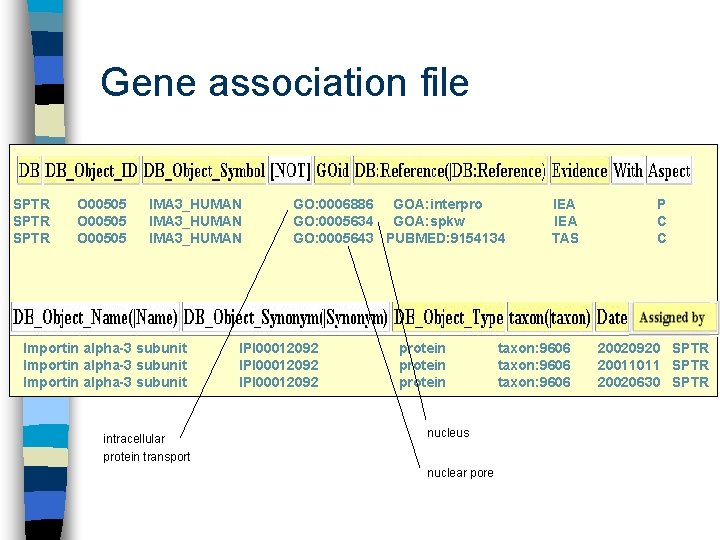
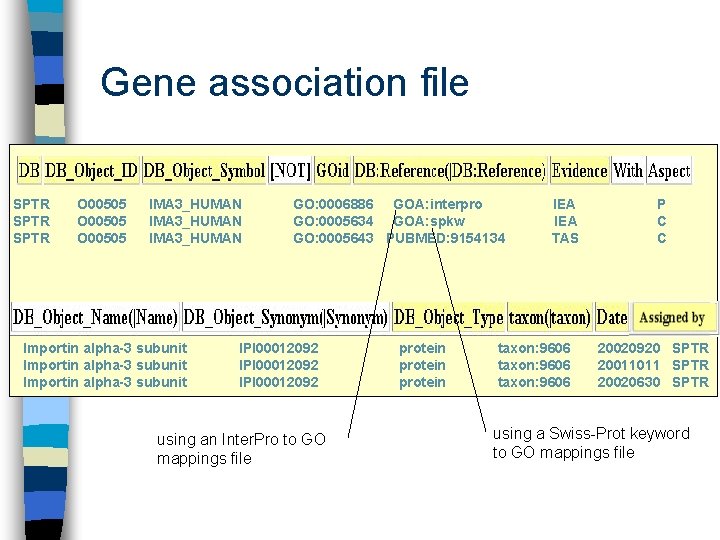
Gene association file SPTR O 00505 IMA 3_HUMAN Importin alpha-3 subunit intracellular protein transport GO: 0006886 GOA: interpro GO: 0005634 GOA: spkw GO: 0005643 PUBMED: 9154134 IPI 00012092 protein nucleus nuclear pore IEA TAS taxon: 9606 P C C 20020920 SPTR 20011011 SPTR 20020630 SPTR

Types of GO annotation: Electronic Annotation Manual Annotation

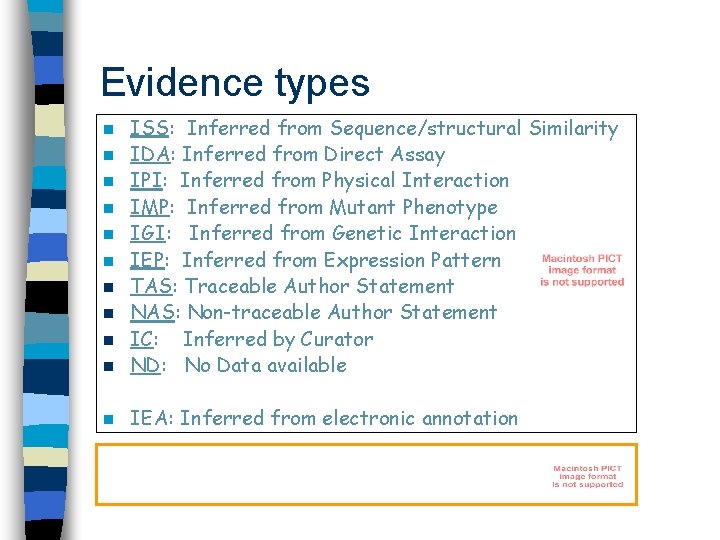
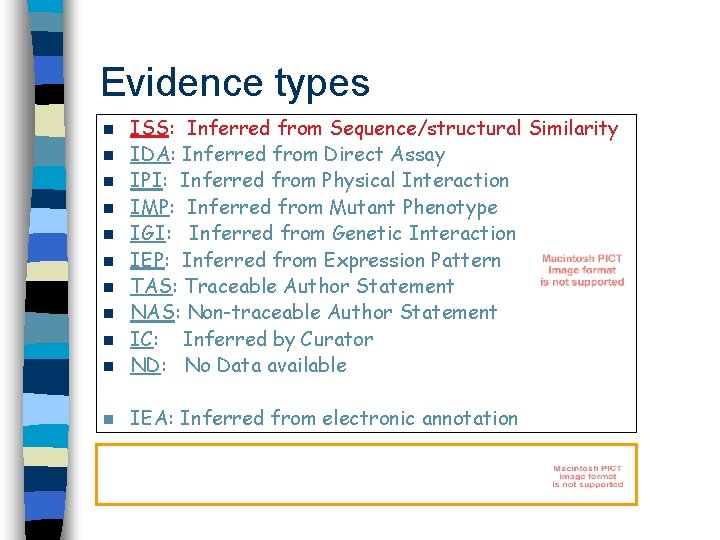
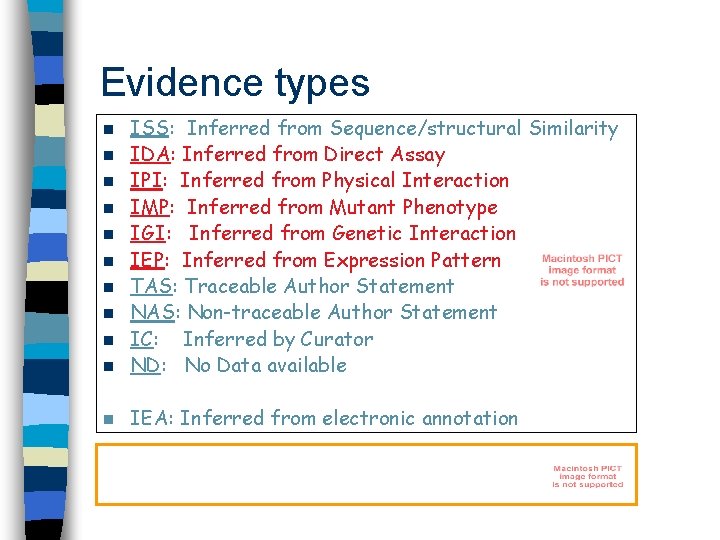
Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

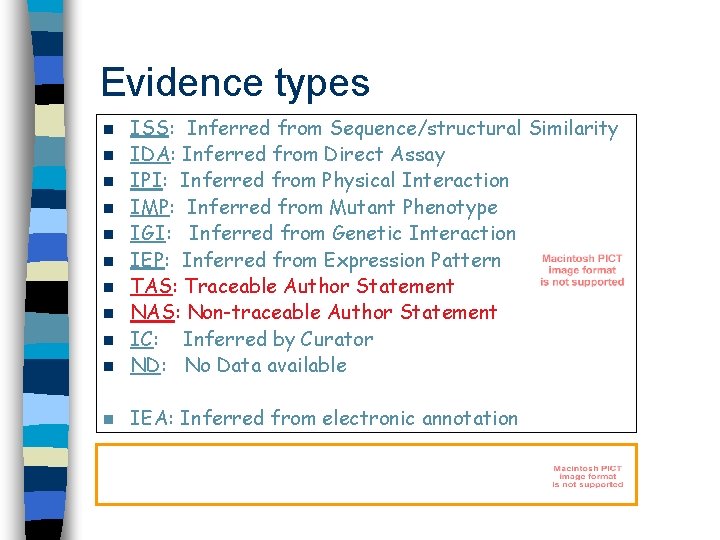
Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

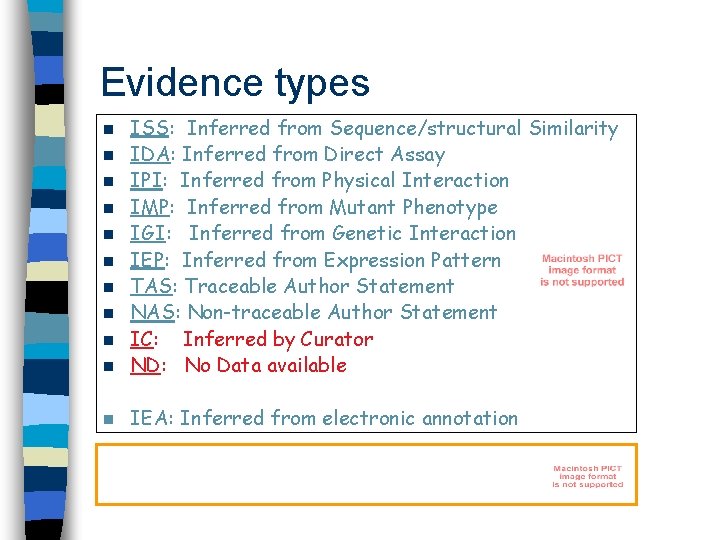
Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

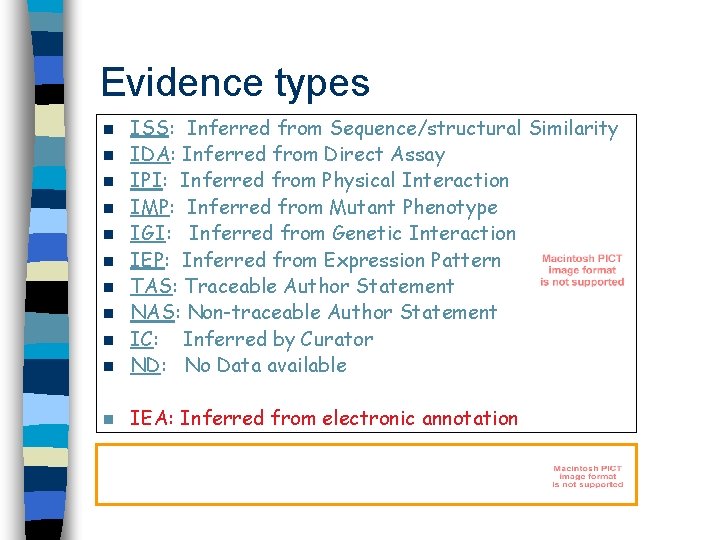
Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

Evidence types n ISS: Inferred from Sequence/structural Similarity IDA: Inferred from Direct Assay IPI: Inferred from Physical Interaction IMP: Inferred from Mutant Phenotype IGI: Inferred from Genetic Interaction IEP: Inferred from Expression Pattern TAS: Traceable Author Statement NAS: Non-traceable Author Statement IC: Inferred by Curator ND: No Data available n IEA: Inferred from electronic annotation n n n n

Inferred by Electronic Annotation n Annotation derived without human validation – mappings file e. g. interpro 2 go, ec 2 go. – Blast search ‘hits’ n Lower ‘quality’ than experimental codes

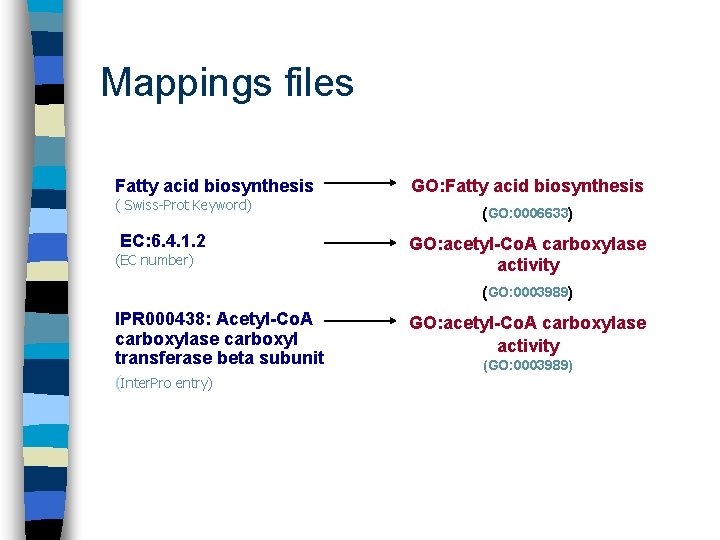
Mappings files Fatty acid biosynthesis ( Swiss-Prot Keyword) EC: 6. 4. 1. 2 (EC number) GO: Fatty acid biosynthesis (GO: 0006633) GO: acetyl-Co. A carboxylase activity (GO: 0003989) IPR 000438: Acetyl-Co. A carboxylase carboxyl transferase beta subunit (Inter. Pro entry) GO: acetyl-Co. A carboxylase activity (GO: 0003989)

Gene association file SPTR O 00505 IMA 3_HUMAN Importin alpha-3 subunit GO: 0006886 GOA: interpro GO: 0005634 GOA: spkw GO: 0005643 PUBMED: 9154134 IPI 00012092 using an Inter. Pro to GO mappings file protein IEA TAS taxon: 9606 P C C 20020920 SPTR 20011011 SPTR 20020630 SPTR using a Swiss-Prot keyword to GO mappings file

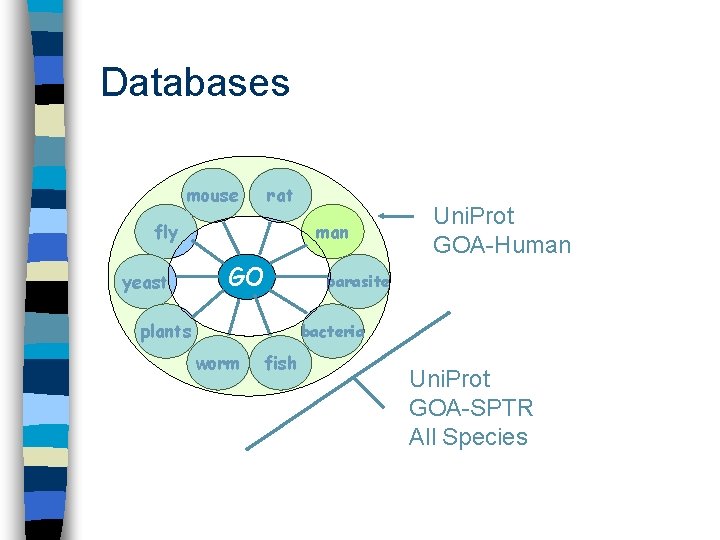
Submitting gene associations n Many model organism databases – Drosophila, mouse, Saccharomyces, rat, zebrafish, prokaryotes, Arabidopsis, slime mould, C. elegans, rice, parasites, viruses n Swiss-Prot (Uni. Prot) – Associations for >8000 species including human

Databases mouse rat fly yeast man GO plants Uni. Prot GOA-Human parasite bacteria worm fish Uni. Prot GOA-SPTR All Species

Finding GO terms In this study, we report the isolation and molecular characterization of the B. napus PERK 1 c. DNA, that is predicted to encode a novel receptor-like kinase. We have shown that like other plant RLKs, the kinase domain of PERK 1 has serine/threonine kinase activity, In addition, the location of a PERK 1 -GTP fusion protein to the plasma membrane supports the prediction that PERK 1 is an integral membrane protein…these kinases have been implicated in early stages of wound response…

GO slims n Restricted view of the ontologies n Give broad view of gene function n Can be organism-specific or generic – plant – mammal – microbe

GO slims

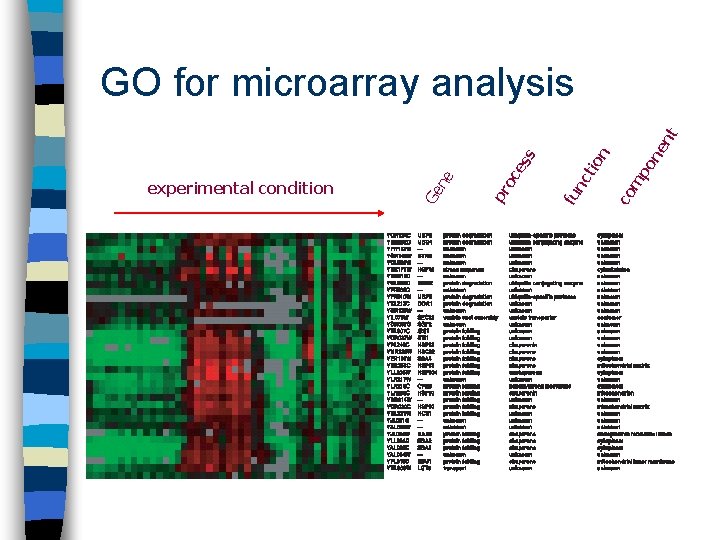
GO for microarray analysis n Annotations give ‘function’ label to genes n Ask meaningful questions of microarray data e. g. – genes involved in the same process, same/different expression patterns?

ne po m co tio n nc fu oc pr Ge experimental condition ne es s nt GO for microarray analysis

The tutorial n Part I – Navigating GO and its annotations using n Part II – Analysing microarray data using GO with
 Gene ontology project
Gene ontology project Gene ontology project
Gene ontology project Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Strategic goals tactical goals operational goals
Strategic goals tactical goals operational goals Dicty base
Dicty base Gene ontology
Gene ontology Gene by gene test results
Gene by gene test results Chapter 17 gene expression from gene to protein
Chapter 17 gene expression from gene to protein General goals and specific goals
General goals and specific goals Motivation in consumer behaviour
Motivation in consumer behaviour What is an ontologist
What is an ontologist Suggested upper merged ontology
Suggested upper merged ontology How to identify epistemology in research
How to identify epistemology in research Pizza ontology
Pizza ontology Ontology epistemology axiology
Ontology epistemology axiology Constructivist epistemology definition
Constructivist epistemology definition Ontology versus epistemology
Ontology versus epistemology Ontology alignment
Ontology alignment Types of ontology
Types of ontology Ontology in biology
Ontology in biology Financial industry business ontology
Financial industry business ontology Barry smith ontology
Barry smith ontology Schema.org ontology
Schema.org ontology Basic formal ontology
Basic formal ontology Ontology kurssi
Ontology kurssi Fibo ontology
Fibo ontology Ontology research methods
Ontology research methods Ontology editors
Ontology editors Ontology creation
Ontology creation Metu class
Metu class