Gene Ontology Driven Classification of Gene Expression Patterns

- Slides: 17

Gene Ontology Driven Classification of Gene Expression Patterns Claudio Lottaz and Rainer Spang Computational Diagnostics Group Computational Molecular Biology Department Max Planck Institute for Molecular Genetics Modelling Untranslated Regions

Overview 03 -Oct-20 Overview • • • Introduction Gene Ontology driven classification of gene expression patterns Preliminary evaluation on leukemia Limitations and future work Conclusions Claudio Lottaz: GO driven classification of gene expression patterns 2

Introduction 03 -Oct-20 Problem Statement Classify gene expression patterns into classes with biological meaning • Typical training data for supervised learning: • Many genes • Few annotated samples • Typical difficulties: • Overfitting • Lack of intuitive rationale for classifications • Claudio Lottaz: GO driven classification of gene expression patterns 3

Introduction 03 -Oct-20 Logistic Regression • Method: • • • Generalised linear statistical model Determine weights for each input variable Signoidal function to map classifier on interval [0, 1] So far we only consider the binary case Limitations • • • Only works with few input variables Troubled by colinear variables No biological knowledge Claudio Lottaz: GO driven classification of gene expression patterns 4

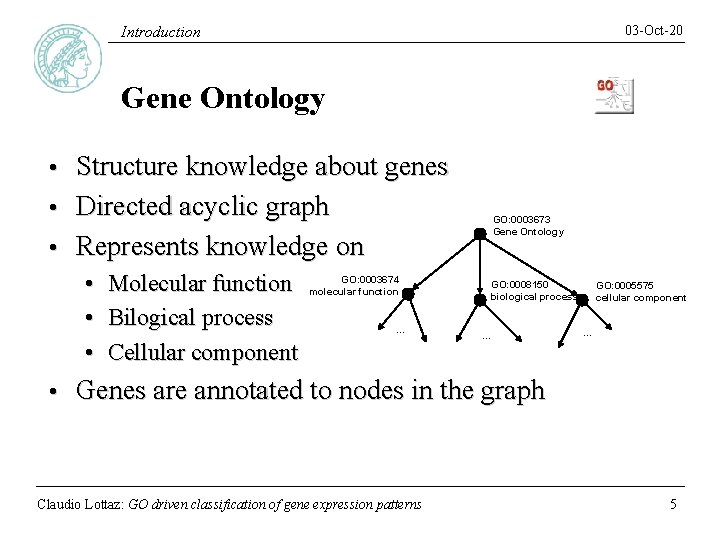
03 -Oct-20 Introduction Gene Ontology Structure knowledge about genes • Directed acyclic graph • Represents knowledge on • • • Molecular function Bilogical process Cellular component GO: 0003673 Gene Ontology GO: 0003674 molecular function . . . GO: 0008150 biological process . . . GO: 0005575 cellular component . . . Genes are annotated to nodes in the graph Claudio Lottaz: GO driven classification of gene expression patterns 5

GO driven gene expression classification 03 -Oct-20 One Classifier per GO-Node • One GO node has • • Identifier, name, description Children (other GO nodes) Probe-set annotations One logistic regression per node • Same classification task in each node • Smaller sets of input variables (directly annotated genes and direct children) Claudio Lottaz: GO driven classification of gene expression patterns 6

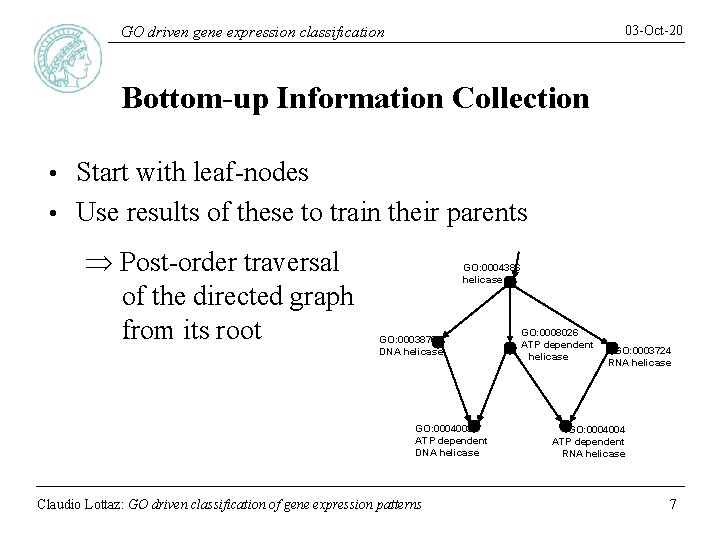
03 -Oct-20 GO driven gene expression classification Bottom-up Information Collection Start with leaf-nodes • Use results of these to train their parents • Þ Post-order traversal of the directed graph from its root GO: 0004386 helicase GO: 0003876 DNA helicase GO: 0004003 ATP dependent DNA helicase Claudio Lottaz: GO driven classification of gene expression patterns GO: 0008026 ATP dependent helicase GO: 0003724 RNA helicase GO: 0004004 ATP dependent RNA helicase 7

GO driven gene expression classification 03 -Oct-20 Explaining Classification • Weights on edges after supervised training: • Which aspects are considered important to support a given hypothesis? • Results in nodes after classification: • Which aspects favour a given hypothesis? • Whcih aspects are missing for a given hypothesis? Claudio Lottaz: GO driven classification of gene expression patterns 8

Preliminary Evaluation 03 -Oct-20 Prototype Implementation • Java-program (by Stefan Bentink) • • • Crawls through the Gene Ontology Annotates probe-sets to GO nodes Generates post-order list of GO-nodes Perl-script translates list of GO-nodes to R • R-program implements training and classification • Perl-scripts generate HTML result listings (planned) • Claudio Lottaz: GO driven classification of gene expression patterns 9

Preliminary Evaluation 03 -Oct-20 Annotating GO-Nodes (Stefan Bentink) 12625 probe-sets on Affymetrix HG-U 95 Av 2 • 7115 probe-sets are annotated • 6310 probe-sets are annotated several times, up to 23 • 2979 nodes have probe-set annotations below them • 50 nodes have more than 100, up to 965 annotations • 33 nodes have more than 10, up to 31 children • Claudio Lottaz: GO driven classification of gene expression patterns 10

Preliminary Evaluation 03 -Oct-20 Preparing Leukemia Expression Data (Stefanie Scheid) • Study on acute lymphoblastic leukemia (ALL) • • 327 patients 12625 genes (Affymetrix HG-U 95 Av 2) Various genetic subtypes of ALLs clinically confirmed 269 patients with follow-up on relapse Gene expression values generated by MAS 4. 0 • Variance stabilisation and normalisation • Attempt for relapse prediction: subtract mean per group • Claudio Lottaz: GO driven classification of gene expression patterns 11

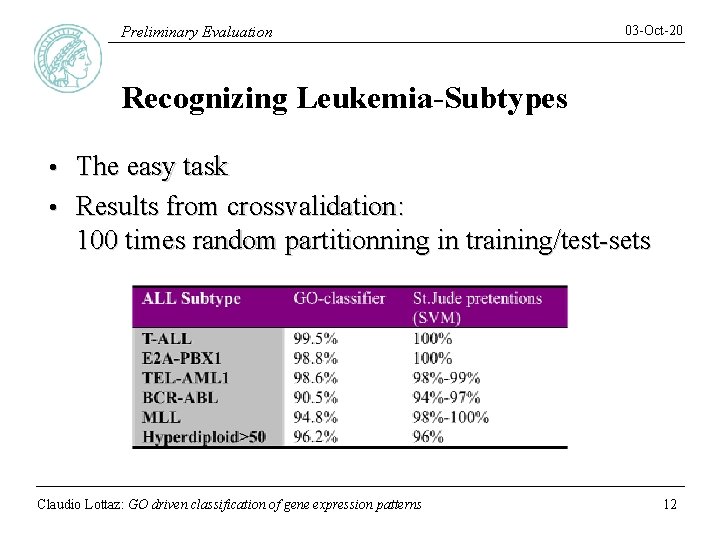
Preliminary Evaluation 03 -Oct-20 Recognizing Leukemia-Subtypes The easy task • Results from crossvalidation: 100 times random partitionning in training/test-sets • Claudio Lottaz: GO driven classification of gene expression patterns 12

Preliminary Evaluation 03 -Oct-20 Recognizing Relapse Cases • • The tough task St. Jude researchers did not find a corresponding signature to detect relapse Our detection rate: 70. 5% 74. 7% of the cases have no relapse 2 nd attempts: filter subtype information by groupwise subtracting mean expression values • 2 nd rate: 49. 9%… • Claudio Lottaz: GO driven classification of gene expression patterns 13

Limitation and future work 03 -Oct-20 Colinear Variables A few dozens of the 23545 weights cannot be determined due to colinear input variables • Reasons for correlation: • • • Perfect classifiers Multiply annotated input variables Current work-around: equally distribute weight across corralated variables • Sometimes logistic regression attributes different weights to correlated variables • Claudio Lottaz: GO driven classification of gene expression patterns 14

Limitations and future work 03 -Oct-20 User-friendly Interface to Explore the Results • HTML browsing • Results per node • Weights per edge • Links along edges in the DAG • Java application for tree-browsing • Collapse and expand branches • Multiple annotations occur several times Claudio Lottaz: GO driven classification of gene expression patterns 15

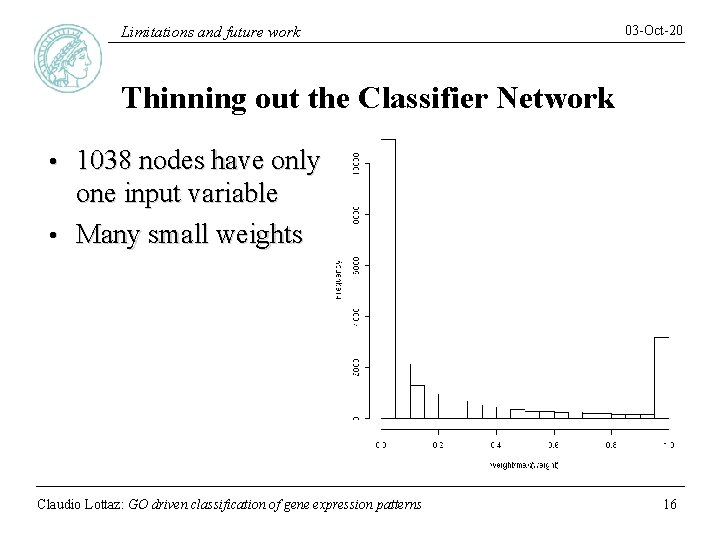
Limitations and future work 03 -Oct-20 Thinning out the Classifier Network 1038 nodes have only one input variable • Many small weights • Claudio Lottaz: GO driven classification of gene expression patterns 16

Conclusions 03 -Oct-20 Conclusions Feature selection is still an issue due to nodes with many probe-sets annotated • Classification accuracy: GO driven classification can compete with support vector machines and the like • A thinned out GO network may provide an intuitive rationale for a classification result • Fine-tuning and improvement of usability still to be developed • Claudio Lottaz: GO driven classification of gene expression patterns 17