Introduction to the Gene Ontology and GO Annotation

































- Slides: 78

Introduction to the Gene Ontology and GO Annotation Resources EBI Bioinformatics Roadshow 13 th June 2012 Rotterdam, Netherlands Duncan Legge EBI is an Outstation of the European Molecular Biology Laboratory.

OUTLINE OF TUTORIAL: PART I: Ontologies and the Gene Ontology (GO) PART II: GO Annotations How to access GO annotations How scientists use GO annotations

PART I: Gene Ontology

What does an ontology provide? 1. Consistent terminology – controlled vocabulary. 2. Relationships between terms – hierarchy.

Controlled vocabulary Q: What is a cell? A: It really depends who you ask!

Different things can be described by the same name

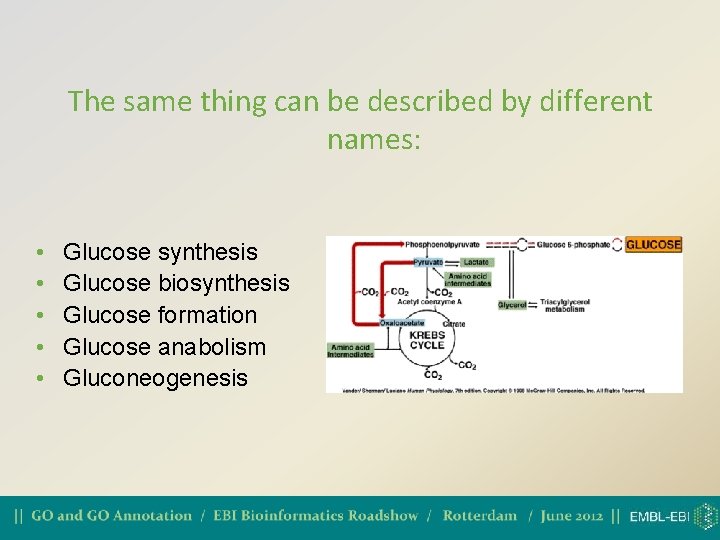
The same thing can be described by different names: • • • Glucose synthesis Glucose biosynthesis Glucose formation Glucose anabolism Gluconeogenesis

Inconsistency in naming of biological concepts • Same name for different concepts • Different names for the same concept à Comparison is difficult – in particular across species or across databases Just one reason why the Gene Ontology (GO) is is needed…

Why do we need GO? • Inconsistency in naming of biological concepts • Large datasets need to be interpreted quickly • Increasing amounts of biological data available • Increasing amounts of biological data to come

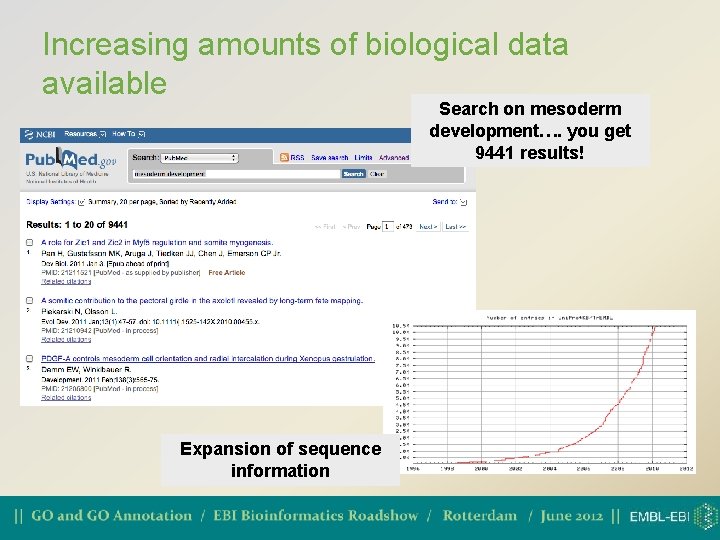
Increasing amounts of biological data available Search on mesoderm development…. you get 9441 results! Expansion of sequence information

1700 s 1606 What is an ontology? • Dictionary: • A branch of metaphysics concerned with the nature and relations of being (philosophy) • A formal representation of the knowledge by a set of concepts within a domain and the relationships between those concepts (computer science) • Barry Smith: • The science of what is, of the kinds and structures of objects, properties, events, processes and relations in every area of reality.

What is an ontology? • More usefully: • An ontology is the representation of something we know about. “Ontologies" consist of a representation of things, that are detectable or directly observable, and the relationships between those things.

What’s in an Ontology?

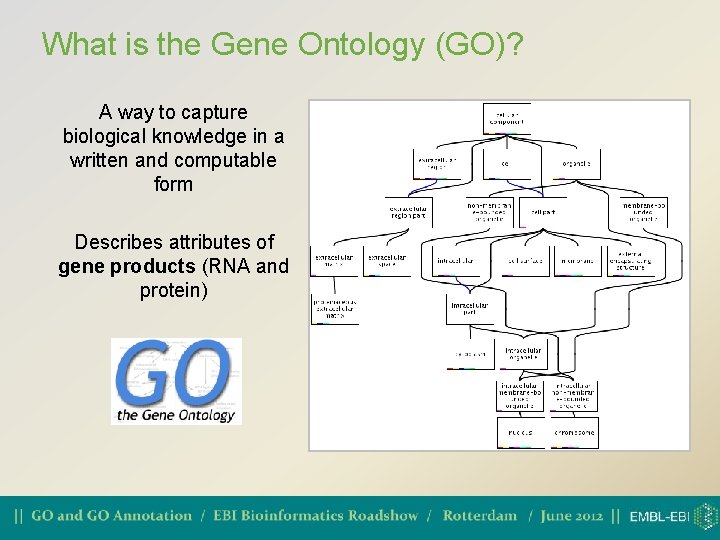
What is the Gene Ontology (GO)? A way to capture biological knowledge in a written and computable form Describes attributes of gene products (RNA and protein)

E. Coli hub http: //www. geneontology. org Reactome

The scope of GO What information might we want to capture about a gene product? • What does the gene product do? • Where does it act? • How does it act?

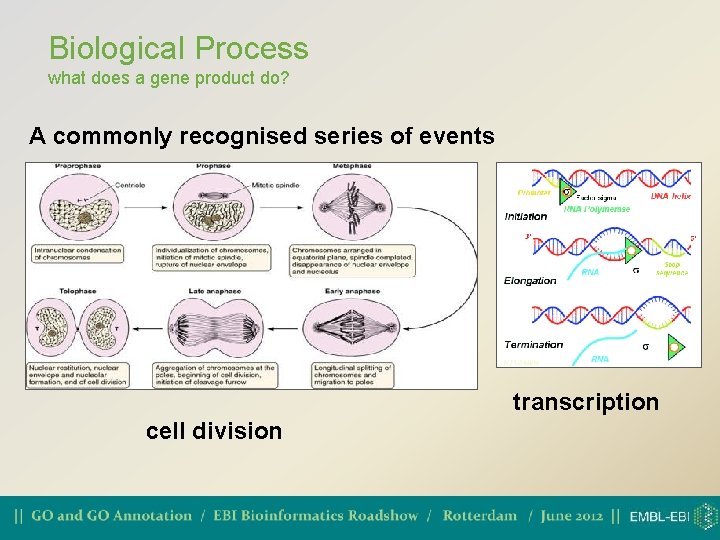
Biological Process what does a gene product do? A commonly recognised series of events transcription cell division

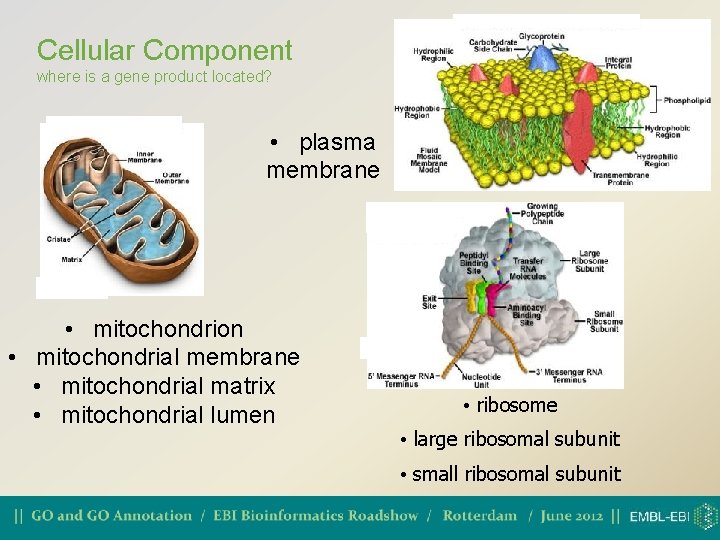
Cellular Component where is a gene product located? • plasma membrane • mitochondrion • mitochondrial membrane • mitochondrial matrix • mitochondrial lumen • ribosome • large ribosomal subunit • small ribosomal subunit

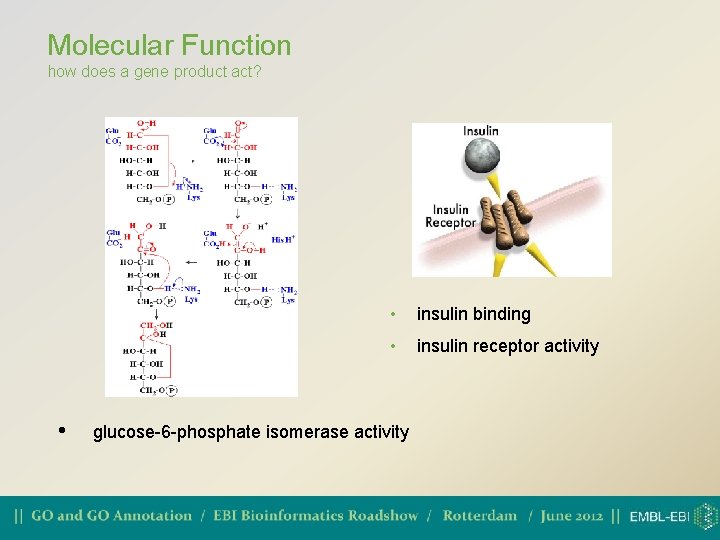
Molecular Function how does a gene product act? • • insulin binding • insulin receptor activity glucose-6 -phosphate isomerase activity

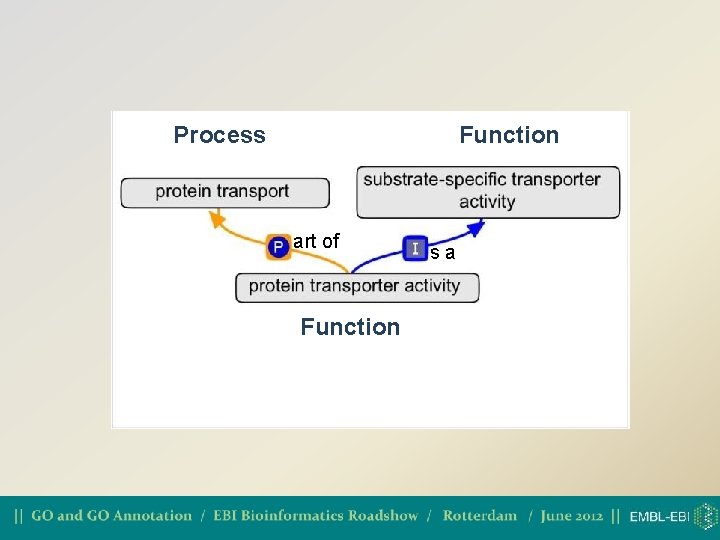
Three separate ontologies or one large one? • GO was originally three completely independent hierarchies, with no relationships between them • As of 2009, GO have started making relationships between biological process and molecular function in the live ontology

Process Function art of Function sa

• GO IS: • species independent • covers normal processes • GO is NOT: • NO pathological/disease processes • NO experimental conditions • NO evolutionary relationships • NOT a nomenclature system

Aims of the GO project • Edit the ontologies • Annotate gene products using ontology terms • Provide a public resource of data and tools

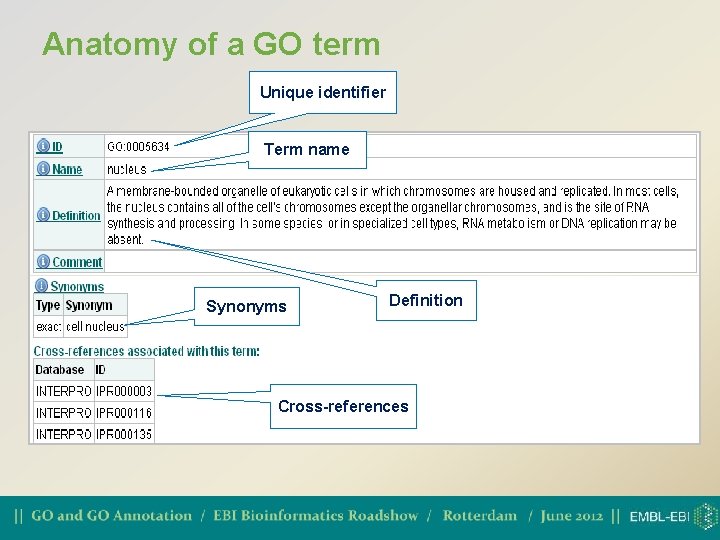
Anatomy of a GO term Unique identifier Term name Synonyms Definition Cross-references

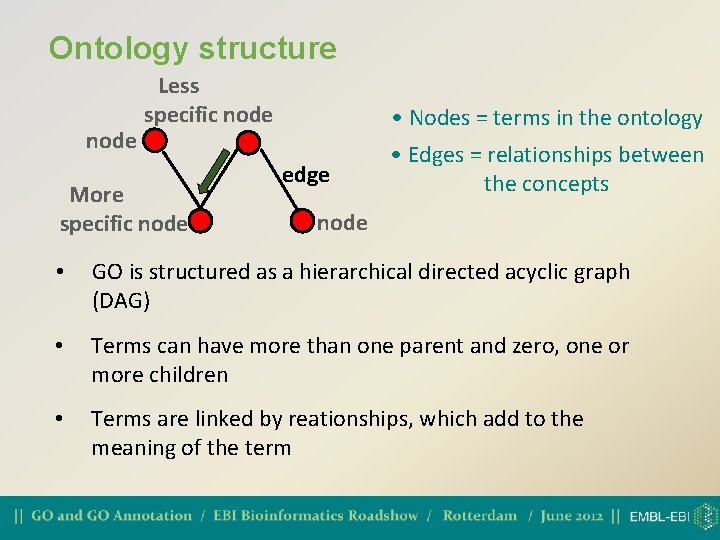
Ontology structure node Less specific node More specific node • Nodes = terms in the ontology edge • Edges = relationships between the concepts node • GO is structured as a hierarchical directed acyclic graph (DAG) • Terms can have more than one parent and zero, one or more children • Terms are linked by reationships, which add to the meaning of the term

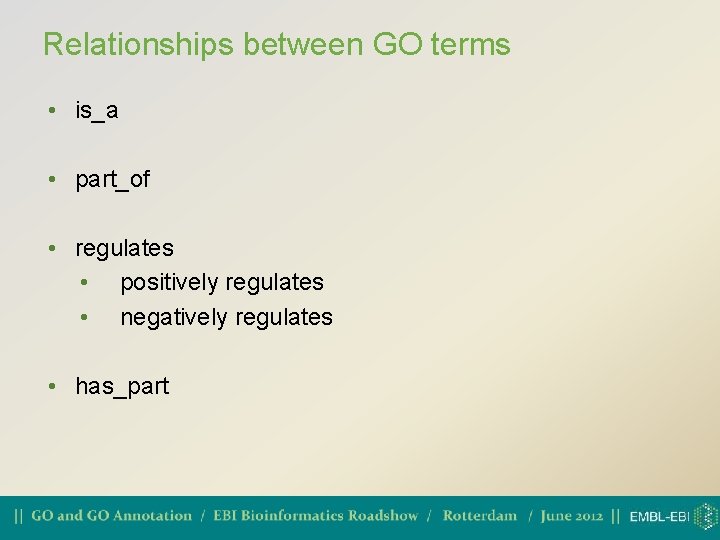
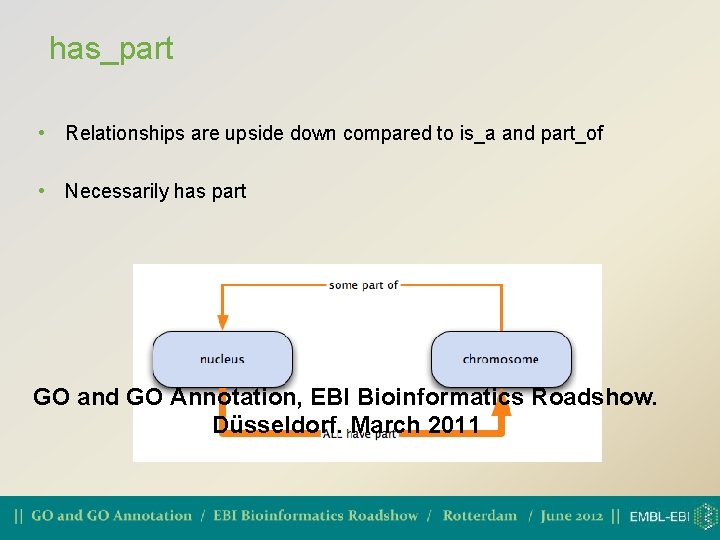
Relationships between GO terms • is_a • part_of • regulates • positively regulates • negatively regulates • has_part

is_a • If A is a B, then A is a subtype of B • mitotic cell cycle is a cell cycle • lyase activity is a catalytic activity. • Transitive relationship: can infer up the graph

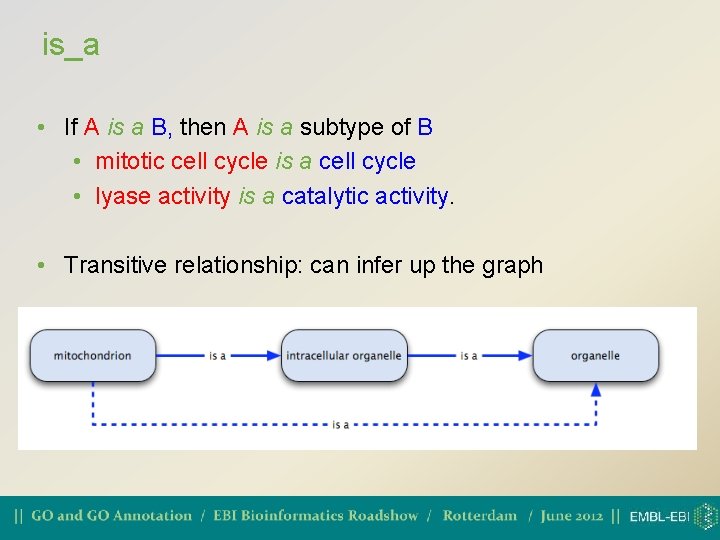
part_of • Necessarily part of • Wherever B exists, it is as part of A. But not all B is part of A. A • Transitive relationship (can infer up the graph) B

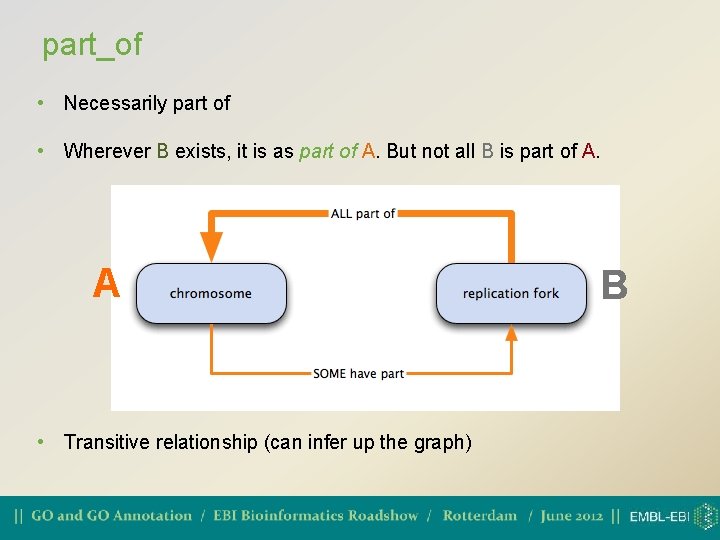
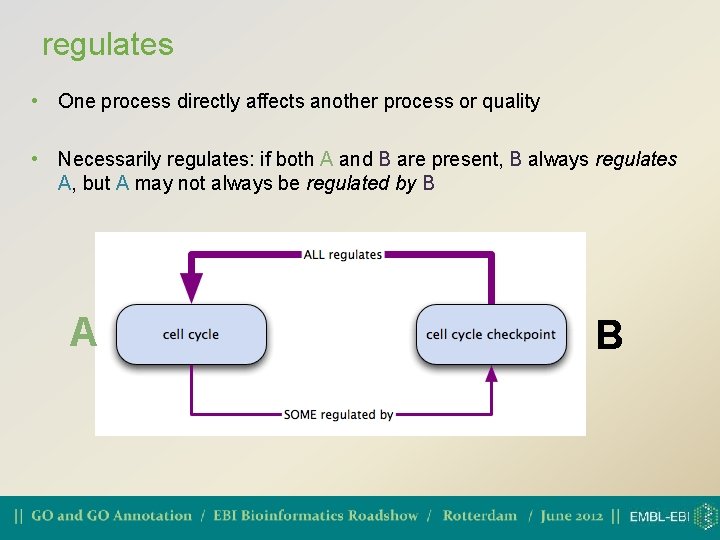
regulates • One process directly affects another process or quality • Necessarily regulates: if both A and B are present, B always regulates A, but A may not always be regulated by B A B

has_part • Relationships are upside down compared to is_a and part_of • Necessarily has part GO and GO Annotation, EBI Bioinformatics Roadshow. Düsseldorf. March 2011

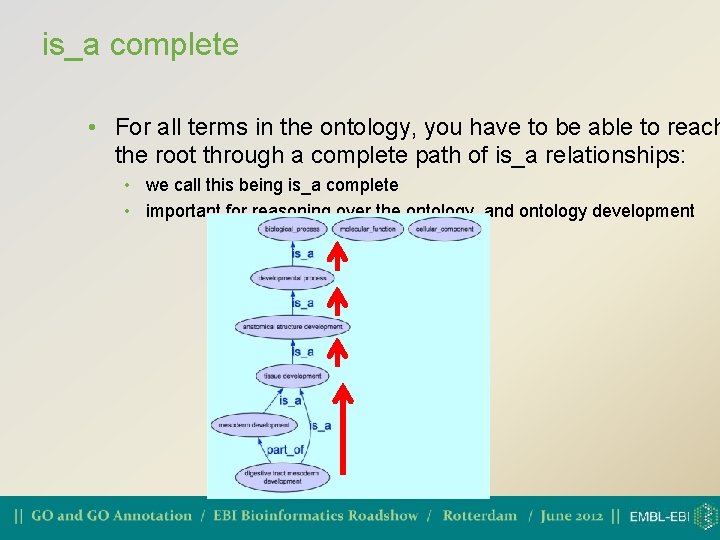
is_a complete • For all terms in the ontology, you have to be able to reach the root through a complete path of is_a relationships: • we call this being is_a complete • important for reasoning over the ontology, and ontology development

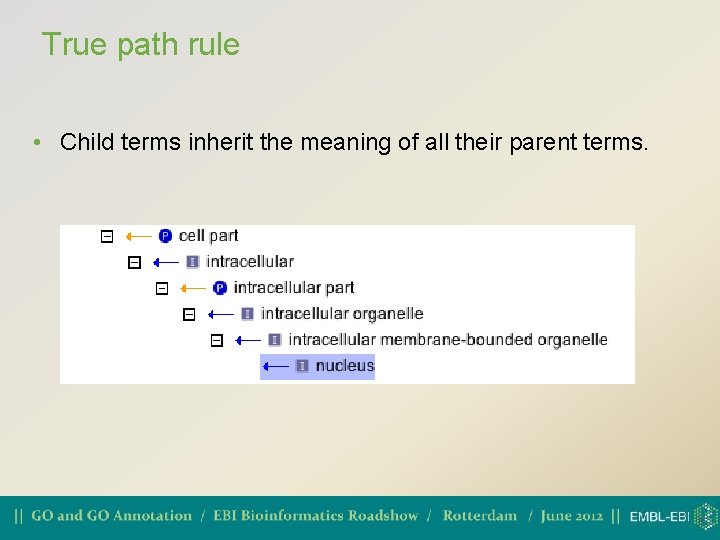
True path rule • Child terms inherit the meaning of all their parent terms.

How is GO maintained? • GO editors and annotators work with experts to remodel specific areas of the ontology • Signaling • Kidney development • Transcription • Pathogenesis • Cell cycle • Deal with requests from the community • database curators, researchers, software developers • Some simple requests can be dealt with automatically • GO Consortium meetings for large changes • Mailing lists, conference calls, content workshops

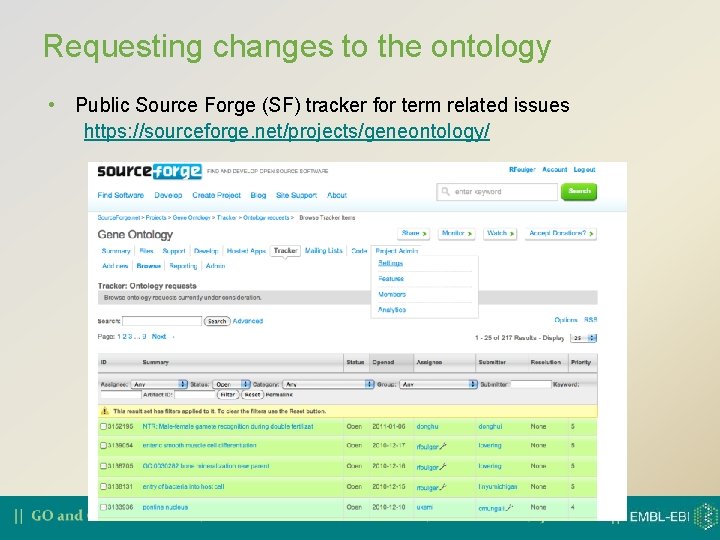
Requesting changes to the ontology • Public Source Forge (SF) tracker for term related issues https: //sourceforge. net/projects/geneontology/

Why modify the GO? • GO reflects current knowledge of biology • Information from new organisms can make existing terms and arrangements incorrect • Not everything perfect from the outset • Improving definitions • Adding in synonyms and extra relationships

Searching for GO terms http: //www. ebi. ac. uk/Quick. GO/ http: //amigo. geneontology. org … there are more browsers available on the GO Tools page: http: //www. geneontology. org/GO. tools. browsers. shtml The latest OBO Gene Ontology file can be downloaded from: http: //www. geneontology. org/ontology/gene_ontology. obo

Exercise Browsing the Gene Ontology using Quick. GO • Exercise 1 15 mins

PART II: GO Annotation

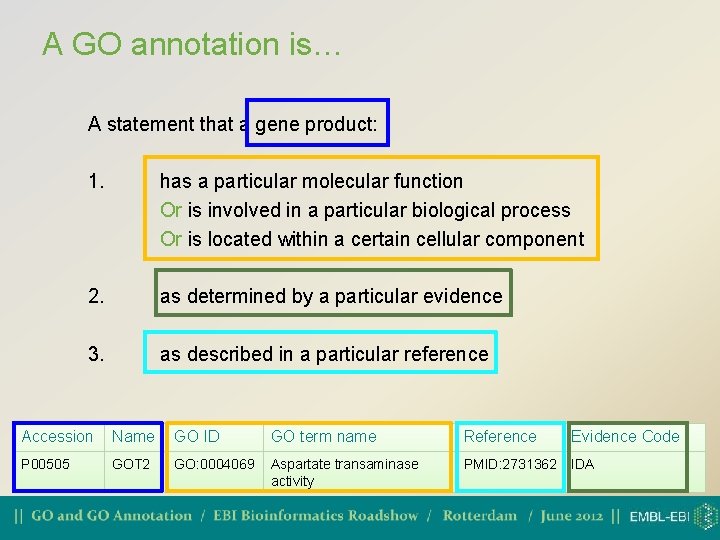
A GO annotation is… A statement that a gene product: 1. has a particular molecular function Or is involved in a particular biological process Or is located within a certain cellular component 2. as determined by a particular evidence 3. as described in a particular reference Accession Name GO ID GO term name Reference Evidence Code P 00505 GOT 2 GO: 0004069 Aspartate transaminase activity PMID: 2731362 IDA

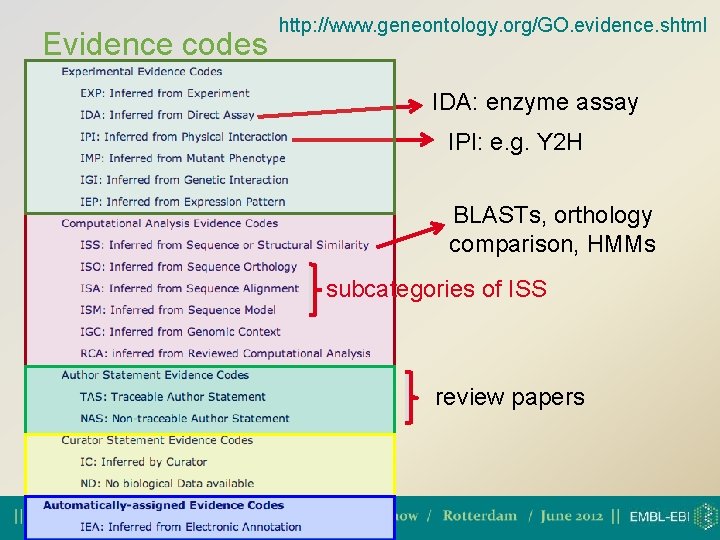
Evidence codes http: //www. geneontology. org/GO. evidence. shtml IDA: enzyme assay IPI: e. g. Y 2 H BLASTs, orthology comparison, HMMs subcategories of ISS review papers

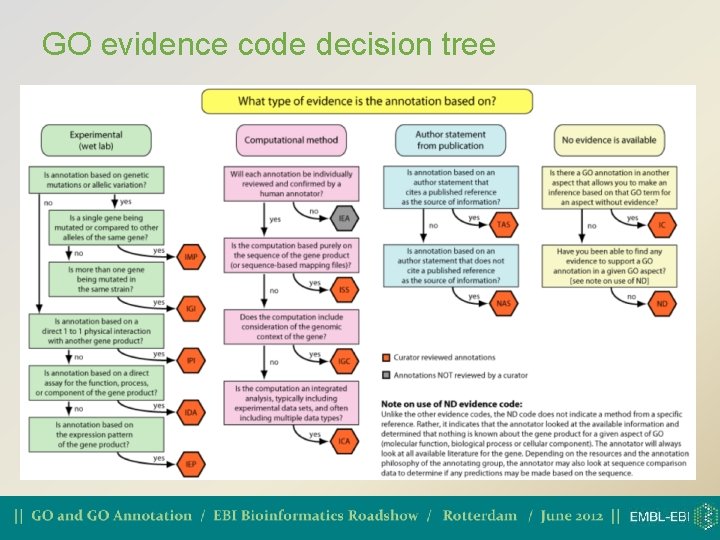
GO evidence code decision tree

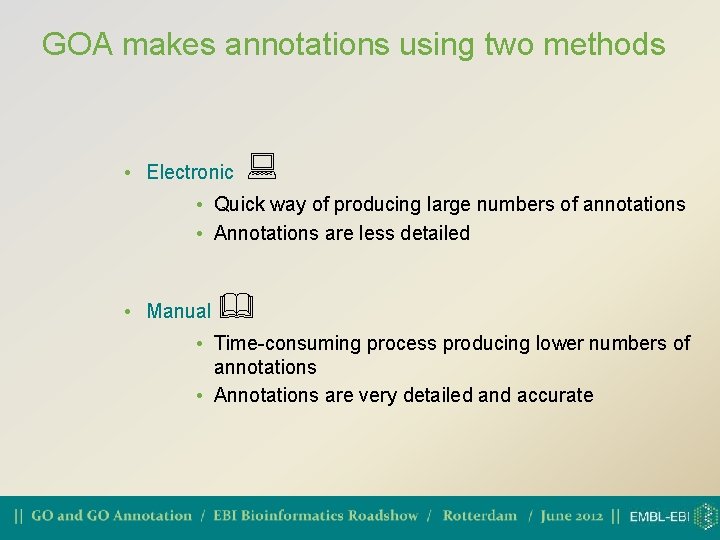
GOA makes annotations using two methods • Electronic • Quick way of producing large numbers of annotations • Annotations are less detailed • Manual • Time-consuming process producing lower numbers of annotations • Annotations are very detailed and accurate

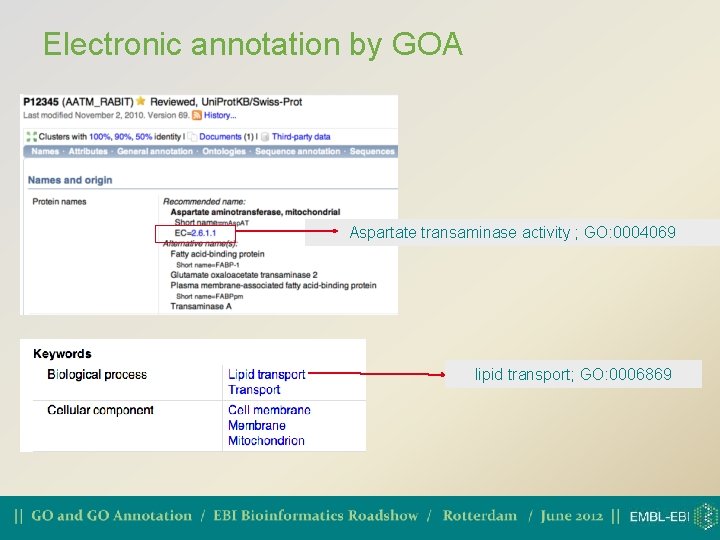
Electronic annotation by GOA • 1. Mapping of external concepts to GO terms • Inter. Pro 2 GO (protein domains) • SPKW 2 GO (Uni. Prot/Swiss-Prot keywords) • HAMAP 2 GO (Microbial protein annotation) • EC 2 GO (Enzyme Commission numbers) • SPSL 2 GO (Swiss-Prot subcellular locations)

Electronic annotation by GOA Aspartate transaminase activity ; GO: 0004069 lipid transport; GO: 0006869

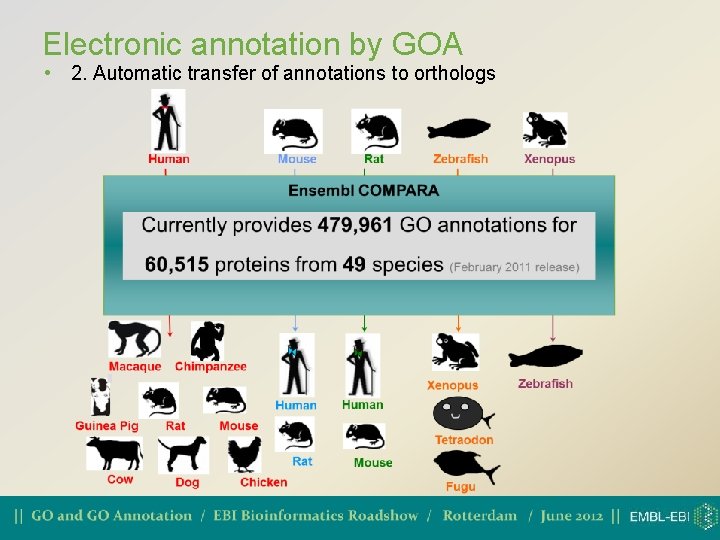
Electronic annotation by GOA • 2. Automatic transfer of annotations to orthologs

Manual annotation by GOA • High-quality, specific annotations using: • Peer-reviewed papers • A range of evidence codes to categorize the types of evidence found in a paper www. ebi. ac. uk/GOA

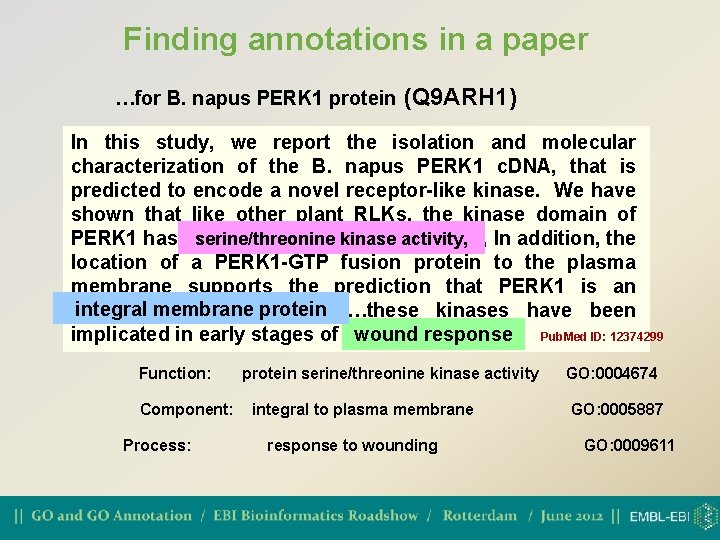
Finding annotations in a paper …for B. napus PERK 1 protein (Q 9 ARH 1) In this study, we report the isolation and molecular characterization of the B. napus PERK 1 c. DNA, that is predicted to encode a novel receptor-like kinase. We have shown that like other plant RLKs, the kinase domain of serine/threonine kinase , In addition, the PERK 1 has serine/threonine kinaseactivity, location of a PERK 1 -GTP fusion protein to the plasma membrane supports the prediction that PERK 1 is an integral membraneprotein…these kinases have been implicated in early stages of woundresponse… response Pub. Med ID: 12374299 Function: Component: Process: protein serine/threonine kinase activity integral to plasma membrane response to wounding GO: 0004674 GO: 0005887 GO: 0009611

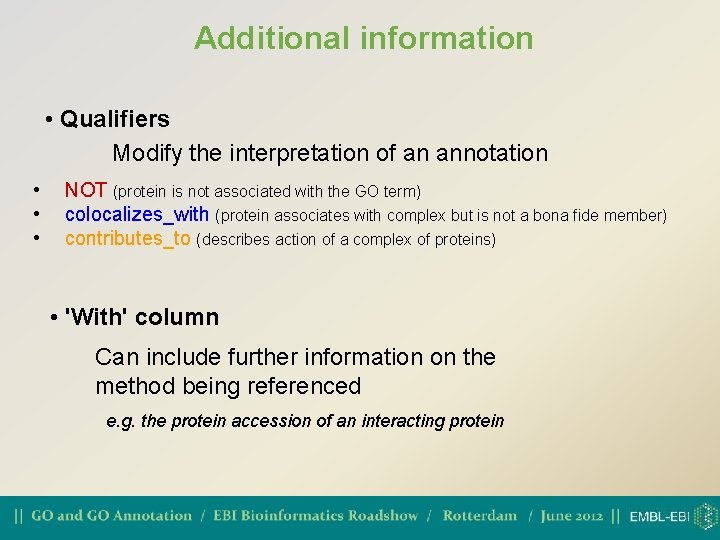
Additional information • Qualifiers Modify the interpretation of an annotation • • • NOT (protein is not associated with the GO term) colocalizes_with (protein associates with complex but is not a bona fide member) contributes_to (describes action of a complex of proteins) • 'With' column Can include further information on the method being referenced e. g. the protein accession of an interacting protein

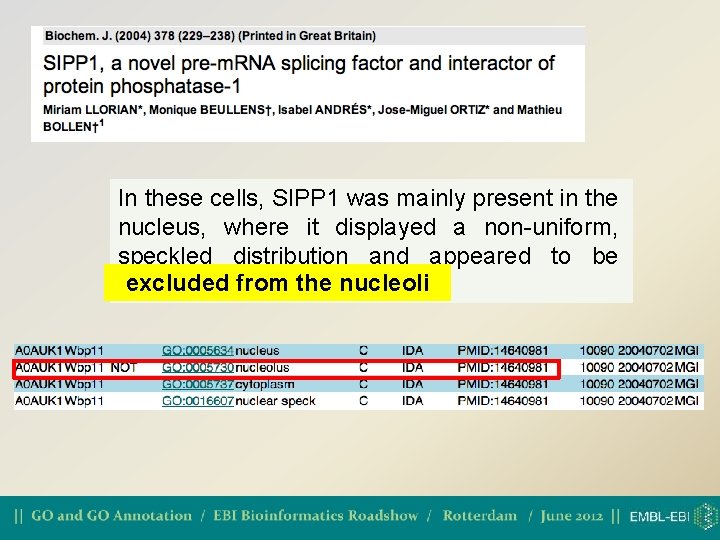
The NOT qualifier • NOT is used to make an explicit note that the gene product is not associated with the GO term • Also used to document conflicting claims in the literature • NOT can be used with ALL three gene ontologies

In these cells, SIPP 1 was mainly present in the nucleus, where it displayed a non-uniform, speckled distribution and appeared to be excludedfrom the nucleoli excluded the nucleoli.

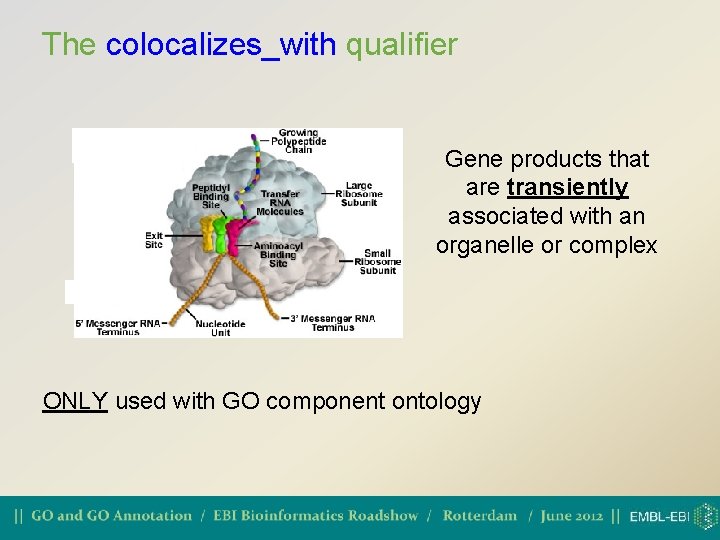
The colocalizes_with qualifier Gene products that are transiently associated with an organelle or complex ONLY used with GO component ontology

The colocalizes_with qualifier Example (from Schizosaccharomyces pombe): Clp 1 (Q 9 P 7 H 1) relocalizes from the nucleolus to the spindle and site of cell division; i. e. it is associated transiently with the contractile ring (evidence from GFP fusion).

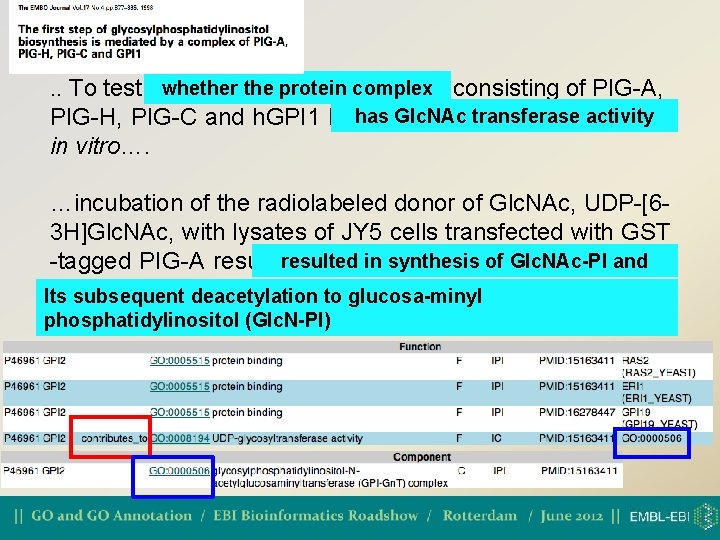
The contributes_to qualifier • Where an individual gene product that is part of a complex can be annotated to terms that describe the action (function or process) of the whole complex • contributes_to is not needed to annotate a catalytic subunit. ONLY used with GO function ontology

whether the protein complex. . To test whether complex consisting of PIG-A, Glc. NActransferase activity PIG-H, PIG-C and h. GPI 1 hashas Glc. NAc activity in vitro…. …incubation of the radiolabeled donor of Glc. NAc, UDP-[63 H]Glc. NAc, with lysates of JY 5 cells transfected with GST resulted in synthesis of Glc. NAc-PI andits -tagged PIG-A resulted in synthesis of Glc. NAc-PI and subsequent deacetylation to glucosaminyl Its subsequent deacetylation to glucosa-minyl phosphatidylinositol (Glc. N-PI)

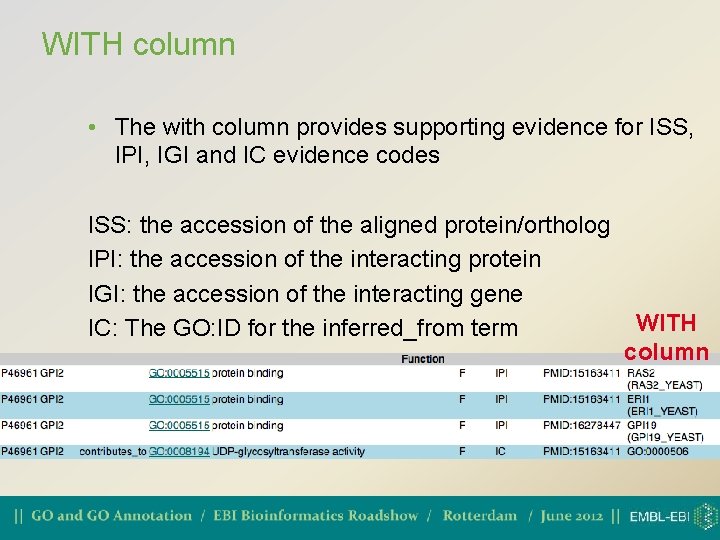
WITH column • The with column provides supporting evidence for ISS, IPI, IGI and IC evidence codes ISS: the accession of the aligned protein/ortholog IPI: the accession of the interacting protein IGI: the accession of the interacting gene IC: The GO: ID for the inferred_from term WITH column

How to access GO annotation data

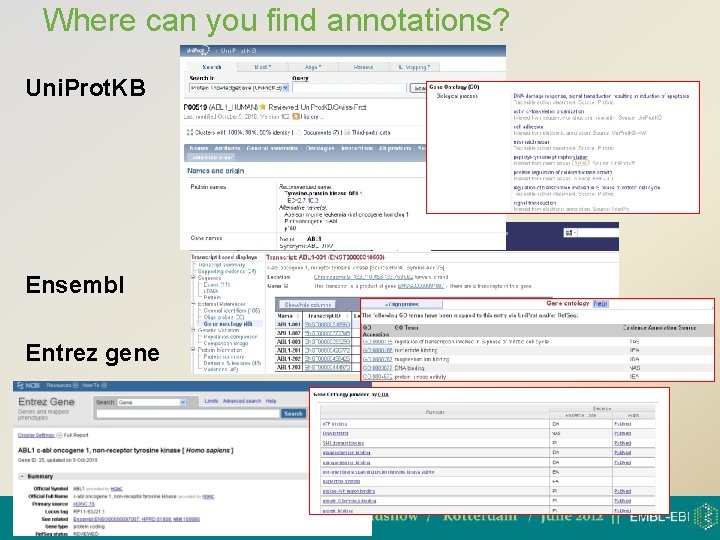
Where can you find annotations? Uni. Prot. KB Ensembl Entrez gene

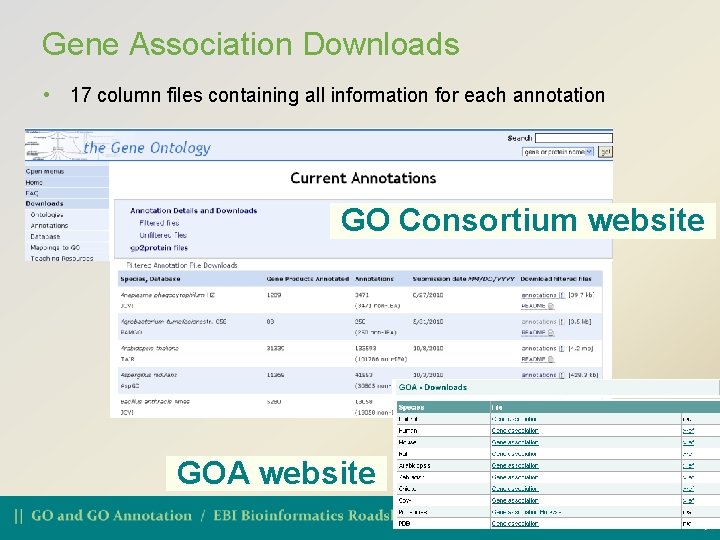
Gene Association Downloads • 17 column files containing all information for each annotation GO Consortium website GOA website

GO browsers

GO Slims

GO slims • Many GO analysis tools use GO slims to give a broad overview of the dataset • GO slims are cut-down versions of the GO and contain a subset of the terms in the whole GO • GO slims usually contain less-specialised GO terms

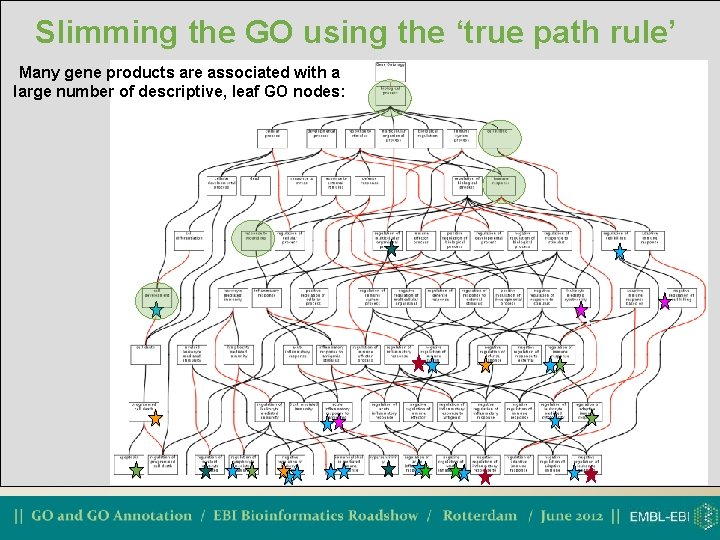
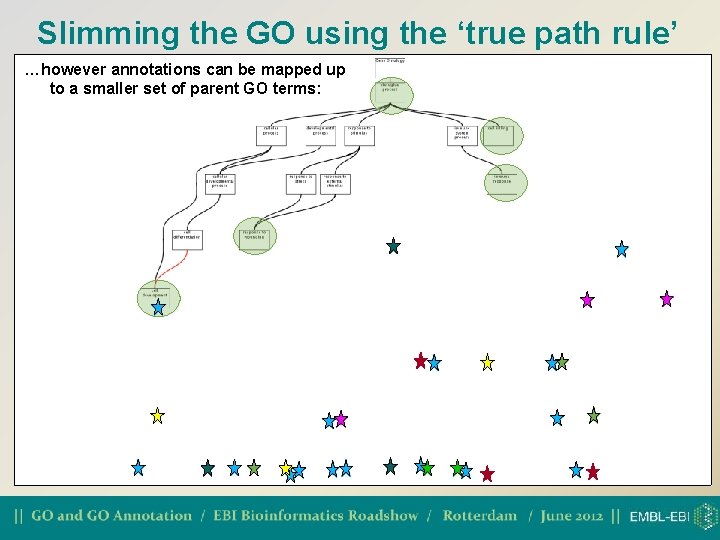
Slimming the GO using the ‘true path rule’ Many gene products are associated with a large number of descriptive, leaf GO nodes:

Slimming the GO using the ‘true path rule’ …however annotations can be mapped up to a smaller set of parent GO terms:

GO slims • Custom slims are available for download; http: //www. geneontology. org/GO. slims. shtml • Or you can make your own using; • Quick. GO • http: //www. ebi. ac. uk/Quick. GO • Ami. GO's GO slimmer • http: //amigo. geneontology. org/cgi-bin/amigo/slimmer

Just some things to be aware of…. • The GO is continually changing • New terms created ontology • Existing terms obsoleted • Re-structured annotation • New annotations being created • ALWAYS use a current version of ontology and annotations • If publishing your analyses, please report the versions/dates you use: http: //www. geneontology. org/GO. cite. shtml • Differences in representation of GO terms may be due to biological phenomenon. But also may be due to annotation-bias or experimental assays • Often better to remove the ‘NOT’ annotations before doing any large-scale analysis, as they can skew the results

How scientists use the GO, and the tools they use for analysis

Source of annotation • If you wanted to find out the role of a gene product manually, you’d have to read an awful lot of papers • But by using GO annotations, this work has already been done for you! GO: 0006915 : apoptosis

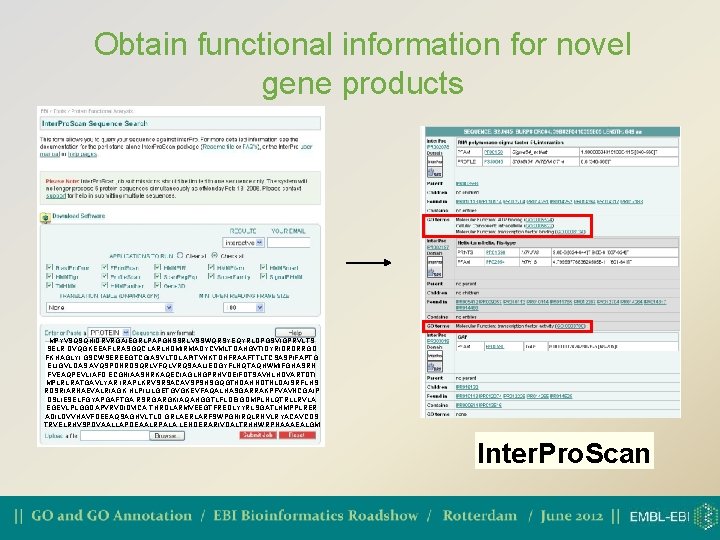
How scientists use the GO • Find out what a gene product does or which genes are involved in a certain biological process/function • Analyse high-throughput genomic or proteomic datasets • Validation of experimental techniques • Get a broad overview of a proteome • Obtain functional information for novel gene products Some examples…

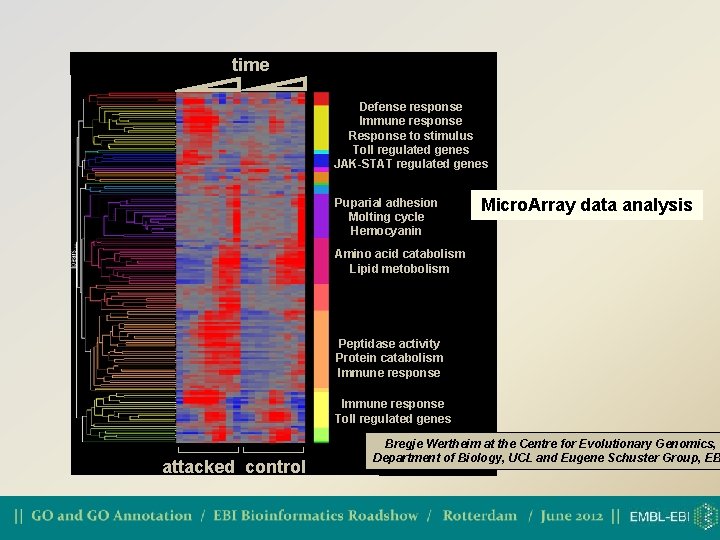
time Defense response Immune response Response to stimulus Toll regulated genes JAK-STAT regulated genes Puparial adhesion Molting cycle Hemocyanin Micro. Array data analysis Amino acid catabolism Lipid metobolism Peptidase activity Protein catabolism Immune response Toll regulated genes attacked control Bregje Wertheim at the Centre for Evolutionary Genomics, Department of Biology, UCL and Eugene Schuster Group, EB

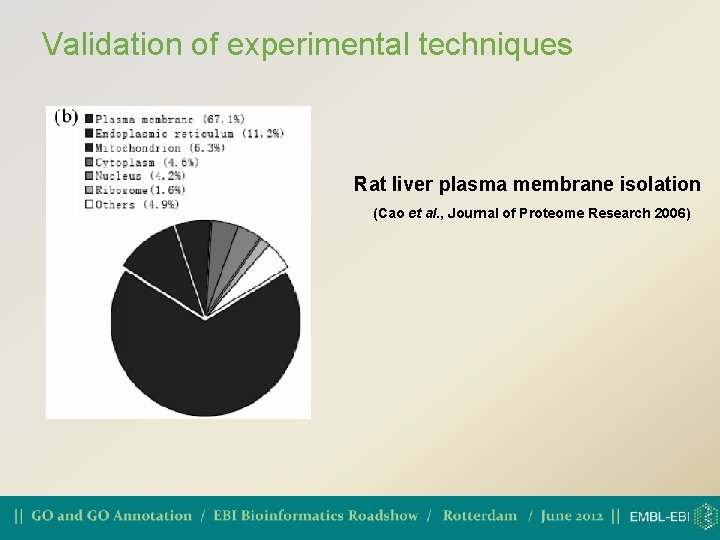
Validation of experimental techniques Rat liver plasma membrane isolation (Cao et al. , Journal of Proteome Research 2006)

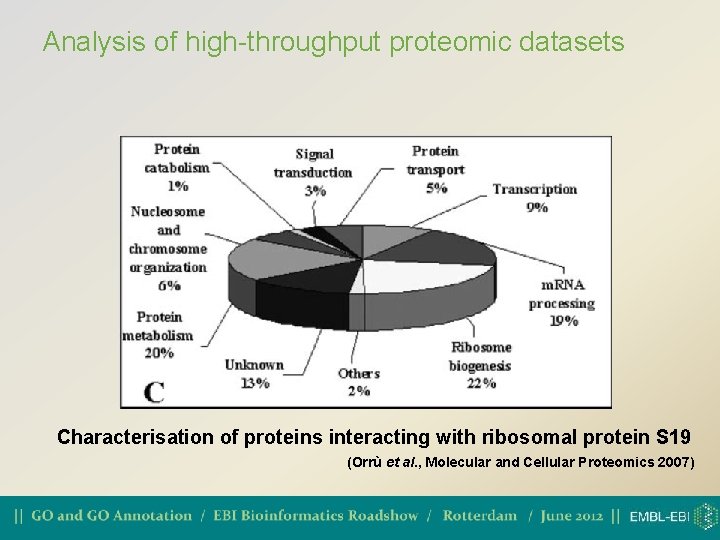
Analysis of high-throughput proteomic datasets Characterisation of proteins interacting with ribosomal protein S 19 (Orrù et al. , Molecular and Cellular Proteomics 2007)

Obtain functional information for novel gene products MPYVSQSQHIDRVRGAIEGRLPAPGNSSRLVSSWQRSYEQYRLDPGSVIGPRVLTS SELR DVQGKEEAFLRASGQCLARLHDMIRMADYCVMLTDAHGVTIDYRIDRDRRGD FKHAGLYI GSCWSEREEGTCGIASVLTDLAPITVHKTDHFRAAFTTLTCSASPIFAPTG ELIGVLDAS AVQSPDNRDSQRLVFQLVRQSAALIEDGYFLNQTAQHWMIFGHASRN FVEAQPEVLIAFD ECGNIAASNRKAQECIAGLNGPRHVDEIFDTSAVHLHDVARTDTI MPLRLRATGAVLYAR IRAPLKRVSRSACAVSPSHSGQGTHDAHNDTNLDAISRFLHS RDSRIARNAEVALRIAGK HLPILILGETGVGKEVFAQALHASGARRAKPFVAVNCGAIP DSLIESELFGYAPGAFTGA RSRGARGKIAQAHGGTLFLDEIGDMPLNLQTRLLRVLA EGEVLPLGGDAPVRVDIDVICA THRDLARMVEEGTFREDLYYRLSGATLHMPPLRER ADILDVVHAVFDEEAQSAGHVLTLD GRLAERLARFSWPGNIRQLRNVLRYACAVCDS TRVELRHVSPDVAALLAPDEAALRPALA LENDERARIVDALTRHHWRPNAAAEALGM Inter. Pro. Scan

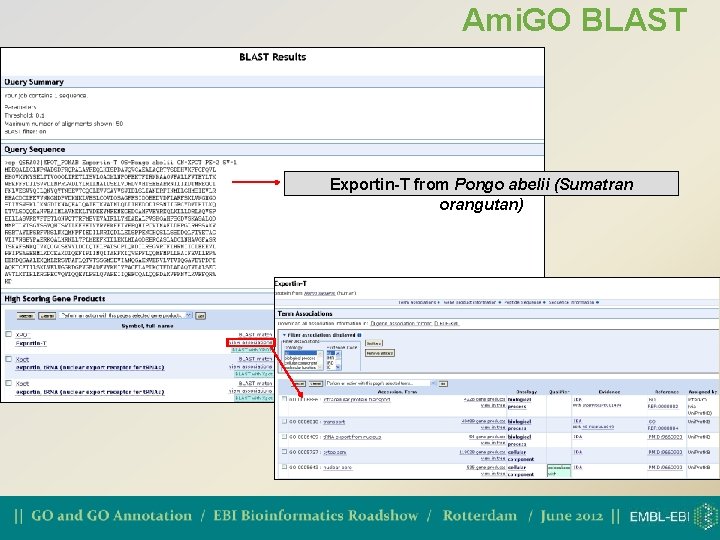
Annotating novel sequences • Can use BLAST queries to find similar sequences with GO annotation which can be transferred to the new sequence • Two tools currently available; • Ami. GO BLAST (from GO Consortium) http: //amigo. geneontology. org/cgi-bin/amigo/blast. cgi • searches the GO Consortium database • BLAST 2 GO (from Babelomics) http: //www. blast 2 go. org/ • searches the NCBI database

Ami. GO BLAST Exportin-T from Pongo abelii (Sumatran orangutan)

Numerous Third Party Tools • Many tools exist that use GO to find common biological functions from a list of genes: http: //www. geneontology. org/GO. tools. microarray. shtml

GO tools: enrichment analysis • Most of these tools work in a similar way: • input a gene list and a subset of ‘interesting’ genes • tool shows which GO categories have most interesting genes associated with them i. e. which categories are ‘enriched’ for interesting genes • tool provides a statistical measure to determine whether enrichment is significant

Exercises Searching for GO annotations in Quick. GO • Exercise 2: using GO terms • Exercise 3: using a protein ID Using Quick. GO to create a tailored set of annotations • Exercise 4: Filtering • Exercise 5: Statistics Map-up annotation using a GO slim • Exercise 6

Thanks for listening EBI is an Outstation of the European Molecular Biology Laboratory.
 Pascale gaudet
Pascale gaudet Gene ontology
Gene ontology Gene ontology project
Gene ontology project Gene ontology project
Gene ontology project Eukaryotic genome
Eukaryotic genome David annotation tool
David annotation tool Gene by gene test results
Gene by gene test results Chapter 17 from gene to protein
Chapter 17 from gene to protein Research ontology and epistemology
Research ontology and epistemology Constructivist epistemology definition
Constructivist epistemology definition Ontology epistemology axiology
Ontology epistemology axiology De novo genome assembly ppt
De novo genome assembly ppt What is an ontology
What is an ontology Suggested upper merged ontology
Suggested upper merged ontology Napolen pizza
Napolen pizza Epistemology vs ontology
Epistemology vs ontology Ontology alignment
Ontology alignment Types of ontology
Types of ontology Ontology in biology
Ontology in biology Financial industry business ontology
Financial industry business ontology Barry smith ontology
Barry smith ontology Schema .org
Schema .org Basic formal ontology
Basic formal ontology Ontology kurssi
Ontology kurssi Fibo ontology
Fibo ontology Ontology research methods
Ontology research methods Ontology editors
Ontology editors Ontology creation
Ontology creation Metu class
Metu class Pizza ontology download
Pizza ontology download Dolce ontology
Dolce ontology Vivo ontology
Vivo ontology Rea ontology
Rea ontology Provi tutorial
Provi tutorial Sims position
Sims position Ontological definition
Ontological definition Business ontology
Business ontology Twinkle helicase
Twinkle helicase Ontology
Ontology Ontology 101
Ontology 101 Ontology schema
Ontology schema Ontology alignment
Ontology alignment Football ontology
Football ontology Business model ontology
Business model ontology Close reading symbols
Close reading symbols Annotating a poem examples
Annotating a poem examples Gcse photography annotation
Gcse photography annotation Scale annotation pipeline
Scale annotation pipeline O valiant cousin worthy gentleman technique
O valiant cousin worthy gentleman technique Acquainted with the night annotation
Acquainted with the night annotation Annotation guide
Annotation guide Bacteriophage annotation
Bacteriophage annotation Acquainted with the night annotation
Acquainted with the night annotation Dull sublunary lovers love meaning
Dull sublunary lovers love meaning What is annotation
What is annotation Living space by imtiaz dharker analysis
Living space by imtiaz dharker analysis Maplex label engine
Maplex label engine Yesterday and those winter sundays comparison
Yesterday and those winter sundays comparison Annotation slows down the reader to deepen understanding
Annotation slows down the reader to deepen understanding Annotation slows down the reader to deepen understanding
Annotation slows down the reader to deepen understanding Alr index
Alr index Amazon data annotation
Amazon data annotation Nothing gold can stay paraphrase
Nothing gold can stay paraphrase Annotating reading strategies
Annotating reading strategies The two terms of comparison in the first two quatrains are
The two terms of comparison in the first two quatrains are Amazon data annotation
Amazon data annotation Landscape with the fall of icarus annotation
Landscape with the fall of icarus annotation Maxims of annotation in corpus linguistics
Maxims of annotation in corpus linguistics The santa ana winds joan didion annotation
The santa ana winds joan didion annotation Annotation toolkit
Annotation toolkit Basking shark annotated
Basking shark annotated Basking shark anatomy
Basking shark anatomy Benjamin banneker letter to thomas jefferson annotation
Benjamin banneker letter to thomas jefferson annotation The most dangerous game annotations
The most dangerous game annotations Morphological annotation
Morphological annotation Eclipse annotation processor
Eclipse annotation processor Catch annotation
Catch annotation Oh captain my captain imagery
Oh captain my captain imagery Critical annotation
Critical annotation