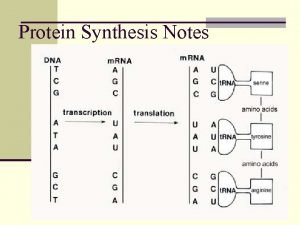
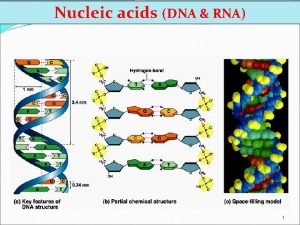
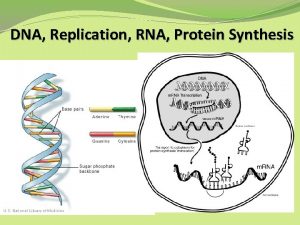
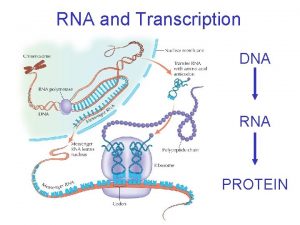
The Gene Ontology Project DNA gene m RNA




![Intersection tags / cross products [Term] id: GO: 0010064 ! embryonic shoot morphogenesis intersection_of: Intersection tags / cross products [Term] id: GO: 0010064 ! embryonic shoot morphogenesis intersection_of:](https://slidetodoc.com/presentation_image/9191d97abd02808a3d3f68481205914f/image-12.jpg)







- Slides: 26

The Gene Ontology Project

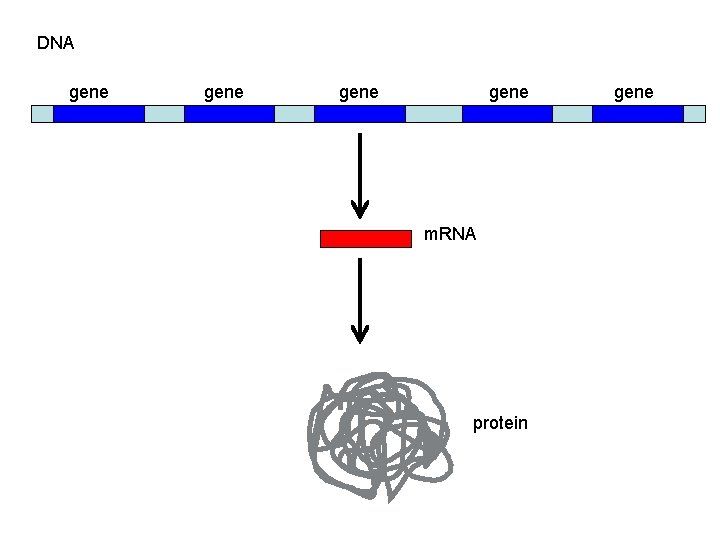
DNA gene m. RNA protein gene

http: //cms. frontiersin. org/content/10. 3389/neuro. 02/007. 2008/ html/fnmol-01 -007/images/article/image_n/fnmol-01 -007 -g 001. gif

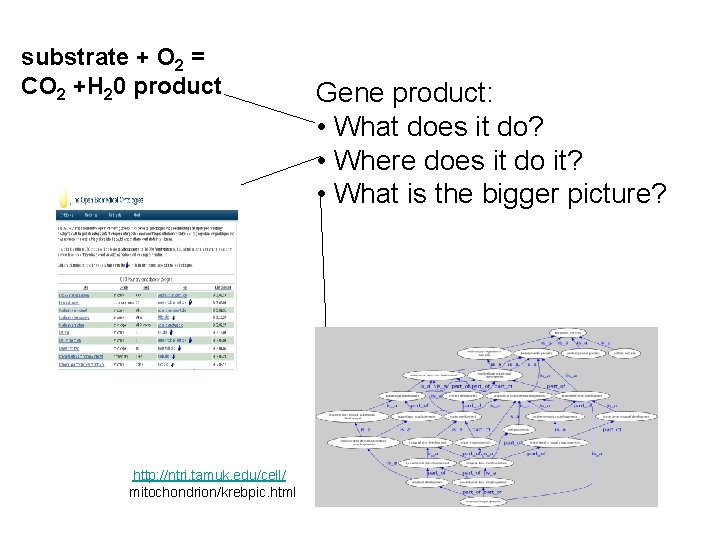
substrate + O 2 = CO 2 +H 20 product http: //ntri. tamuk. edu/cell/ mitochondrion/krebpic. html Gene product: • What does it do? • Where does it do it? • What is the bigger picture?

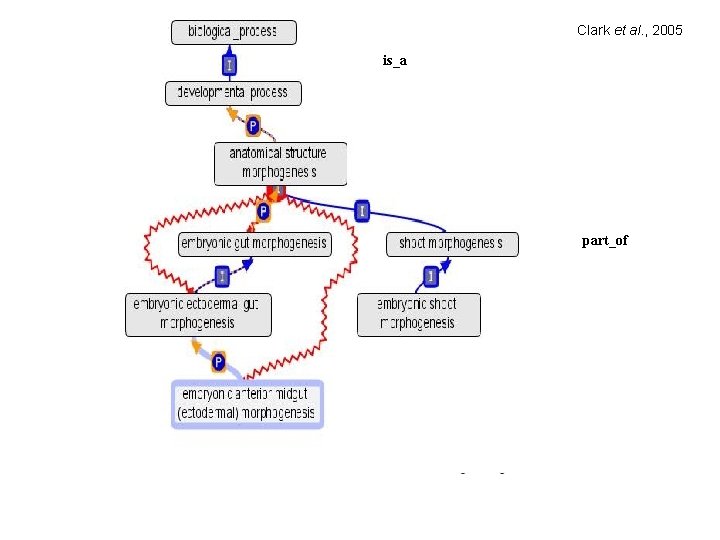
Clark et al. , 2005 is_a part_of

Clark et al. , 2005 is_a part_of

Whole genome analysis (J. D. Munkvold et al. , 2004)

See which processes are upregulated or downregulated. time Defense response Immune response Response to stimulus Toll regulated genes JAK-STAT regulated genes Puparial adhesion Molting cycle hemocyanin Amino acid catabolism Lipid metobolism Peptidase activity Protein catabolism Immune response Toll regulated genes attacked control Bregje Wertheim at the Centre for Evolutionary Genomics, Department of Biology, UCL and Eugene Schuster Group, EBI.

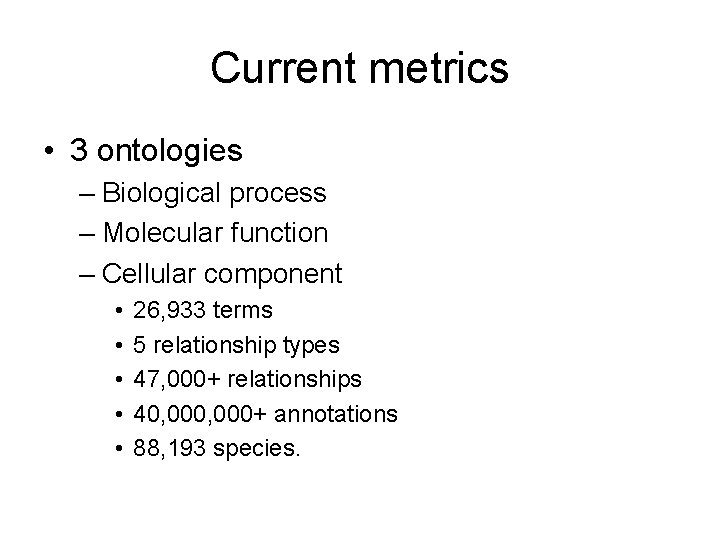
Current metrics • 3 ontologies – Biological process – Molecular function – Cellular component • • • 26, 933 terms 5 relationship types 47, 000+ relationships 40, 000+ annotations 88, 193 species.

Future plans • Cross products • Function-process links

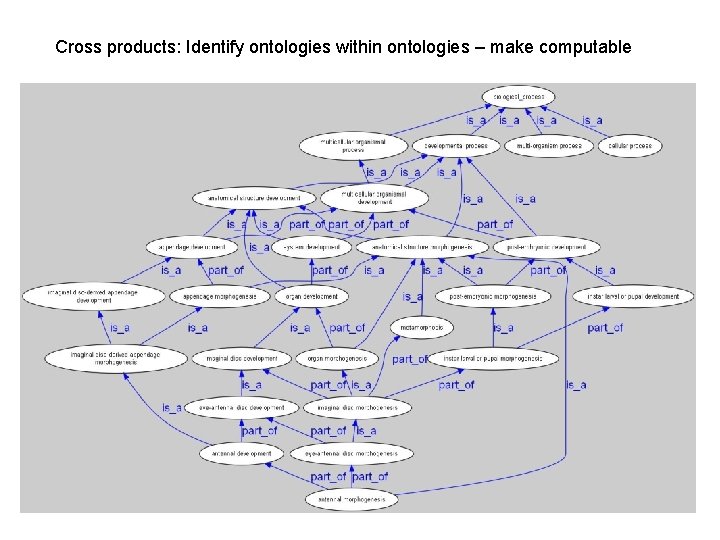
Cross products: Identify ontologies within ontologies – make computable
![Intersection tags cross products Term id GO 0010064 embryonic shoot morphogenesis intersectionof Intersection tags / cross products [Term] id: GO: 0010064 ! embryonic shoot morphogenesis intersection_of:](https://slidetodoc.com/presentation_image/9191d97abd02808a3d3f68481205914f/image-12.jpg)
Intersection tags / cross products [Term] id: GO: 0010064 ! embryonic shoot morphogenesis intersection_of: GO: 0010016 ! shoot morphogenesis intersection_of: part_of GO: 0009790 ! embryonic development [Term] id: GO: 0010101 ! post-embryonic root morphogenesis intersection_of: GO: 0010015 ! root morphogenesis intersection_of: part_of GO: 0009791 ! post-embryonic development

Cross products (xp) in progress • XPs being set up amongst GO ontologies. • Also with: cell type ontology, uberon (anatomy) ontology, Ch. EBI (chemical) ontology. • 170+ new relationship types planned.

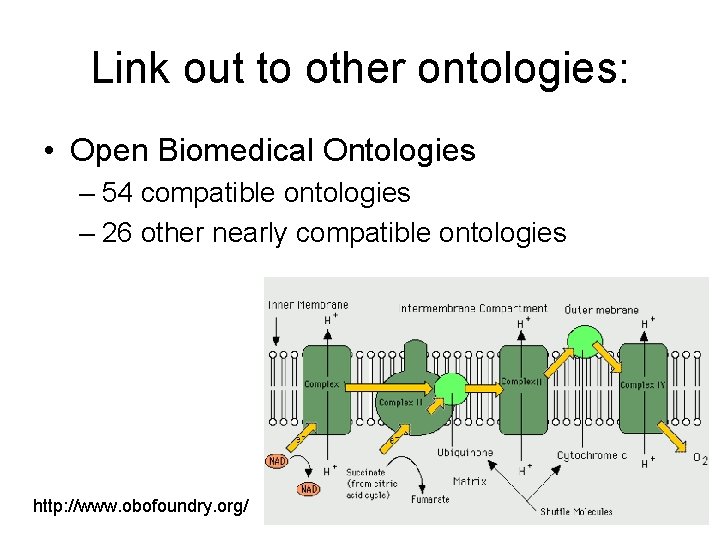
Link out to other ontologies: • Open Biomedical Ontologies – 54 compatible ontologies – 26 other nearly compatible ontologies http: //www. obofoundry. org/

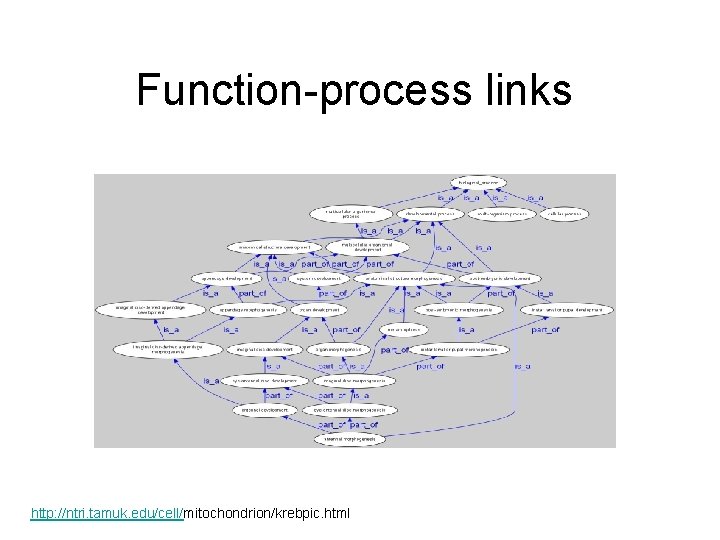
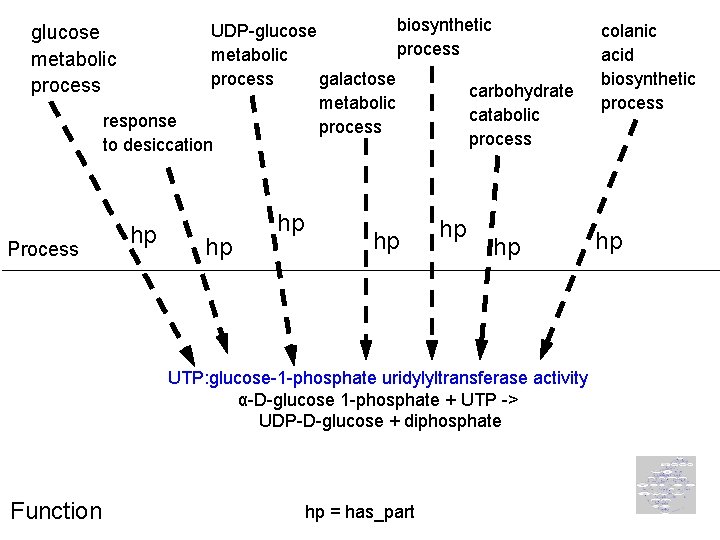
Function-process links http: //ntri. tamuk. edu/cell/mitochondrion/krebpic. html

biosynthetic UDP-glucose process metabolic process galactose carbohydrate metabolic catabolic response process to desiccation glucose metabolic process Process hp hp hp UTP: glucose-1 -phosphate uridylyltransferase activity α-D-glucose 1 -phosphate + UTP -> UDP-D-glucose + diphosphate Function hp = has_part colanic acid biosynthetic process hp

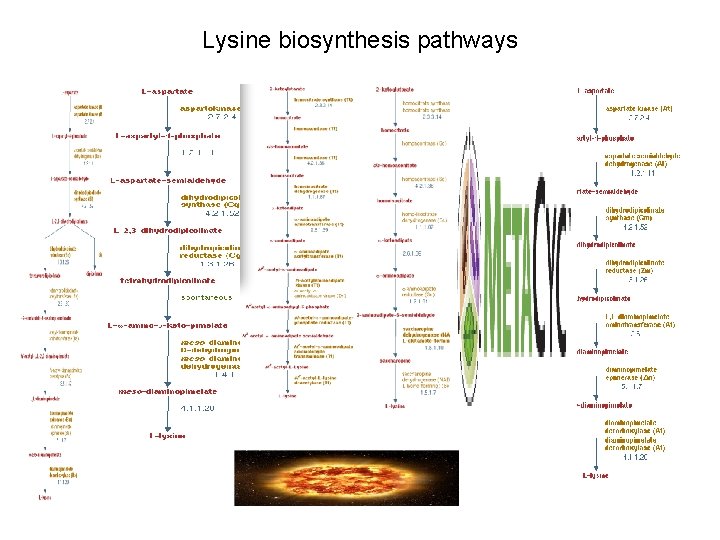
Lysine biosynthesis pathways

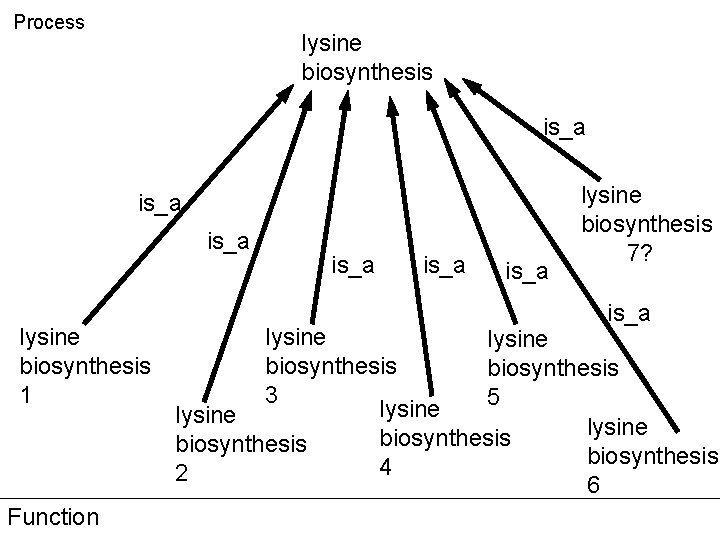
Process lysine biosynthesis is_a lysine biosynthesis 1 Function is_a lysine biosynthesis 7? is_a lysine biosynthesis 3 5 lysine biosynthesis 4 2 6

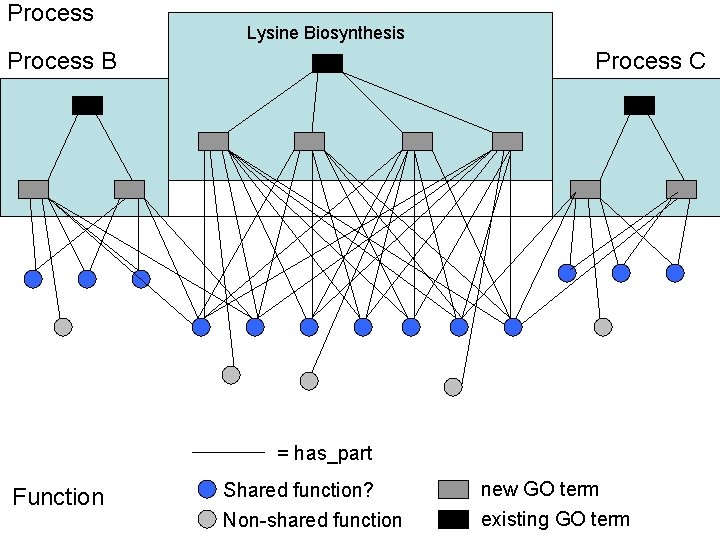
Process Lysine Biosynthesis Process B Process C = has_part Function Shared function? new GO term Non-shared function existing GO term

Process B Function Lysine Biosynthesis Relationship explosion (or Editorial office explosion) Process C

Tools

OBO-Edit: Ontology Editor Tool

Reasoner finds missing and redundant relationships.

2006 Consortium Meeting, St. Croix, U. S. Virgin Islands, March 30 - April 3, 2006

E. Coli hub http: //www. geneontology. org Reactome

Particular thanks to: Chris Mungall - OBOL, Rule Based Reasoner Cross product system Function process link plan – The electron transport working group OBO-Edit: John Day-Richter, Nomi Harris, Amina Abdulla:
 Gene ontology project
Gene ontology project Gene ontology project
Gene ontology project Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Dicty base
Dicty base Gene ontology
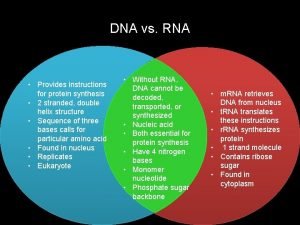
Gene ontology Unlike dna, rna
Unlike dna, rna Amoeba sisters mutations worksheet
Amoeba sisters mutations worksheet Venn diagram rna and dna
Venn diagram rna and dna Dna and rna concept map
Dna and rna concept map Dna to rna rules
Dna to rna rules Replication vs transcription venn diagram
Replication vs transcription venn diagram Dna and rna coloring worksheet
Dna and rna coloring worksheet Dna number of strands
Dna number of strands Dna rna protein central dogma
Dna rna protein central dogma Rna or dna
Rna or dna Rna types
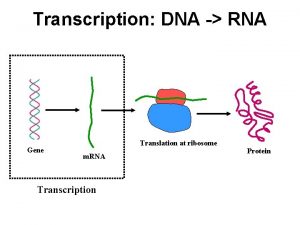
Rna types Dna to rna transcription
Dna to rna transcription Rna or dna
Rna or dna Big q
Big q Phosphodiester bond
Phosphodiester bond Rnabases
Rnabases Dna rna protein central dogma
Dna rna protein central dogma Dna rna and proteins study guide answers
Dna rna and proteins study guide answers Fraction
Fraction Lenine
Lenine Chapter 12 dna and rna
Chapter 12 dna and rna Chapter 12 section 3 dna rna and protein
Chapter 12 section 3 dna rna and protein