Clin Gen The Clinical Genome Resource Sharing Clinical











- Slides: 32

Clin. Gen The Clinical Genome Resource Sharing Clinical Genetic Knowledge HL 7 Clinical Genomics Weekly Call – Dec 12, 2017 Larry Babb, Clin. Gen Data Model WG co-chair larry. babb@sunquestinfo. com

Disclosure • Larry Babb is employed by Mito. Gen/Gene. Insight, a Sunquest Information Systems company. Sunquest is a commercial laboratory software vendor.

Today’s Objective Stimulate discussion on the need for sharing clinical grade genetic knowledge that is not directly associated to a patient finding. • What type of data are we talking about? • Is this within the scope of HL 7? • Might it impact the current HL 7 CG models?

g in r a h S n o i t a Cur on i t a in m e s Dis Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

Clinical Genome Resource • Funded by the United States’ National Institutes of Health • Initiated in September, 2013 • Represents >570 collaborators from 230 institutions Image courtesy of Erin Currey and Erin Ramos, NHGRI Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

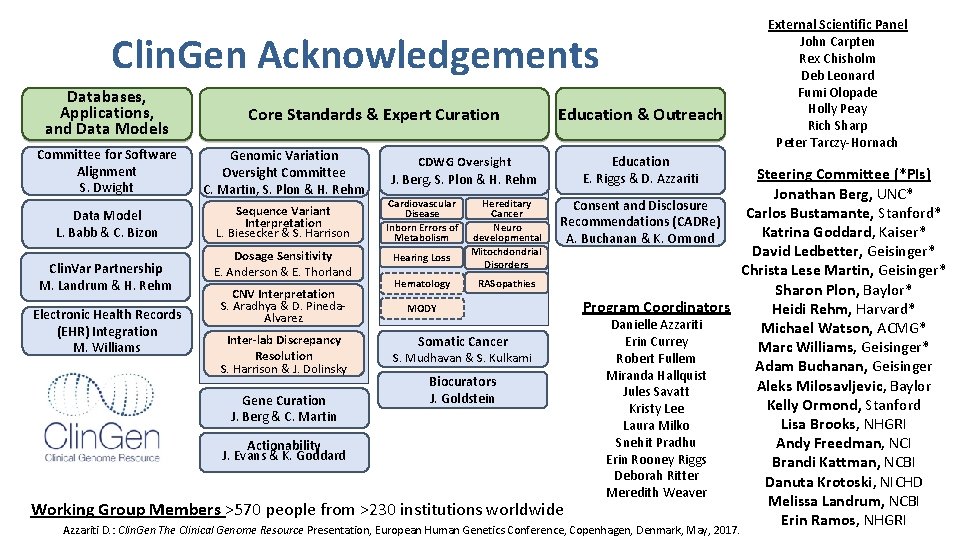
Clin. Gen Acknowledgements Databases, Applications, and Data Models Committee for Software Alignment S. Dwight Core Standards & Expert Curation Genomic Variation Oversight Committee C. Martin, S. Plon & H. Rehm CDWG Oversight J. Berg, S. Plon & H. Rehm Education & Outreach Education E. Riggs & D. Azzariti External Scientific Panel John Carpten Rex Chisholm Deb Leonard Fumi Olopade Holly Peay Rich Sharp Peter Tarczy-Hornach Steering Committee (*PIs) Jonathan Berg, UNC* Hereditary Cardiovascular Consent and Disclosure Sequence Variant Cancer Disease Carlos Bustamante, Stanford* Data Model Recommendations (CADRe) Interpretation Neuro Inborn Errors of L. Biesecker & S. Harrison Katrina Goddard, Kaiser* L. Babb & C. Bizon developmental Metabolism A. Buchanan & K. Ormond Mitochdondrial David Ledbetter, Geisinger* Dosage Sensitivity Hearing Loss Disorders Clin. Var Partnership Christa Lese Martin, Geisinger* E. Anderson & E. Thorland RASopathies Hematology M. Landrum & H. Rehm Sharon Plon, Baylor* CNV Interpretation S. Aradhya & D. Pineda. Program Coordinators MODY Heidi Rehm, Harvard* Electronic Health Records Alvarez Danielle Azzariti Michael Watson, ACMG* (EHR) Integration Inter-lab Discrepancy Somatic Cancer Erin Currey Marc Williams, Geisinger* M. Williams Resolution S. Mudhavan & S. Kulkarni Robert Fullem Adam Buchanan, Geisinger S. Harrison & J. Dolinsky Miranda Hallquist Biocurators Aleks Milosavljevic, Baylor Jules Savatt J. Goldstein Gene Curation Kelly Ormond, Stanford Kristy Lee J. Berg & C. Martin Lisa Brooks, NHGRI Laura Milko Snehit Pradhu Andy Freedman, NCI Actionability J. Evans & K. Goddard Erin Rooney Riggs Brandi Kattman, NCBI Deborah Ritter Danuta Krotoski, NICHD Meredith Weaver Melissa Landrum, NCBI Working Group Members >570 people from >230 institutions worldwide Erin Ramos, NHGRI Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

Data Sharing is Critical to Precision Medicine (not just patient data!) • Updates to clinical-grade knowledge repositories and sources inform physician treatment plans. • Providing standard, accurate and precise association of knowledge and genetic results is essential to applying that knowledge in clinical care. • Both lab and physician systems need the ability to provide access to data in a timely and context appropriate manner • Clinicians and researchers alike need to be able to safely and effectively share new findings in order to speed discovery which benefits all.

What kinds of data are we talking about? • Standardized Variant Representation • Sequence Variants– allele, haplotype, genotype (precise DNA and AA variants) • Structural Variants – CNVs, gene fusions, translocations, inversions, others… • Standardized Gene and Variant Interpretation • • • ACMG Sequence Variant Interpretation CAP/MVLD Somatic Interpretations PGx metabolism/efficacy interpretations Gene Validity / Gene Dosage Maps Actionability

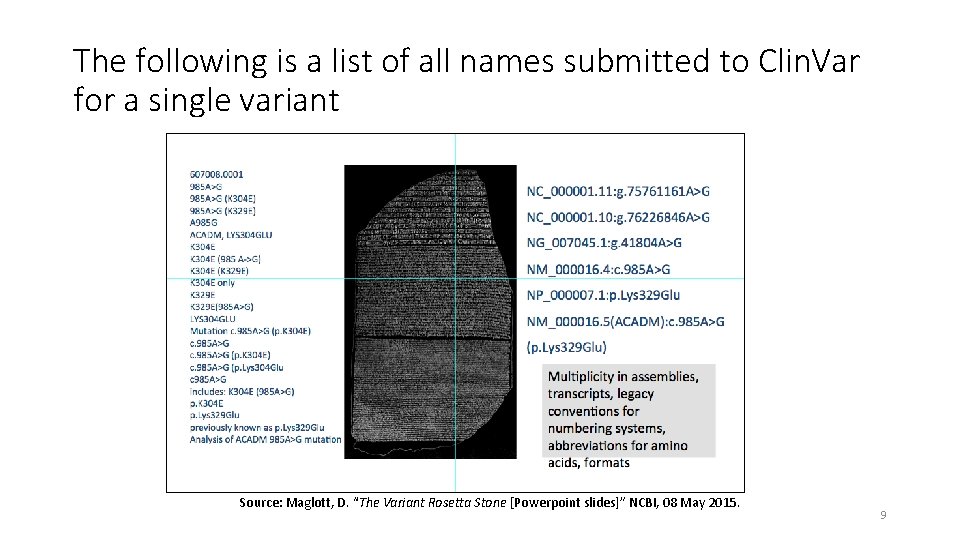
The following is a list of all names submitted to Clin. Var for a single variant Source: Maglott, D. “The Variant Rosetta Stone [Powerpoint slides]” NCBI, 08 May 2015. 9

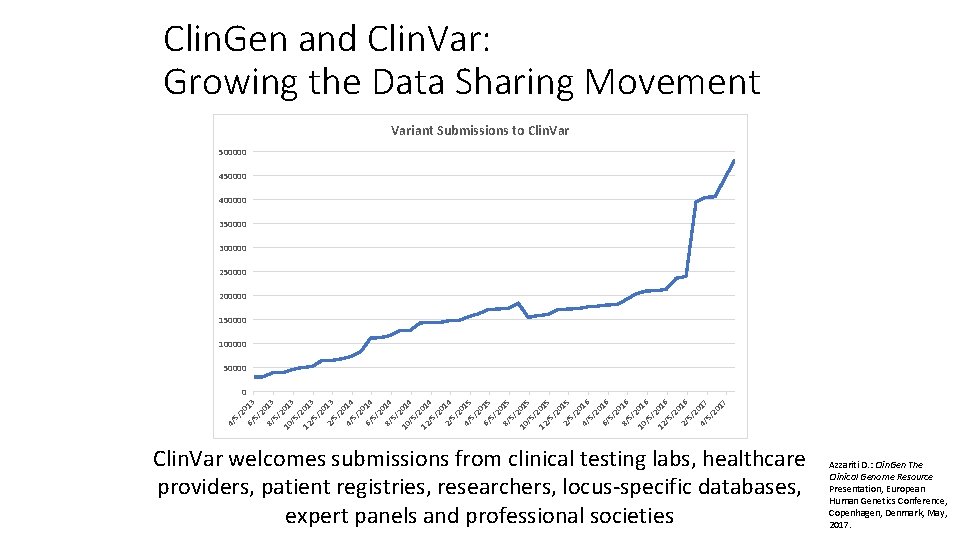
Clin. Gen and Clin. Var: Growing the Data Sharing Movement Variant Submissions to Clin. Var 500000 450000 400000 350000 300000 250000 200000 150000 100000 50000 4/ 5/ 20 6/ 13 5/ 20 8/ 13 5/ 2 10 013 /5 /2 12 01 /5 3 /2 0 2/ 13 5/ 20 4/ 14 5/ 20 6/ 14 5/ 20 8/ 14 5/ 2 10 014 /5 /2 12 01 /5 4 /2 0 2/ 14 5/ 20 4/ 15 5/ 20 6/ 15 5/ 20 8/ 15 5/ 2 10 015 /5 /2 12 01 /5 5 /2 0 2/ 15 5/ 20 4/ 16 5/ 20 6/ 16 5/ 20 8/ 16 5/ 2 10 016 /5 /2 12 01 /5 6 /2 0 2/ 16 5/ 20 4/ 17 5/ 20 17 0 Clin. Var welcomes submissions from clinical testing labs, healthcare providers, patient registries, researchers, locus-specific databases, expert panels and professional societies Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

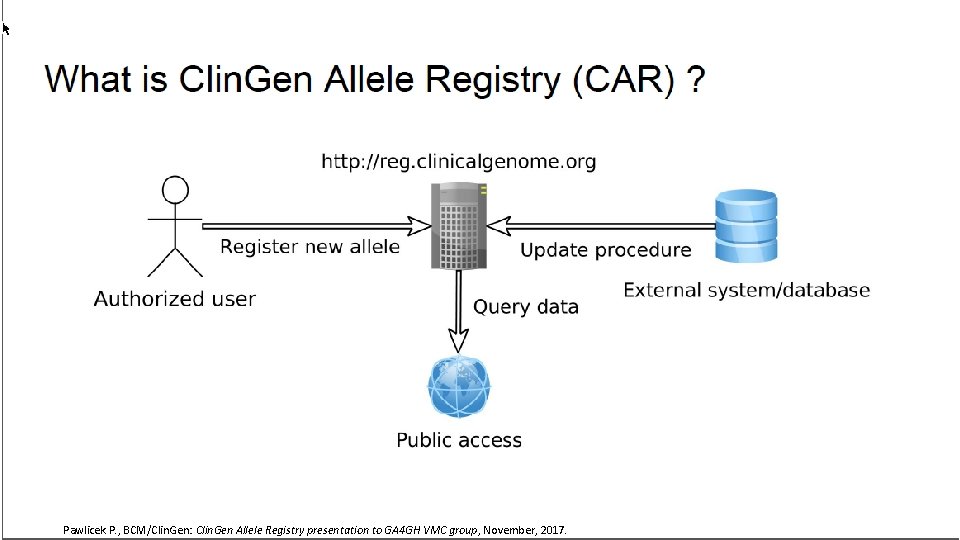
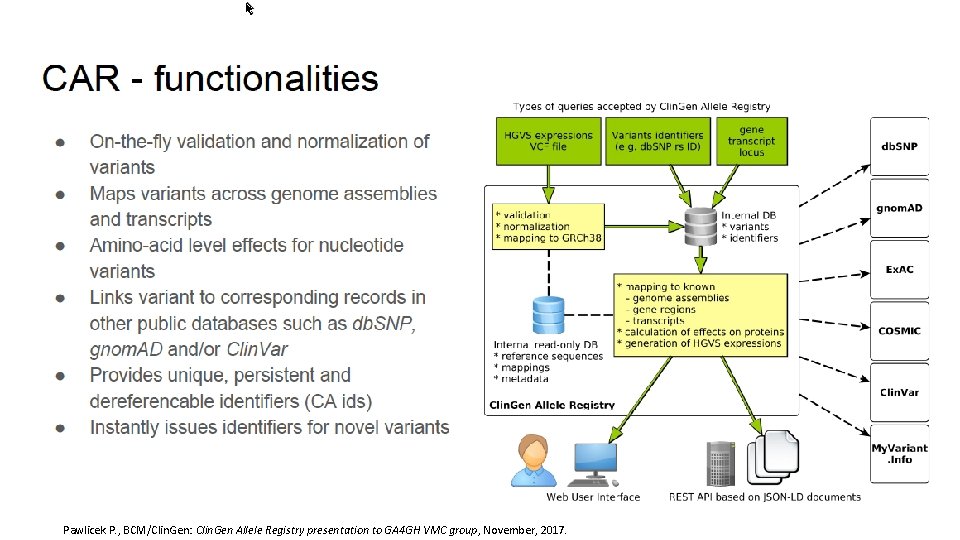
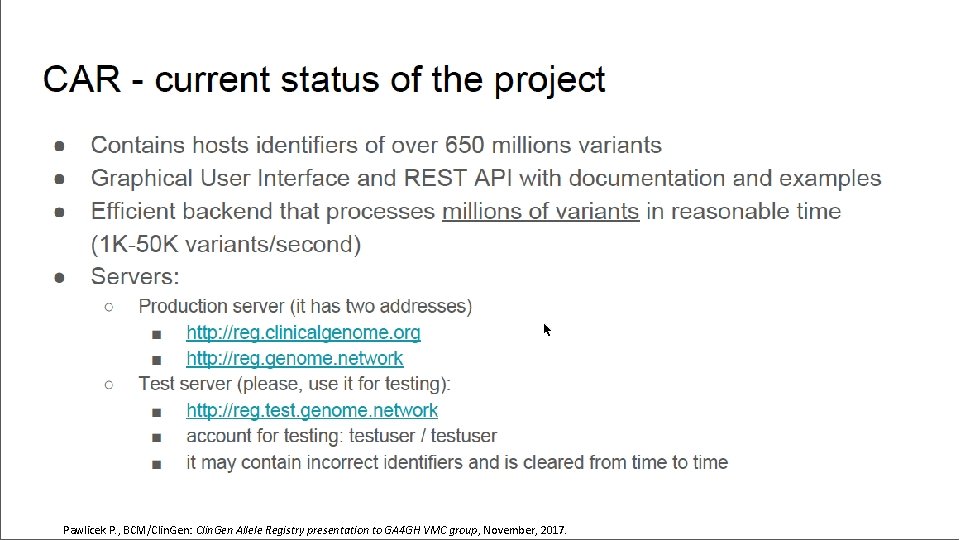
Pawlicek P. , BCM/Clin. Gen: Clin. Gen Allele Registry presentation to GA 4 GH VMC group, November, 2017.

Data Model WG Roadmap • Variant Modeling • Clin. Gen Baylor Allele Registry www. • Interpretation Modeling Pawlicek P. , BCM/Clin. Gen: Clin. Gen Allele Registry presentation to GA 4 GH VMC group, November, 2017.

Pawlicek P. , BCM/Clin. Gen: Clin. Gen Allele Registry presentation to GA 4 GH VMC group, November, 2017.

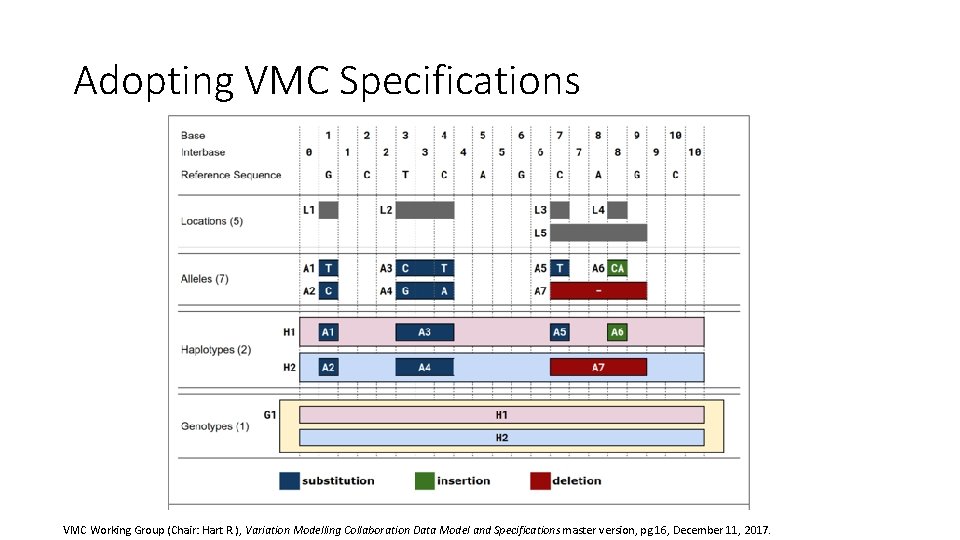
Adopting VMC Specifications VMC Working Group (Chair: Hart R. ), Variation Modelling Collaboration Data Model and Specifications master version, pg 16, December 11, 2017.

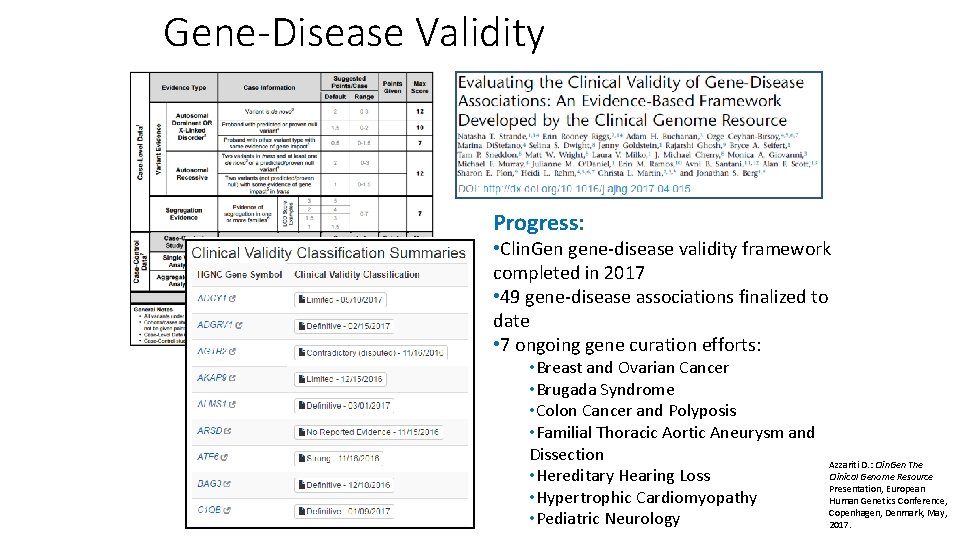
Gene-Disease Validity Progress: • Clin. Gen gene-disease validity framework completed in 2017 • 49 gene-disease associations finalized to date • 7 ongoing gene curation efforts: • Breast and Ovarian Cancer • Brugada Syndrome • Colon Cancer and Polyposis • Familial Thoracic Aortic Aneurysm and Dissection • Hereditary Hearing Loss • Hypertrophic Cardiomyopathy • Pediatric Neurology Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

Dosage Sensitivity www. ncbi. nlm. nih. gov/projects/dbvar/clingen/ Progress: • Continuing efforts of the International Standards of Cytogenomic Arrays Consortium (ISCA) • 1, 324 genes reviewed for dosage sensitivity to date • Goal: to create a genome-wide dosage sensitivity map Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

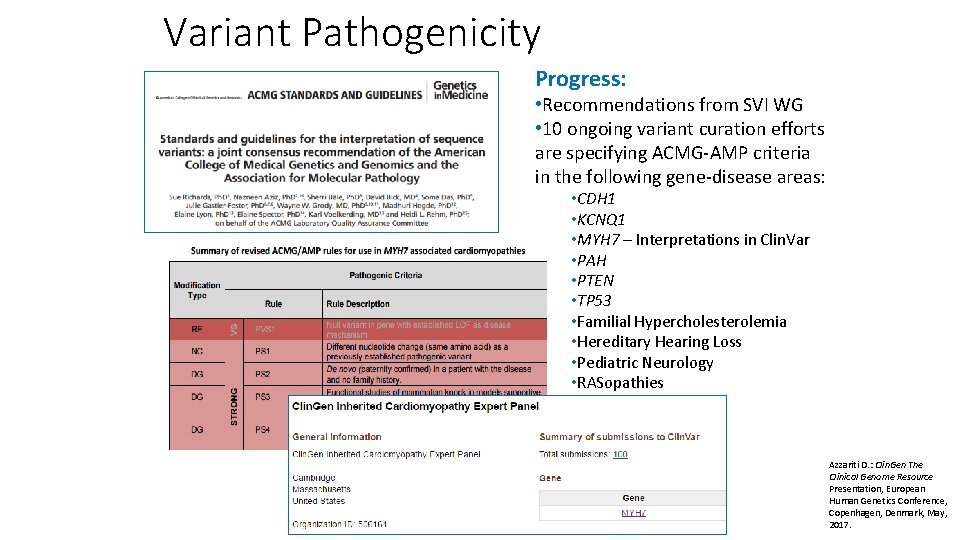
Variant Pathogenicity Progress: • Recommendations from SVI WG • 10 ongoing variant curation efforts are specifying ACMG-AMP criteria in the following gene-disease areas: • CDH 1 • KCNQ 1 • MYH 7 – Interpretations in Clin. Var • PAH • PTEN • TP 53 • Familial Hypercholesterolemia • Hereditary Hearing Loss • Pediatric Neurology • RASopathies Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

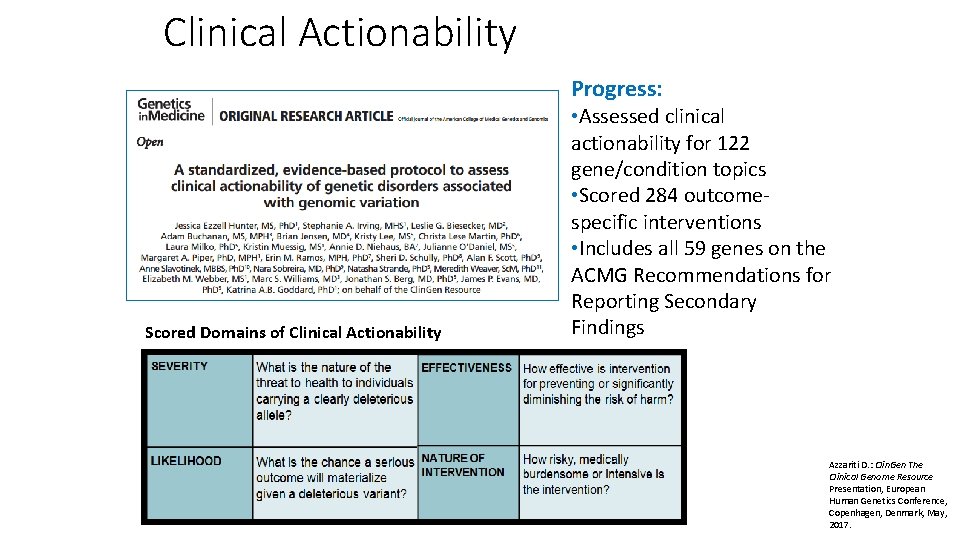
Clinical Actionability Progress: Scored Domains of Clinical Actionability • Assessed clinical actionability for 122 gene/condition topics • Scored 284 outcomespecific interventions • Includes all 59 genes on the ACMG Recommendations for Reporting Secondary Findings Azzariti D. : Clin. Gen The Clinical Genome Resource Presentation, European Human Genetics Conference, Copenhagen, Denmark, May, 2017.

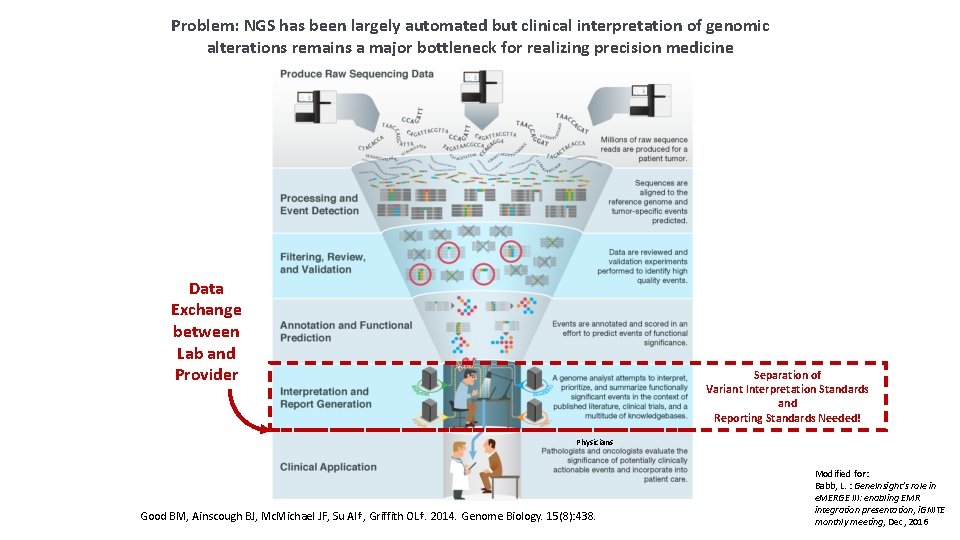
Problem: NGS has been largely automated but clinical interpretation of genomic alterations remains a major bottleneck for realizing precision medicine Data Exchange between Lab and Provider Separation of Variant Interpretation Standards and Reporting Standards Needed! Physicians Good BM, Ainscough BJ, Mc. Michael JF, Su AI†, Griffith OL†. 2014. Genome Biology. 15(8): 438. Modified for: Babb, L. : Gene. Insight’s role in e. MERGE III: enabling EMR integration presentation, i. GNITE monthly meeting, Dec, 2016

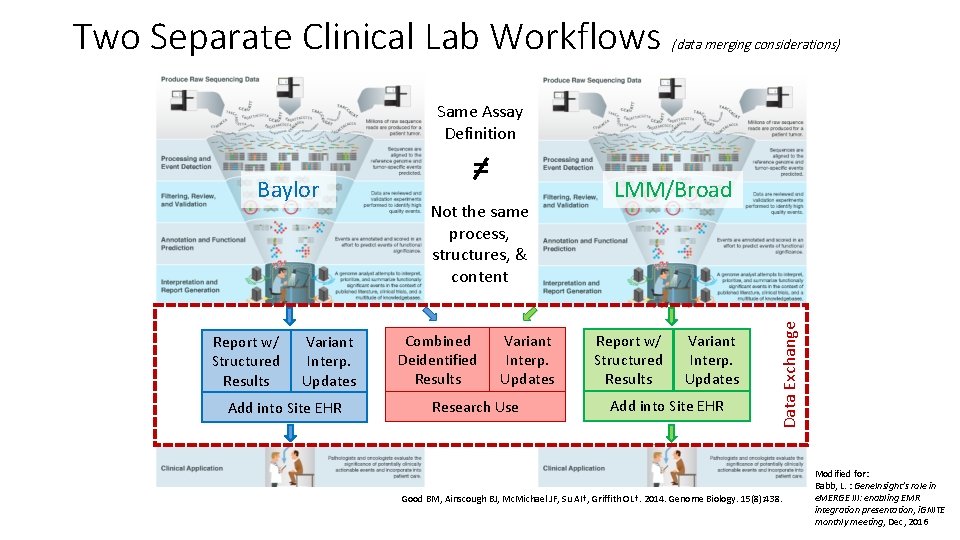
Two Separate Clinical Lab Workflows (data merging considerations) Same Assay Definition Report w/ Structured Results Variant Interp. Updates Add into Site EHR Not the same process, structures, & content Combined Deidentified Results Variant Interp. Updates Research Use LMM/Broad Report w/ Structured Results Variant Interp. Updates Add into Site EHR Data Exchange Baylor = Good BM, Ainscough BJ, Mc. Michael JF, Su AI†, Griffith OL†. 2014. Genome Biology. 15(8): 438. Modified for: Babb, L. : Gene. Insight’s role in e. MERGE III: enabling EMR integration presentation, i. GNITE monthly meeting, Dec, 2016

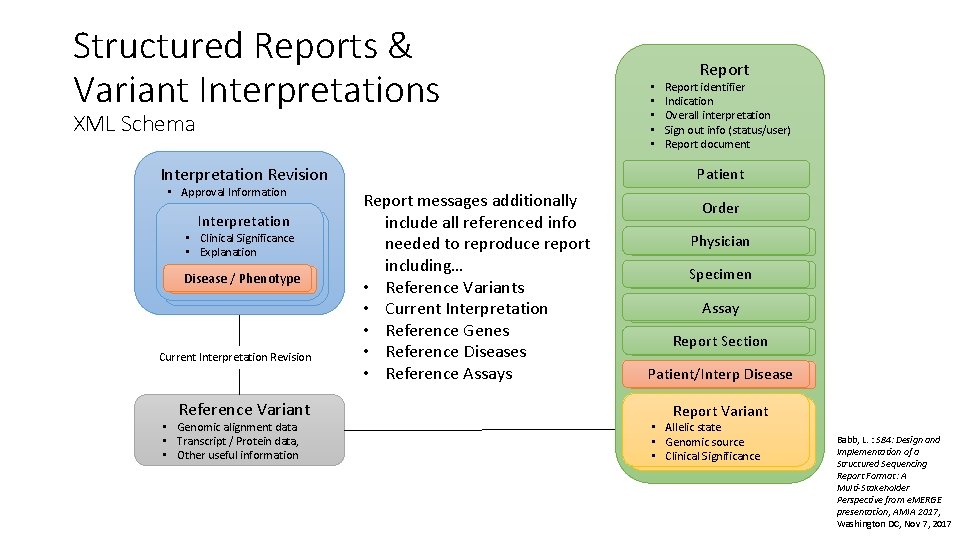
Structured Reports & Variant Interpretations XML Schema Interpretation Revision • Approval Information Interpretation • Clinical Significance • Explanation Disease / Phenotype Current Interpretation Revision Reference Variant • Genomic alignment data • Transcript / Protein data, • Other useful information • • • Report identifier Indication Overall interpretation Sign out info (status/user) Report document Patient Report messages additionally include all referenced info needed to reproduce report including… • Reference Variants • Current Interpretation • Reference Genes • Reference Diseases • Reference Assays Order Physician Specimen Assay Report Section Patient/Interp Disease Report Variant • Allelic state • Genomic source • Clinical Significance Babb, L. : S 84: Design and Implementation of a Structured Sequencing Report Format: A Multi-Stakeholder Perspective from e. MERGE presentation, AMIA 2017, Washington DC, Nov 7, 2017

Review of Core Concepts • Identifiers / Codes • Standard HL 7 -like namespace model (system + code/id + text/display name) • Variant Representation (reported and reference) • Sequence Variants - genomic coordinates as a baseline (grch 37) • CNVs – no common computational approach • PGx – star alleles – not computationally ideal (i. e. CYP 2 C 9*2) • Disease Coding • 39 pre-determined indication / patient disease values, required. • OMIM, Orphanet, Disease Ontology (align with Monarch Initiative ontology) • Interpretation / Clinical Significance • ACMG LOINC codes (Pathogenic, Likely Path, Uncertain Sig, Likely Ben, Benign) • Common structure (variant, disease, inheritance, clinical significance, explanation) • Assay coverage • Single assay for both SCs, some differences however. • No computational representation of assay coverage or methodology or other aspects • Patients • Identifiers, Demographics (DOB, race-coded, sex-coded, …) Babb, L. : S 84: Design and Implementation of a Structured Sequencing Report Format: A Multi-Stakeholder Perspective from e. MERGE presentation, AMIA 2017, Washington DC, Nov 7, 2017

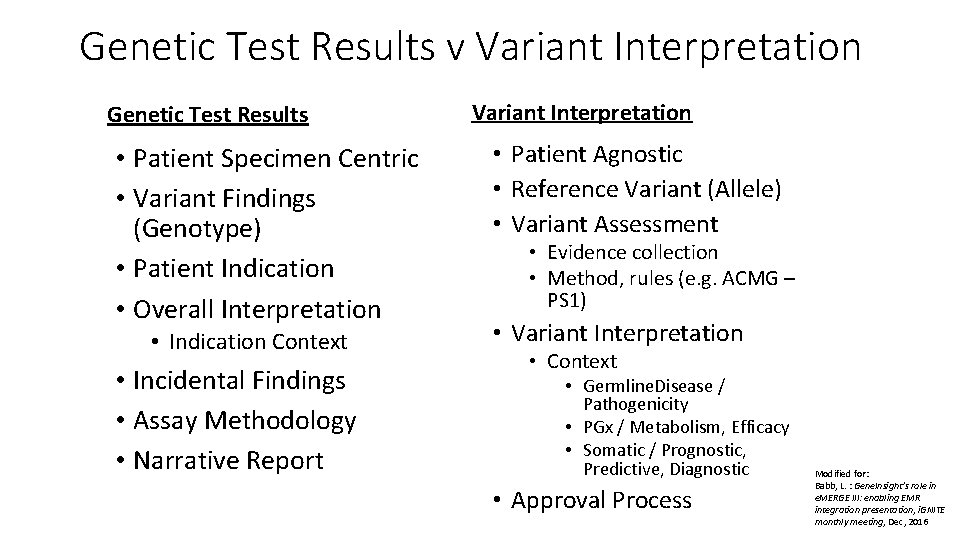
Genetic Test Results v Variant Interpretation Genetic Test Results • Patient Specimen Centric • Variant Findings (Genotype) • Patient Indication • Overall Interpretation • Indication Context • Incidental Findings • Assay Methodology • Narrative Report Variant Interpretation • Patient Agnostic • Reference Variant (Allele) • Variant Assessment • Evidence collection • Method, rules (e. g. ACMG – PS 1) • Variant Interpretation • Context • Germline. Disease / Pathogenicity • PGx / Metabolism, Efficacy • Somatic / Prognostic, Predictive, Diagnostic • Approval Process Modified for: Babb, L. : Gene. Insight’s role in e. MERGE III: enabling EMR integration presentation, i. GNITE monthly meeting, Dec, 2016

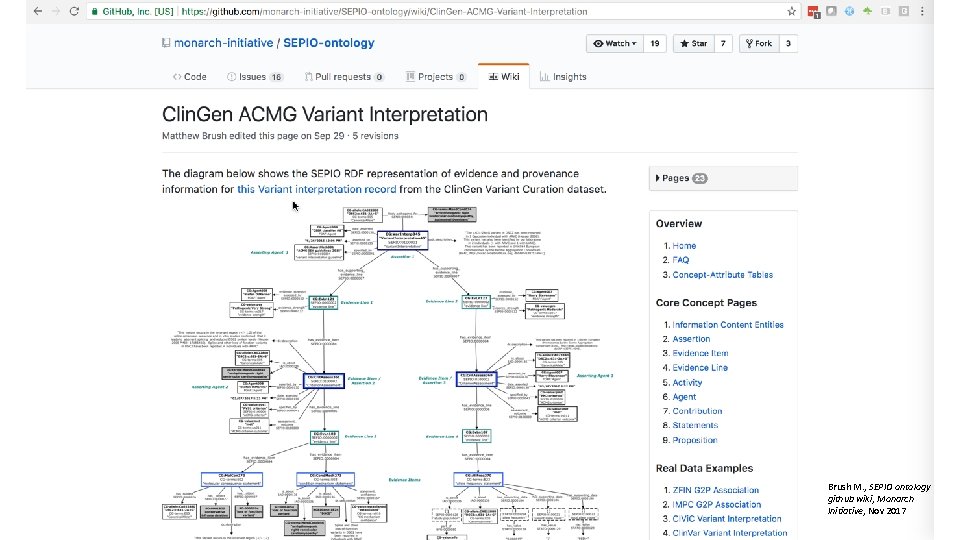
Clin. Gen-SEPIO Interpretation Model Monarch – Clin. Gen Collaboration

Brush M. , SEPIO ontology github wiki, Monarch Initiative, Nov 2017

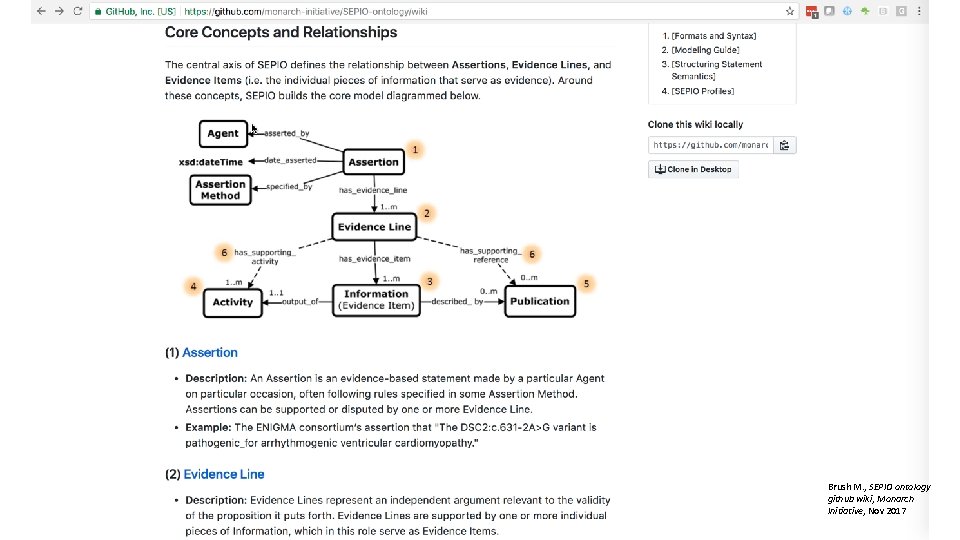
Brush M. , SEPIO ontology github wiki, Monarch Initiative, Nov 2017

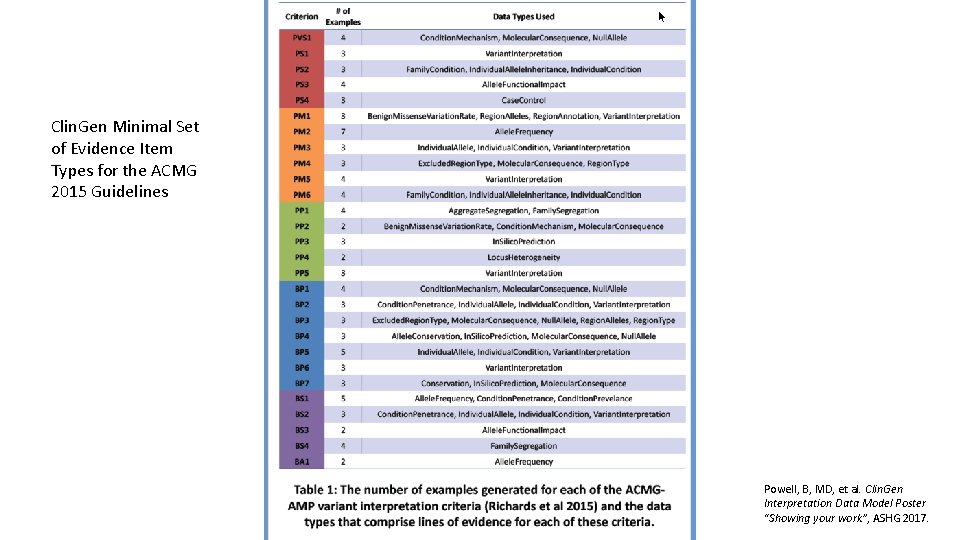
Clin. Gen Minimal Set of Evidence Item Types for the ACMG 2015 Guidelines Powell, B, MD, et al. Clin. Gen Interpretation Data Model Poster “Showing your work”, ASHG 2017.

Brush M. , SEPIO ontology github wiki, Monarch Initiative, Nov 2017

Clin. Gen Interpretation Model Status • Fully harmonized with SEPIO using Clin. Gen defined terms • Defined 23 minimal evidence concepts for v 1 (ACMG guidelines) • Defined 33 value sets – controlled vocabulary for key attributes • Used a comprehensive set of real world examples to validate & document • Initiate a "scripted" approach for generating a Clin. Var submission using the model • Next steps • Extend to Gene validity, Actionability, and other assertion-based models • Implement models in Data Exchange for internal and public use Babb, L. : Data Model WG Status Report, Clin. Gen Steering Committee meeting, Wash. DC, Nov, 2017

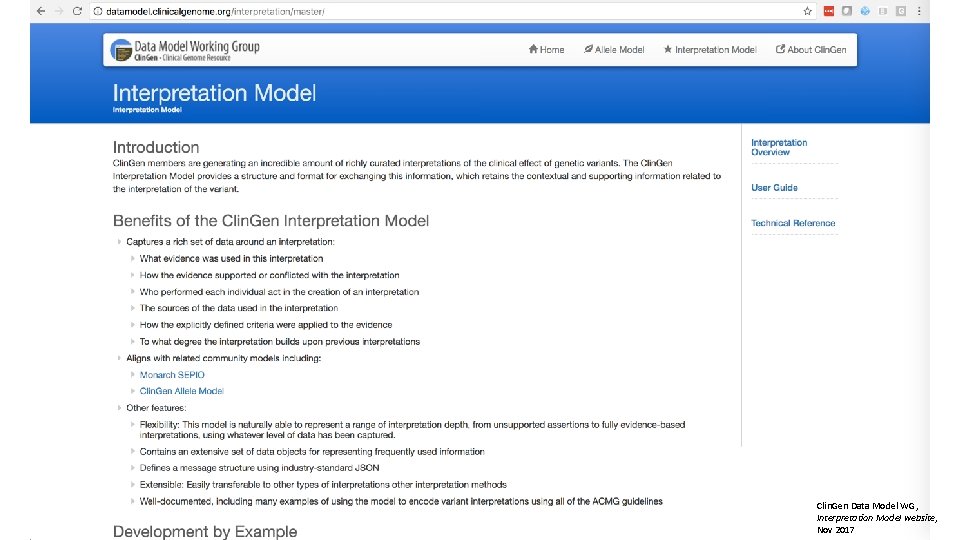
Clin. Gen Data Model WG, Interpretation Model website, Nov 2017

Related web sites and resources • Clin. Gen – http: //clinicalgenome. org • Clin. Gen Data Model – http: //datamodel. clinicalgenome. org • Clin. Gen Allele Registry –http: //reg. clinicalgenome. org/ • Test instance – http: //reg. test. genome. network • API docs - http: //reg. clinicalgenome. org/doc/Allele. Registry_0. 12. xx_api_v 2. pdf • SEPIO wiki - https: //github. com/monarch-initiative/SEPIO-ontology/wiki • Clin. Var – http: //www. ncbi. nlm. nih. gov/clinvar/ • GA 4 GH VMC Specification - https: //docs. google. com/document/d/12 E 8 Wb. Qlvf. ZWk 5 Nrxw. Lytmymp. Pby 6 vsv 60 Rx. Ce. D 5 wc 1 E • VMC github project - https: //github. com/ga 4 gh/vmc

Acknowledgements • Clin. Gen Grant • Working group chairs • Program coordinators • Steering committee members • Variation Modeling Collaboration Working Group (GA 4 GH) • Monarch Initiative / SEPIO (Matt Brush) • e. MERGE Grant – Phase III • Clin. Var - Clinical Variant Archive (NCBI)
 Clin gen
Clin gen Clin gen
Clin gen Semi-global alignment
Semi-global alignment Clin var
Clin var Research card
Research card Clin chest med
Clin chest med Pengertian resource sharing
Pengertian resource sharing Ncsbn next generation nclex
Ncsbn next generation nclex Resource leveling is the approach to even out the peaks of
Resource leveling is the approach to even out the peaks of Perbedaan antara resource loading dan resource levelling
Perbedaan antara resource loading dan resource levelling National human genome research institute
National human genome research institute Genome is
Genome is History of sequencing
History of sequencing Shotgun sequencing
Shotgun sequencing Chapter 13 section 3 the human genome
Chapter 13 section 3 the human genome Mash bioinformatics
Mash bioinformatics Img genome
Img genome Marc fiume
Marc fiume Alternate splicing
Alternate splicing Human genome size
Human genome size National human genome research institute
National human genome research institute Patric genome
Patric genome Biologists search the volumes of the human genome using
Biologists search the volumes of the human genome using Tomato genome browser
Tomato genome browser Per partes
Per partes Innovation genome project
Innovation genome project Genome
Genome Plant genome research program
Plant genome research program Chapter 15 the human genome answer key
Chapter 15 the human genome answer key Savant genome browser
Savant genome browser Genome sequencing
Genome sequencing Human genome size
Human genome size Mqall
Mqall