Burkhard Morgenstern Institut fr Mikrobiologie und Genetik Grundlagen

Burkhard Morgenstern Institut für Mikrobiologie und Genetik Grundlagen der Bioinformatik Multiples Sequenzalignment Juni 2007

`Progressive´ Alignment Most popular approach to (global) multiple sequence alignment: Progressive Alignment Since mid-Eighties: Feng/Doolittle, Higgins/Sharp, Taylor, …

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN WWRLNDKEGYVPRNLLGLYP AVVIQDNSDIKVVPKAKIIRD YAVESEAHPGSFQPVAALERIN WLNYNETTGERGDFPGTYVEYIGRKKISP
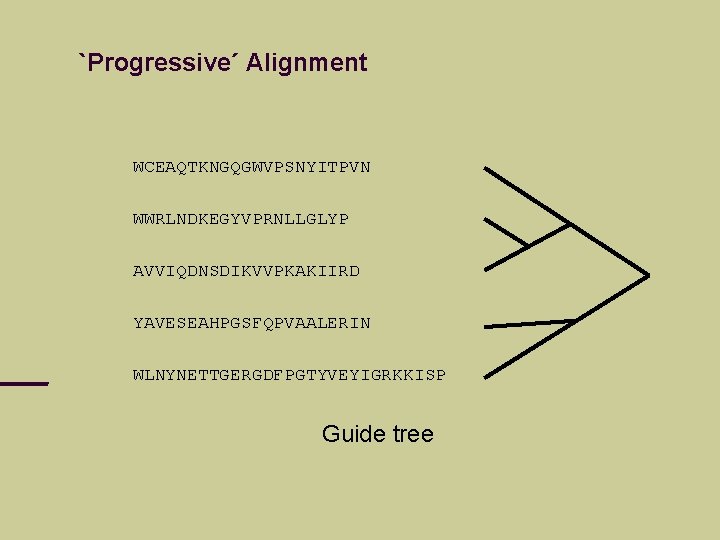
`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN WWRLNDKEGYVPRNLLGLYP AVVIQDNSDIKVVPKAKIIRD YAVESEAHPGSFQPVAALERIN WLNYNETTGERGDFPGTYVEYIGRKKISP Guide tree

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN WW--RLNDKEGYVPRNLLGLYPAVVIQDNSDIKVVP--KAKIIRD YAVESEASFQPVAALERIN WLNYNEERGDFPGTYVEYIGRKKISP Profile alignment, “once a gap - always a gap”

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN WW--RLNDKEGYVPRNLLGLYPAVVIQDNSDIKVVP--KAKIIRD YAVESEASVQ--PVAALERIN-----WLN-YNEERGDFPGTYVEYIGRKKISP Profile alignment, “once a gap - always a gap”

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVNWW--RLNDKEGYVPRNLLGLYPAVVIQDNSDIKVVP--KAKIIRD YAVESEASVQ--PVAALERIN-----WLN-YNEERGDFPGTYVEYIGRKKISP Profile alignment, “once a gap - always a gap”

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN-------WW--RLNDKEGYVPRNLLGLYP-------AVVIQDNSDIKVVP--KAKIIRD------YAVESEA---SVQ--PVAALERIN-----WLN-YNE---ERGDFPGTYVEYIGRKKISP Profile alignment, “once a gap - always a gap”

`Progressive´ Alignment WCEAQTKNGQGWVPSNYITPVN-------WW--RLNDKEGYVPRNLLGLYP-------AVVIQDNSDIKVVP--KAKIIRD------YAVESEA---SVQ--PVAALERIN-----WLN-YNE---ERGDFPGTYVEYIGRKKISP Most important implementation: CLUSTAL W
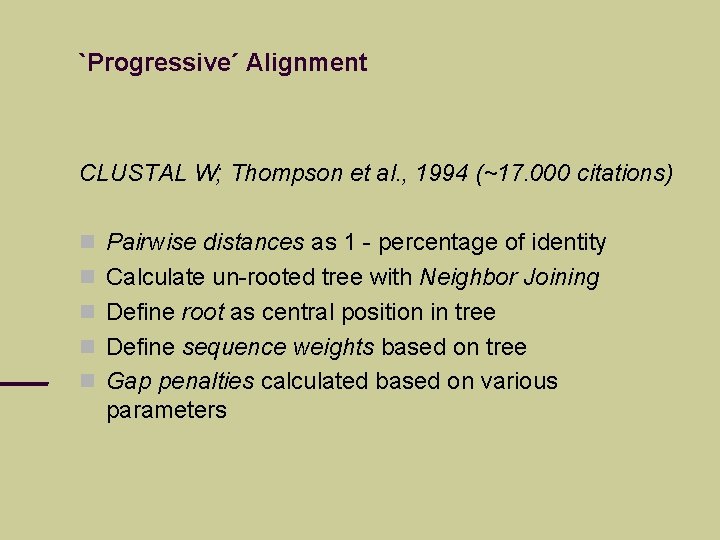
`Progressive´ Alignment CLUSTAL W; Thompson et al. , 1994 (~17. 000 citations) Pairwise distances as 1 - percentage of identity Calculate un-rooted tree with Neighbor Joining Define root as central position in tree Define sequence weights based on tree Gap penalties calculated based on various parameters
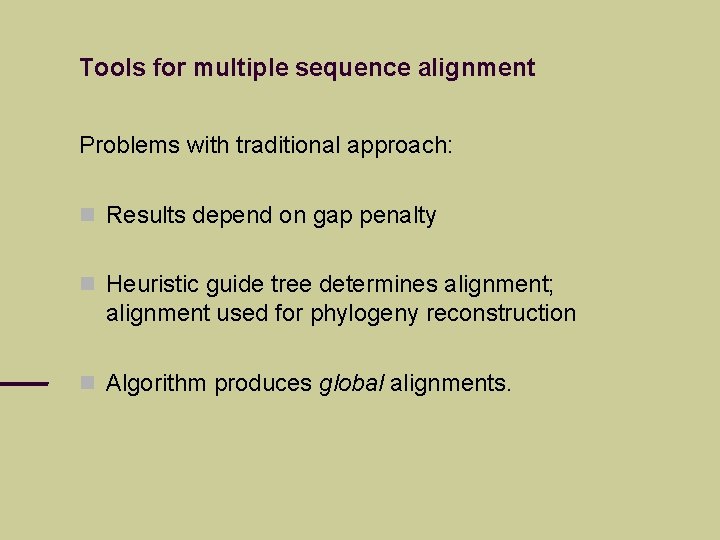
Tools for multiple sequence alignment Problems with traditional approach: Results depend on gap penalty Heuristic guide tree determines alignment; alignment used for phylogeny reconstruction Algorithm produces global alignments.

Tools for multiple sequence alignment Problems with traditional approach: But: Many sequence families share only local similarity E. g. sequences share one conserved motif
Local sequence alignment EYENS ERYAS Find common motif in sequences; ignore the rest
Local sequence alignment E-YENS ERYA-S Find common motif in sequences; ignore the rest

Local sequence alignment E-YENS ERYA-S Find common motif in sequences; ignore the rest – Local alignment

Local sequence alignment Traditional alignment approaches: Either global or local methods!

New question: sequence families with multiple local similarities Neither local nor global methods appliccable
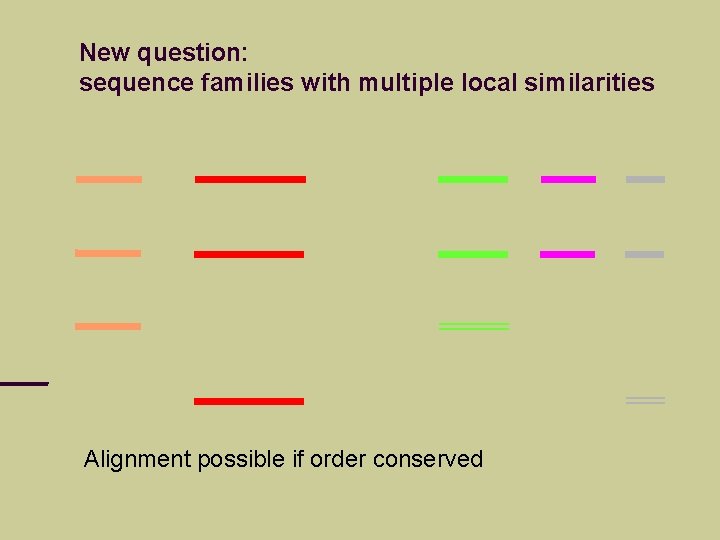
New question: sequence families with multiple local similarities Alignment possible if order conserved

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach Consistency!

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

The DIALIGN approach

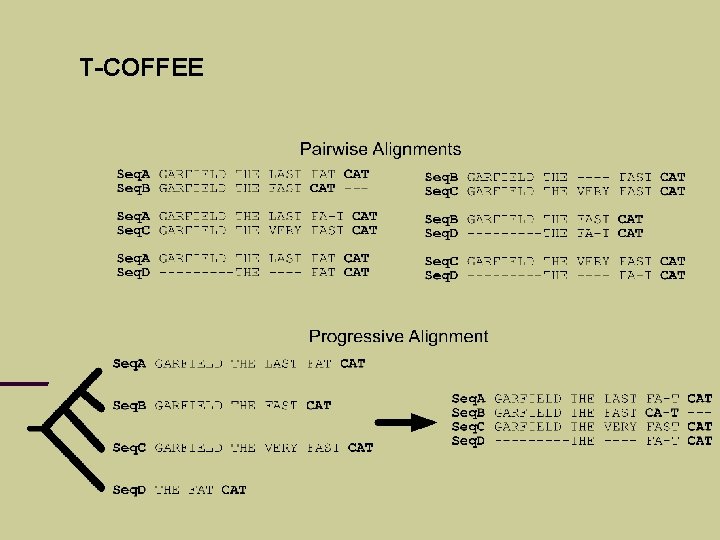
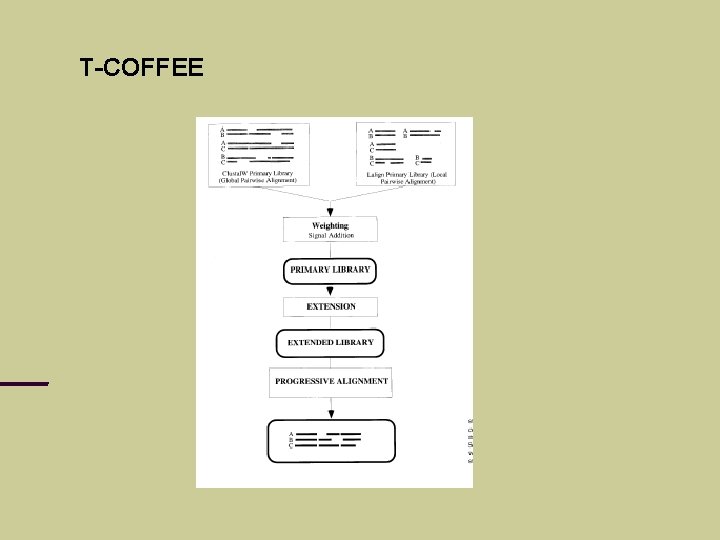
T-COFFEE C. Notredame, D. Higgins, J. Heringa (2000), T-Coffee: A novel algorithm for multiple sequence alignment, J. Mol. Biol. Problem: progressive alignment can go wrong if mistakes are made at an early stage. Example …

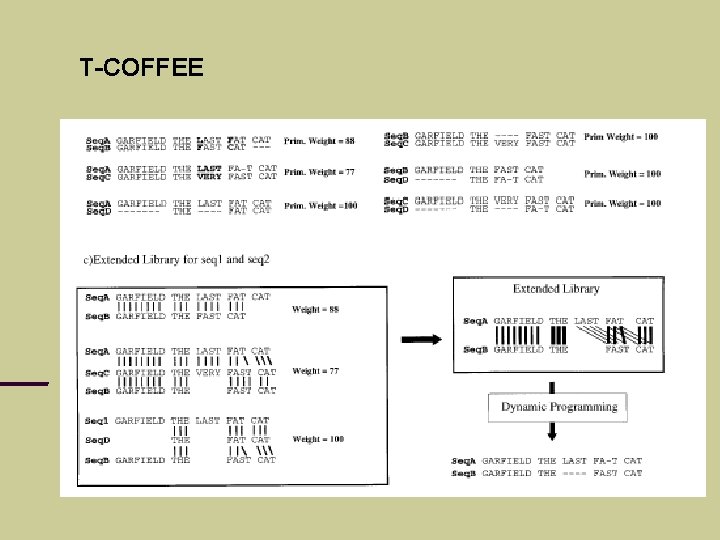
T-COFFEE Seq. A Seq. B Seq. C Seq. D GARFIELD THE LAST FAT CAT GARFIELD THE FAST CAT GARFIELD THE VERY FAST CAT THE FAT CAT

T-COFFEE Seq. A Seq. B Seq. C Seq. D GARFIELD THE LAST FAT CAT GARFIELD THE FAST CAT GARFIELD THE VERY FAST CAT THE FAT CAT
T-COFFEE

T-COFFEE Idea: consider different pairwise alignments (local and global) check how these alignments support each other
T-COFFEE
T-COFFEE

T-COFFEE Less sensitive to spurious pairwise similarities Can handle local homologies better than CLUSTAL

Evaluation of multi-alignment methods Alignment evaluation by comparison to trusted benchmark alignments. `True’ alignment known by information about structure or evolution.
Evaluation of multi-alignment methods For protein alignment: M. Mc. Clure et al. (1994): 4 protein families, known functional sites J. Thompson et al. (1999): Benchmark data base, 130 known 3 D structures (BAli. BASE) T. Lassmann & E. Sonnhammer (2002): BAli. BASE + simulated evolution (ROSE)

Evaluation of multi-alignment methods

Evaluation of multi-alignment methods Alignment evaluation by comparison to trusted benchmark alignments. `True’ alignment known by information about structure or evolution.
Evaluation of multi-alignment methods
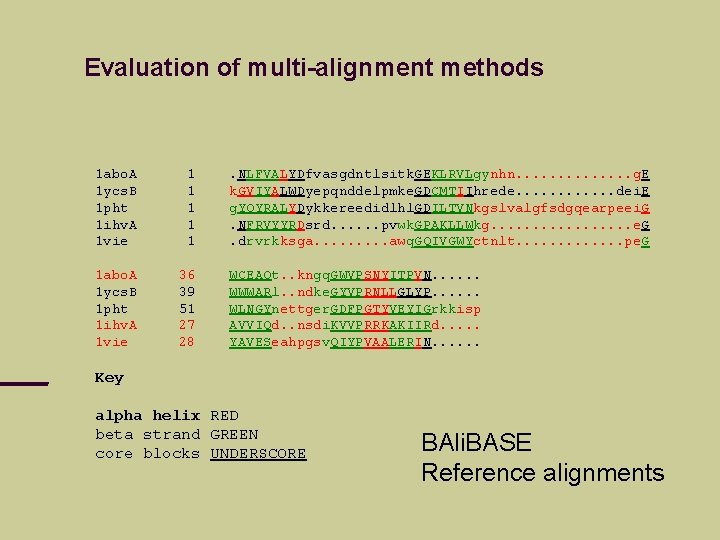
Evaluation of multi-alignment methods 1 abo. A 1 ycs. B 1 pht 1 ihv. A 1 vie 1 1 1 abo. A 1 ycs. B 1 pht 1 ihv. A 1 vie 36 39 51 27 28 . NLFVALYDfvasgdntlsitk. GEKLRVLgynhn. . . g. E k. GVIYALWDyepqnddelpmke. GDCMTIIhrede. . . dei. E g. YQYRALYDykkereedidlhl. GDILTVNkgslvalgfsdgqearpeei. G. NFRVYYRDsrd. . . pvwk. GPAKLLWkg. . . . e. G. drvrkksga. . awq. GQIVGWYctnlt. . . pe. G WCEAQt. . kngq. GWVPSNYITPVN. . . WWWARl. . ndke. GYVPRNLLGLYP. . . WLNGYnettger. GDFPGTYVEYIGrkkisp AVVIQd. . nsdi. KVVPRRKAKIIRd. . . YAVESeahpgsv. QIYPVAALERIN. . . Key alpha helix RED beta strand GREEN core blocks UNDERSCORE BAli. BASE Reference alignments
Evaluation of multi-alignment methods 5 categories of benchmark sequences (globally related, internal gaps, end gaps) CLUSTAL W, RPPR perform well on globally related sequences, DIALIGN superior for local similarities Conclusion: no single best multi alignment program!

Evaluation of multi-alignment methods T. Lassmann & E. Sonnhammer (2002): BAli. BASE + simulated evolution (ROSE)

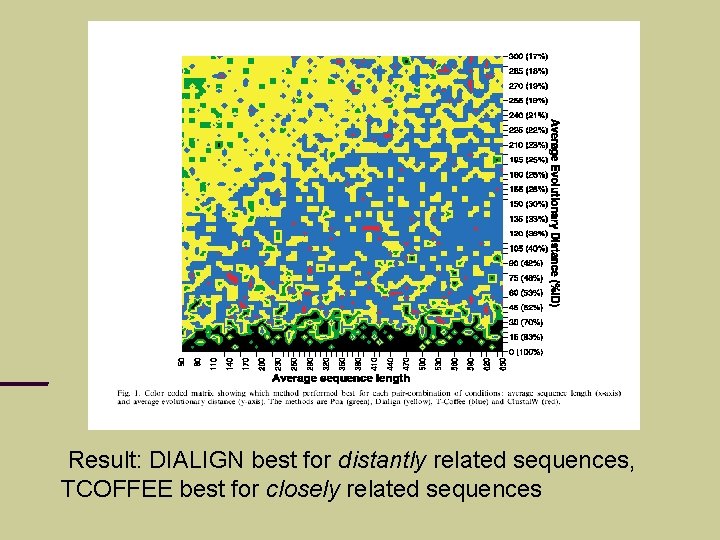
Result: DIALIGN best for distantly related sequences, TCOFFEE best for closely related sequences
- Slides: 66