Tumor Genome Sequencing Xiaole Shirley Liu STAT 115215






- Slides: 35

Tumor Genome Sequencing Xiaole Shirley Liu STAT 115/215, BIO/BST 282

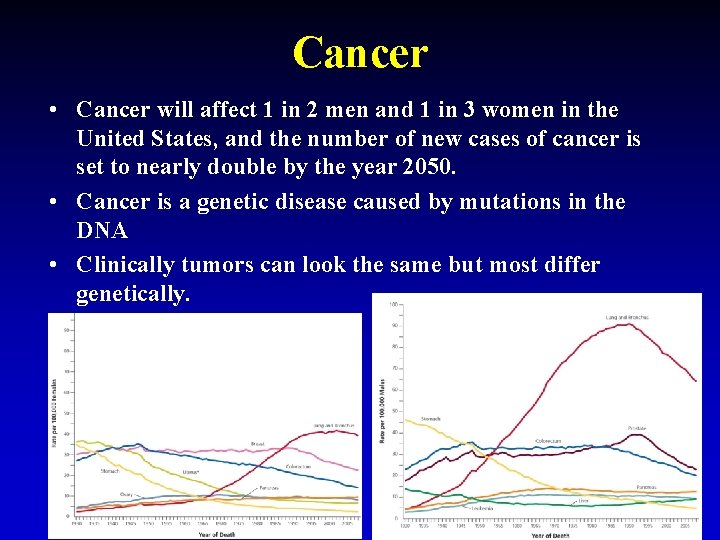
Cancer • Cancer will affect 1 in 2 men and 1 in 3 women in the United States, and the number of new cases of cancer is set to nearly double by the year 2050. • Cancer is a genetic disease caused by mutations in the DNA • Clinically tumors can look the same but most differ genetically. 2

Different Sequencing Approaches • Capture-seq (~$500) – Could focus on well known mutations • Exome-seq ($700 -1200) – All the exons in genes; promoters and Lnc. RNA genes? • RNA-seq ($300 -1000) – Expression and mutations together, miss anything? • Whole genome sequencing ($1500 -2500) – Mutations in non-coding, function unknown – Better at detecting structural changes (translocations, fusions) • Cost-vs-benefit balance 3

Two Major Cancer Genome Projects • TCGA: The Cancer Genome Atlas (US) – – > 30 cancer types and > 10 K tumor samples Primary tumors, fewer death events Genome, transcriptome, DNA methylome, proteomics Rigorous tumor sample QC, consistent profiling platform • ICGC: International Cancer Genome Consortium (11 countries) – 20 cancer types * 500 tumor samples each 4

Tumor Gene Expression • Microarrays or RNA-seq • Data analysis? • Differential expression between cancer and normal • Cluster the tumor samples into sub-types – Consensus clustering: sampling genes or tumors, get robust clustering • Predict patient outcome (survival or recurrence) Break 5

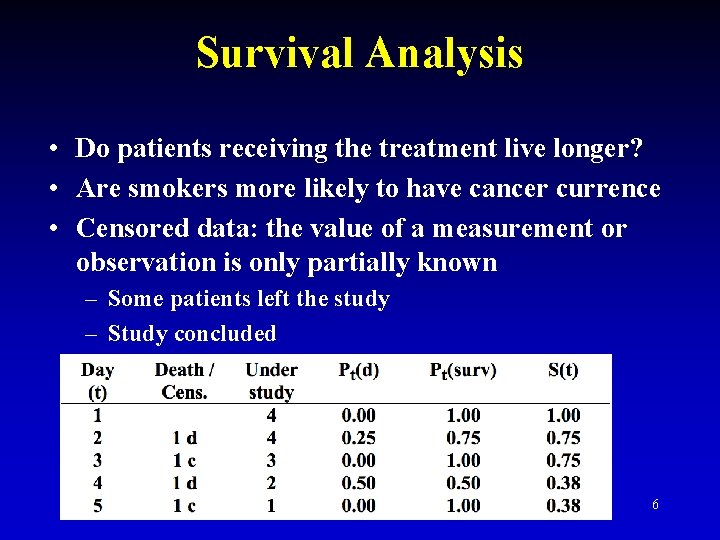
Survival Analysis • Do patients receiving the treatment live longer? • Are smokers more likely to have cancer currence • Censored data: the value of a measurement or observation is only partially known – Some patients left the study – Study concluded 6

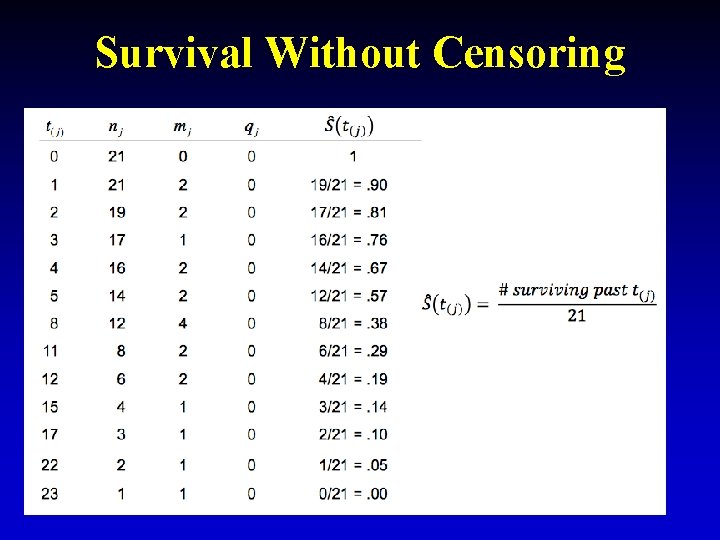
Survival Without Censoring 7

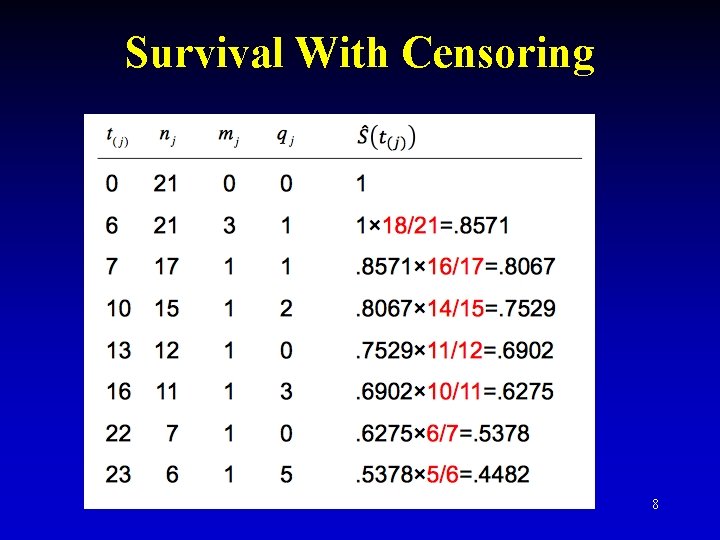
Survival With Censoring 8

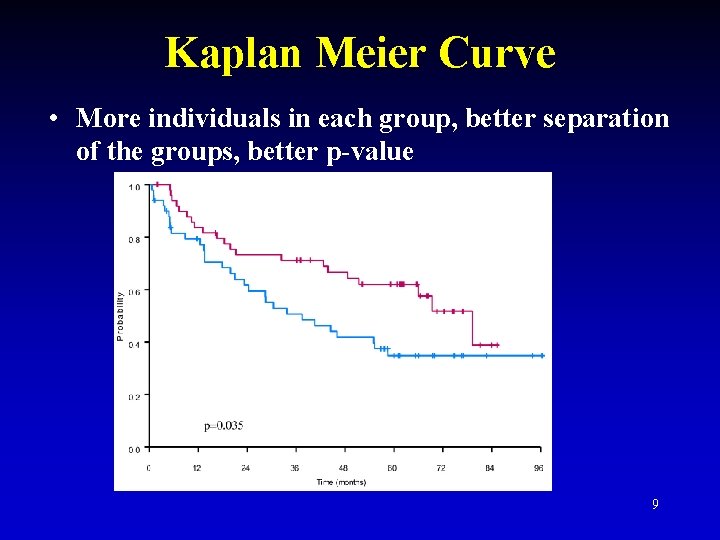
Kaplan Meier Curve • More individuals in each group, better separation of the groups, better p-value 9

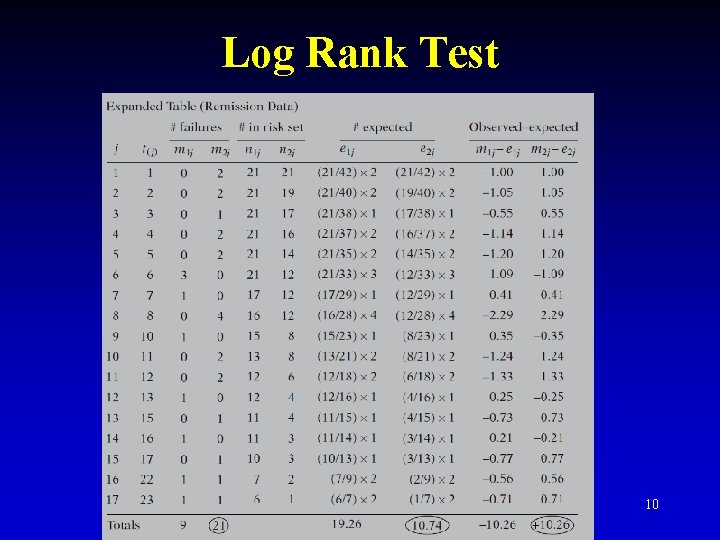
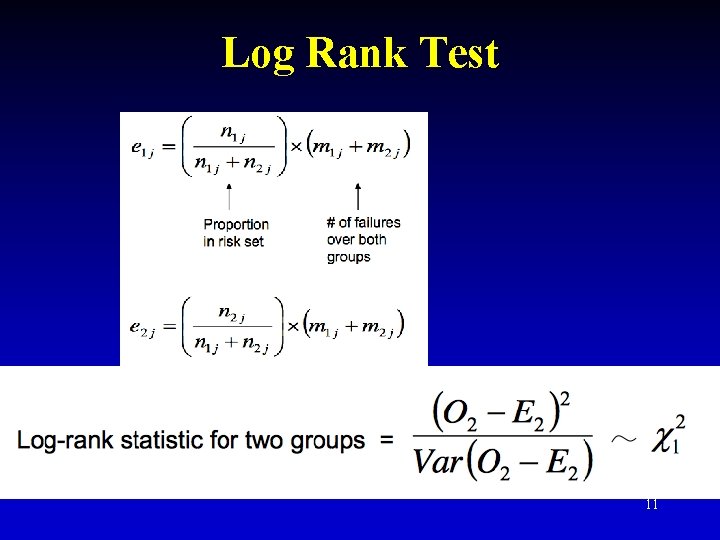
Log Rank Test 10

Log Rank Test 11

More Variables • 50 -signature? • Logistic regression: – Estimate odds ratio: ratio of proportions – Linear combination of all the genes to separate outcome (0, 1). • Cox Regression – Estimate hazard ratio: ratio of incidence rates – Models the effect of covariates on the hazard rate but leaves the baseline hazard rate unspecified 12

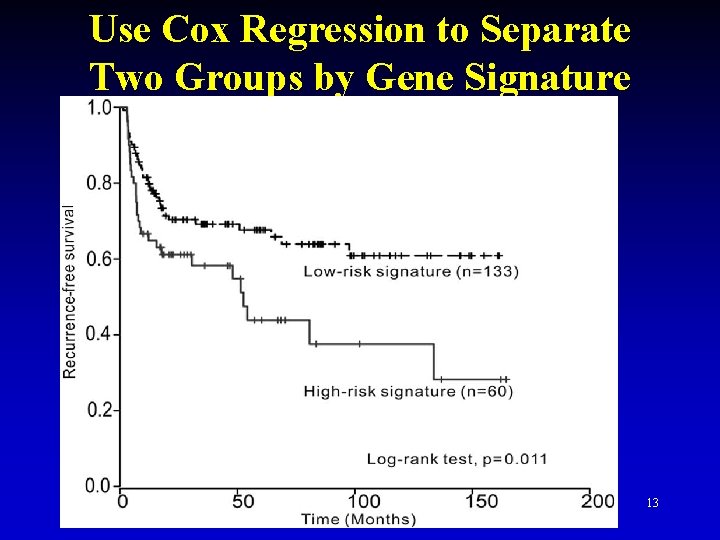
Use Cox Regression to Separate Two Groups by Gene Signature 13

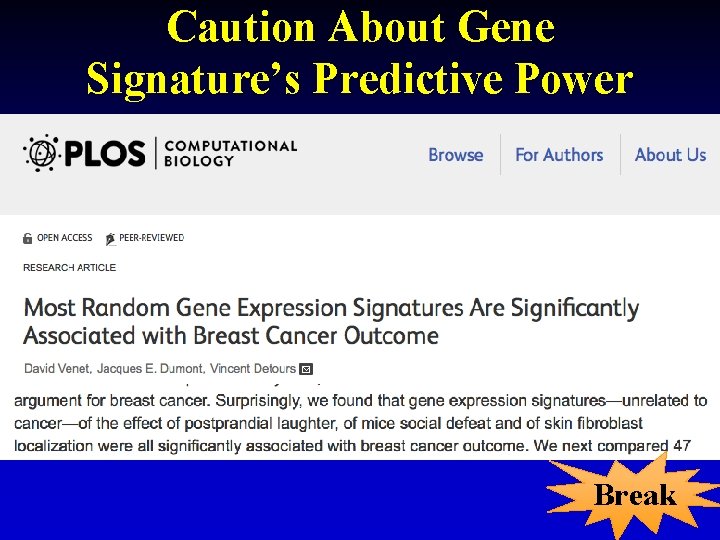
Caution About Gene Signature’s Predictive Power Break 14

Mutations in the Tumor Genome • Help us identify important genes for tumorigenesis and cancer progression • Drivers – a. k. a gatekeepers, mutations that cause and accelerate cancers • Passengers – Accidental by-products and thwarted DNA-repair mechanisms • Recurrent mutations on genes or pathways are likely drivers 15

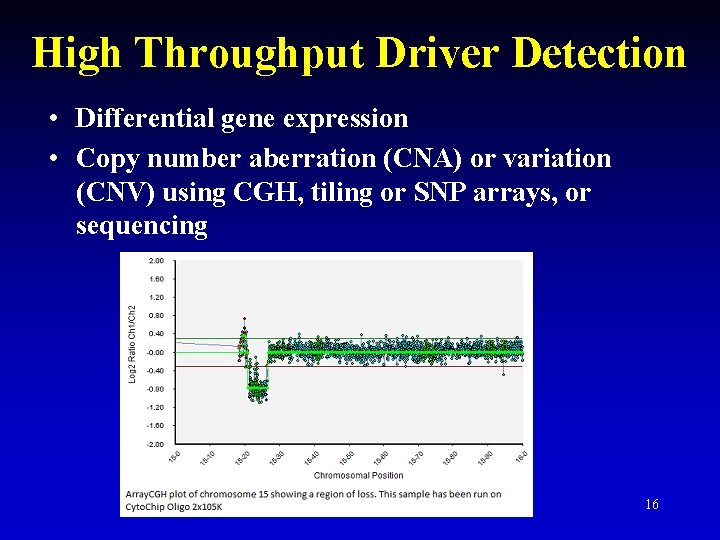
High Throughput Driver Detection • Differential gene expression • Copy number aberration (CNA) or variation (CNV) using CGH, tiling or SNP arrays, or sequencing 16

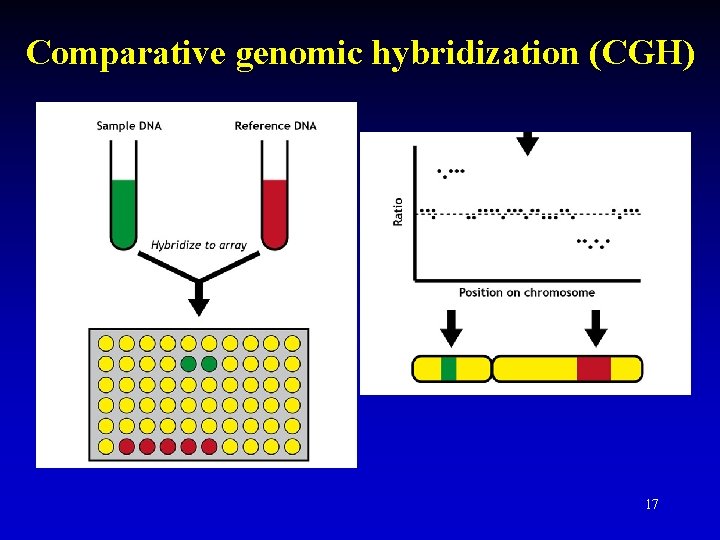
Comparative genomic hybridization (CGH) 17

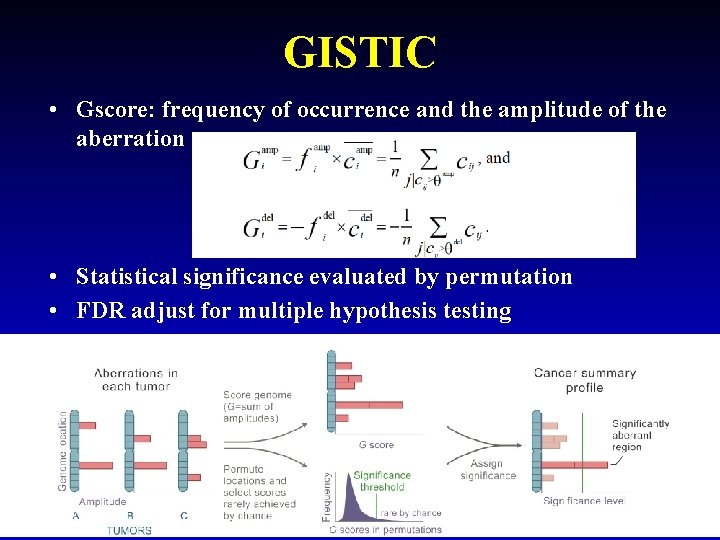
GISTIC • Gscore: frequency of occurrence and the amplitude of the aberration • Statistical significance evaluated by permutation • FDR adjust for multiple hypothesis testing 18

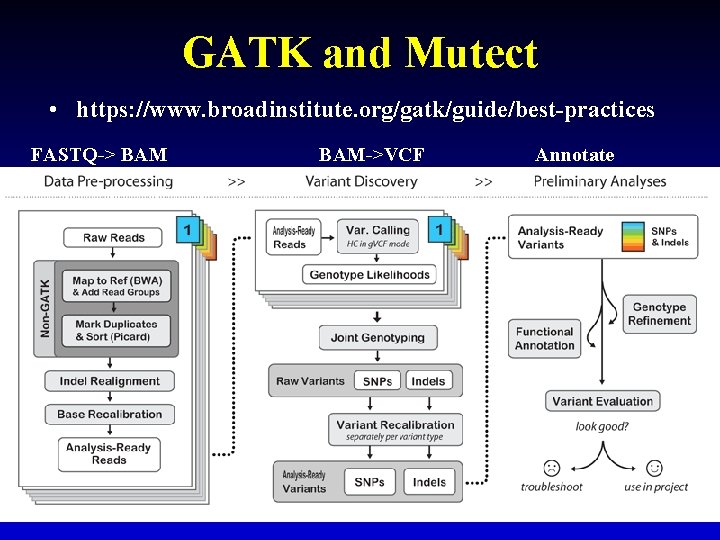
GATK and Mutect • https: //www. broadinstitute. org/gatk/guide/best-practices FASTQ-> BAM->VCF Annotate 19

MAF and VCF Formats • VCF (GWAS format) and MAF (TCGA format) • Both can annotate somatic mutations and germline variants • Tab delimited text file • CHROM, POS, ID (SNP id, gene symbol, or ENTREZ gene id), REF (reference seq), ALT (altered sequence), QUAL (quality score), FILTER (PASS vs “q 10; s 50” quality <=10, <=50% samples have data here), INFO (allele counts, total counts, number of samples with data, somatic or not, validated, etc) 20

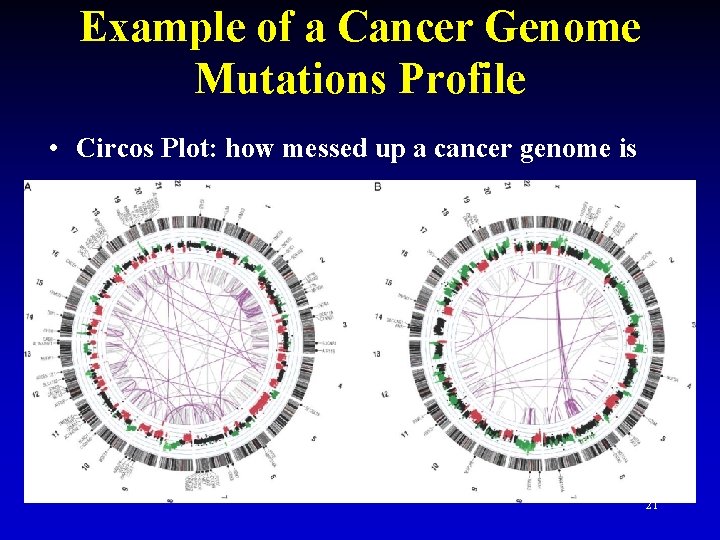
Example of a Cancer Genome Mutations Profile • Circos Plot: how messed up a cancer genome is 21

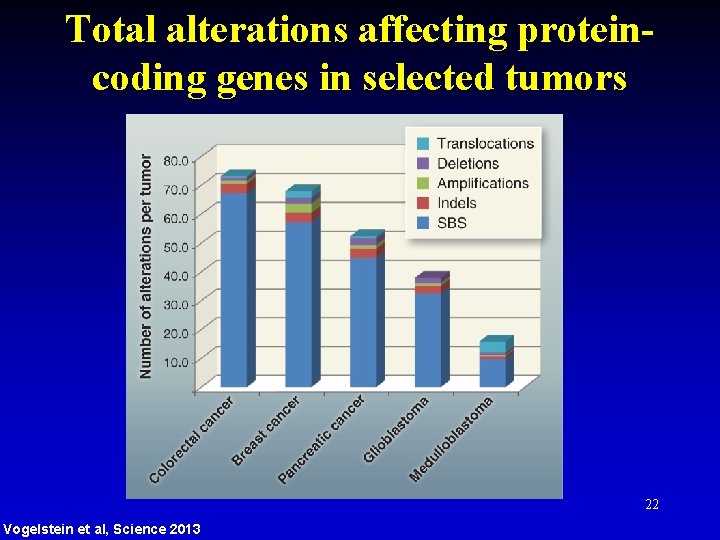
Total alterations affecting proteincoding genes in selected tumors 22 Vogelstein et al, Science 2013

Somatic Mutation Frequency in 3 K Tumor-Normal Pairs • More mutations for tumors facing outside 23

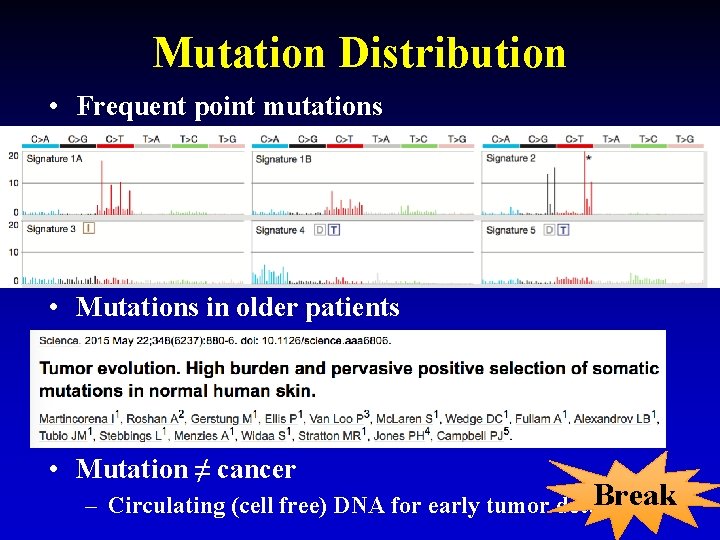
Mutation Distribution • Frequent point mutations • Mutations in older patients • Mutation ≠ cancer Break 24 – Circulating (cell free) DNA for early tumor detection?

TS vs Oncogenes, Go. F vs Lo. F • Tumor suppressors vs oncogenes • Gain of Function (Go. F) or Loss of Function (Lo. F) mutations – Phenotypes • How to tell? – From mutation patterns – From expression patterns – Functional studies • Some genes can be both TS and oncogenes 25

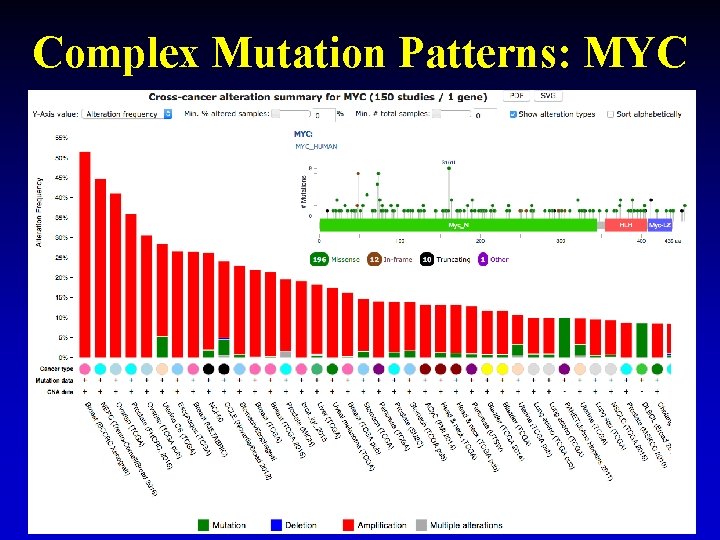
Complex Mutation Patterns: MYC 26

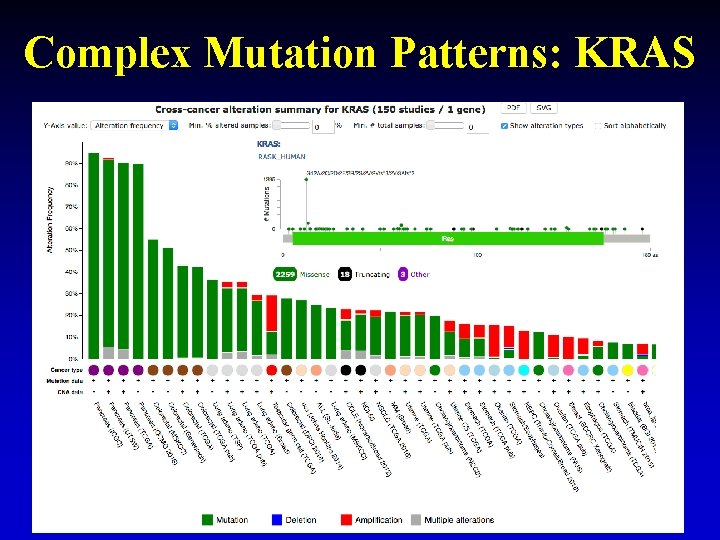
Complex Mutation Patterns: KRAS 27

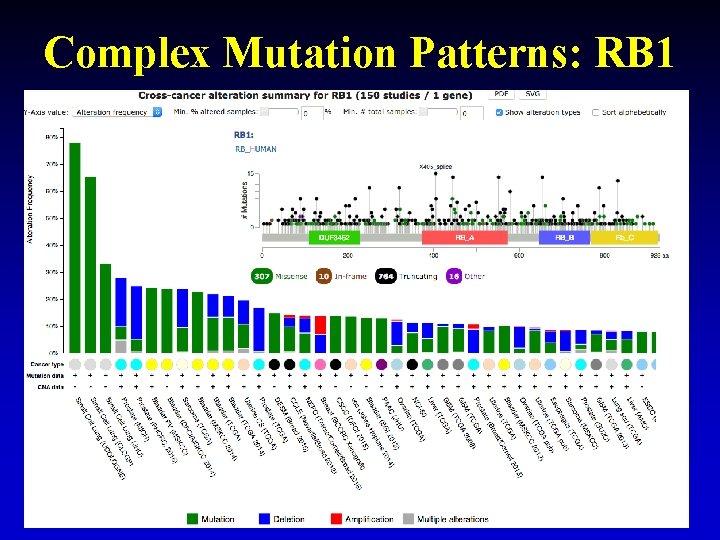
Complex Mutation Patterns: RB 1 28

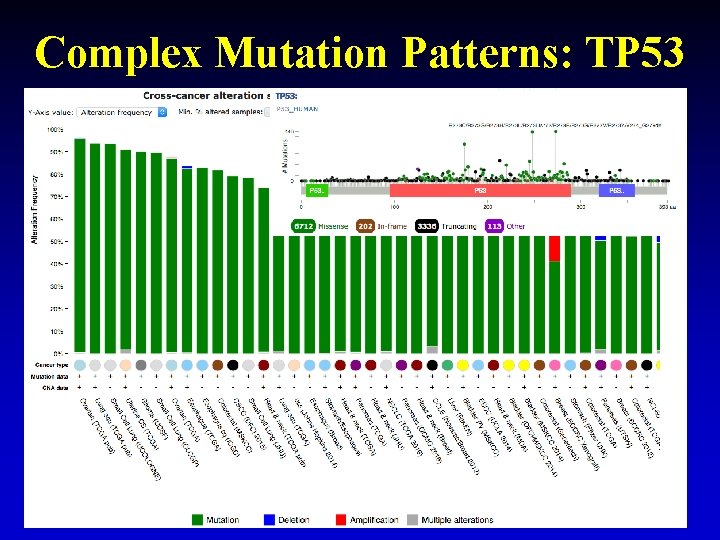
Complex Mutation Patterns: TP 53 29

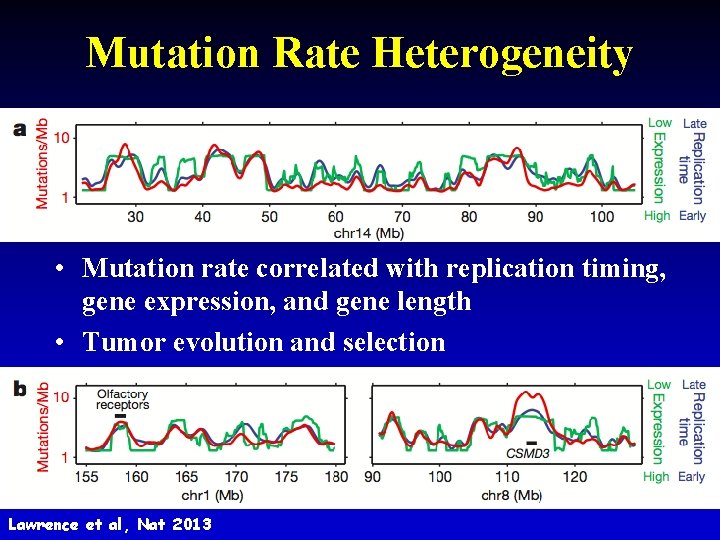
Mutation Rate Heterogeneity • Mutation rate correlated with replication timing, gene expression, and gene length • Tumor evolution and selection 30 Lawrence et al, Nat 2013

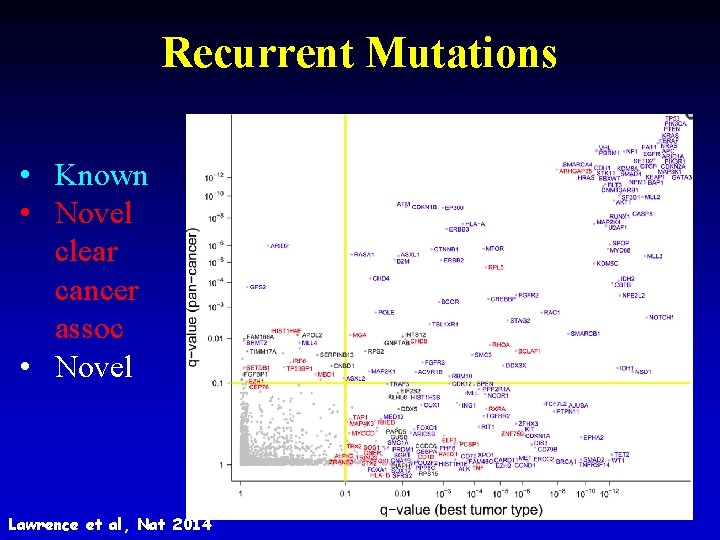
Recurrent Mutations • Known • Novel clear cancer assoc • Novel 31 Lawrence et al, Nat 2014

How Much Should We Sequence? • Need ~200 patients for 20% mutation rate, ~550 pts for 10%, ~1200 pts for 5% mutation rate. • Most driver mutations have been found, pressing need in basic cancer research to study their function • Biggest surprise: mutations on chromatin regulators – – > 50% new and strong cancer driver genes Oncogenes: DNMT 3 A, IDH 1 Tumor Suppressor: MLL, ATRX, ARID 1 A, SNF 5 Both: EZH 2 • Sequencing metastasized or drug resistant tumors might yield insights on tumor progression 32

Resources • MSKCC CBio. Portal – GUI interface for experimental biologists • Broad Fire. Hose – API for accessing processed TCGA data • UCSC CGHub – API for accessing raw and processed cancer data • Sanger COSMIC – Catalog of Somatic Mutations in Cancer • Many also provide software tools 33

Summary • • • Different sequencing approaches Gene Expression, tumor sub-typing Survival analysis: KM vs Cox Regression Different mutation types and distributions Gain or loss of function mutations Tumor suppressor vs oncogenes 34

Acknolwedgement • • • Aleksandar Milosavljevic Kristin Sainani Linda Staub & Alexandros Gekenidis Yin Bun Cheung, Paul Yip John Pack Cheng Li Xujun Wang Bo Li Peng Jiang 35
 Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Genome-to-genome distance calculator
Genome-to-genome distance calculator Human genome project code
Human genome project code Hierarchical shotgun sequencing vs whole genome
Hierarchical shotgun sequencing vs whole genome Genome sequencing
Genome sequencing Per partes
Per partes Genome sequencing
Genome sequencing Shotgun sequencing
Shotgun sequencing Alex liu cecilia liu
Alex liu cecilia liu Líu líu lo lo ta ca hát say sưa
Líu líu lo lo ta ca hát say sưa Genome adalah
Genome adalah Savant genome browser
Savant genome browser Human genome features
Human genome features Artemis genome
Artemis genome 1000 genome project
1000 genome project Translation
Translation Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Plant genome research program
Plant genome research program Human genome size
Human genome size Epidna
Epidna Genome.gov
Genome.gov Genome mapping
Genome mapping Genome
Genome National human genome research institute
National human genome research institute Human genome project
Human genome project Vntrs vs strs
Vntrs vs strs Raspberry pi 3 model b specifications pdf
Raspberry pi 3 model b specifications pdf Genome project
Genome project Genome research limited
Genome research limited Genome assembly
Genome assembly Patric bioinformatics
Patric bioinformatics Euphenics
Euphenics Mash genome
Mash genome Biology dna
Biology dna