Evolution and Population Genetics Xiaole Shirley Liu STAT






- Slides: 31

Evolution and Population Genetics Xiaole Shirley Liu STAT 115 / STAT 215

Evolution • Evolution is a gradual change in genetic makeup from one generation to the next • Evolution: Nonrandom • Natural Selection process • Mutation Random • Genetic Drift processes … • Natural selection and genetic drift are the two most important causes of allele substitution in populations 2

Evolution • Evolution creates species-specific and population-specific differences • Are they all selected for advantages to the species or population? Some definitions: • Locus: position on chromosome where a sequence or a gene is located • Allele: alternative form of DNA on a locus • Written as A vs a, or A vs B 3

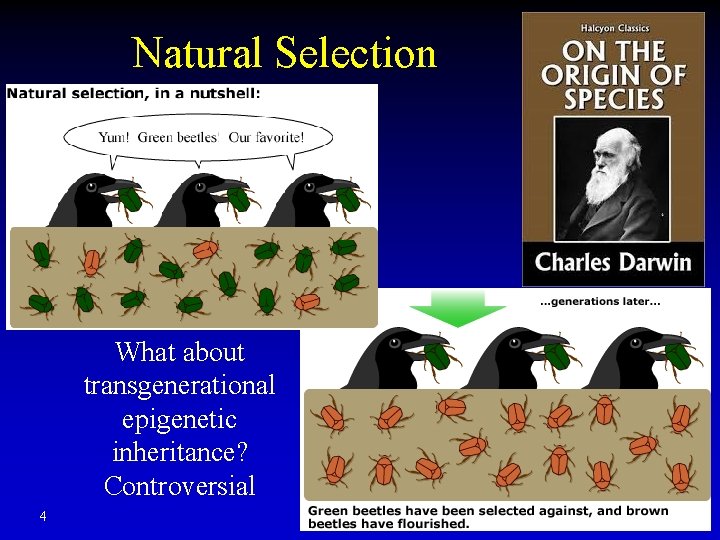
Natural Selection What about transgenerational epigenetic inheritance? Controversial 4

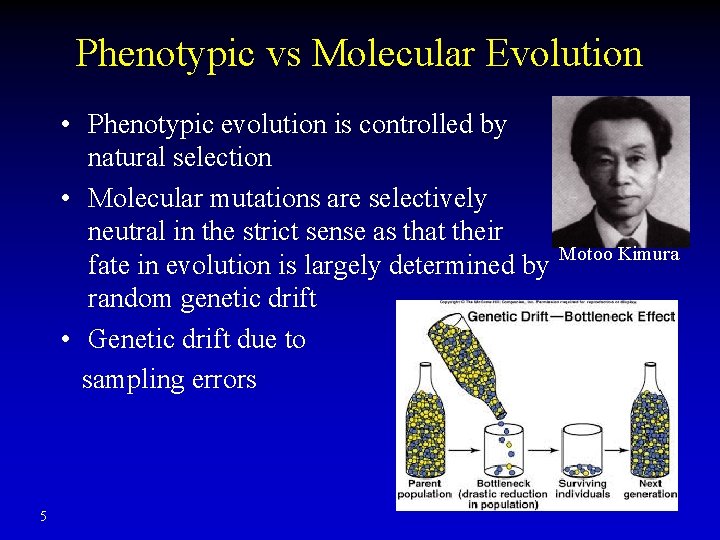
Phenotypic vs Molecular Evolution • Phenotypic evolution is controlled by natural selection • Molecular mutations are selectively neutral in the strict sense as that their fate in evolution is largely determined by random genetic drift • Genetic drift due to sampling errors 5 Motoo Kimura

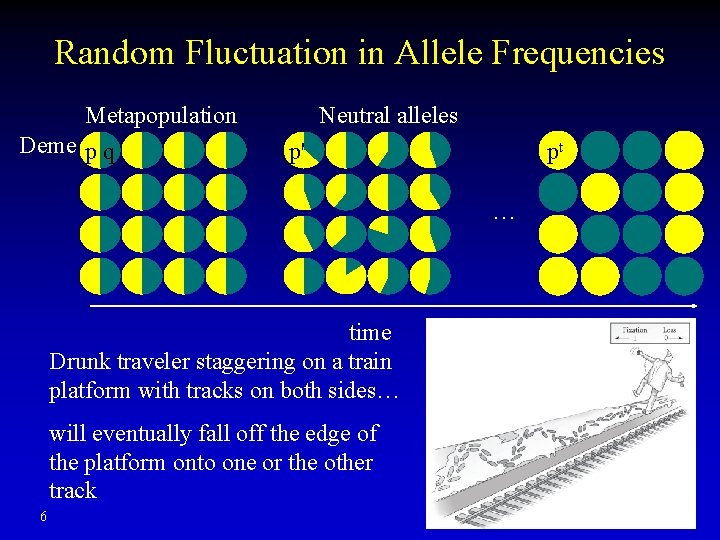
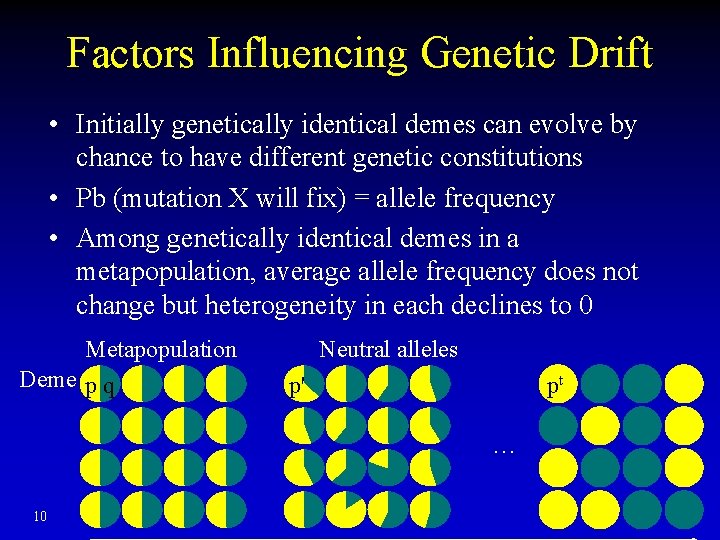
Random Fluctuation in Allele Frequencies Metapopulation Deme p q Neutral alleles p' pt … time Drunk traveler staggering on a train platform with tracks on both sides… will eventually fall off the edge of the platform onto one or the other track 6

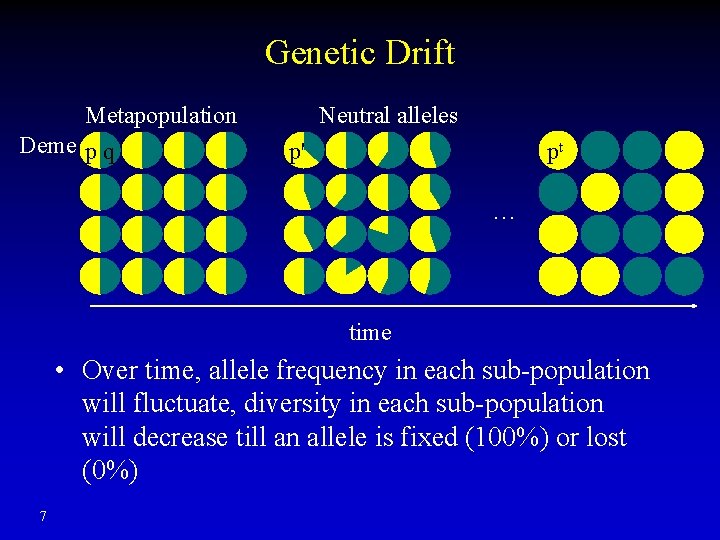
Genetic Drift Metapopulation Deme p q Neutral alleles p' pt … time • Over time, allele frequency in each sub-population will fluctuate, diversity in each sub-population will decrease till an allele is fixed (100%) or lost (0%) 7

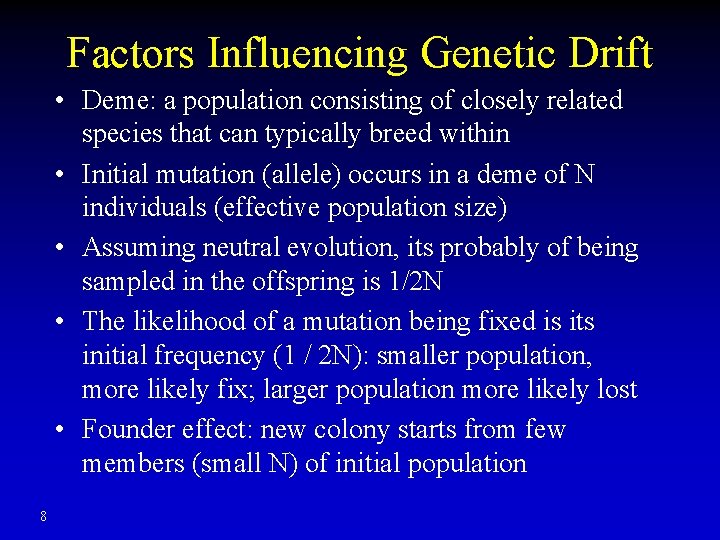
Factors Influencing Genetic Drift • Deme: a population consisting of closely related species that can typically breed within • Initial mutation (allele) occurs in a deme of N individuals (effective population size) • Assuming neutral evolution, its probably of being sampled in the offspring is 1/2 N • The likelihood of a mutation being fixed is its initial frequency (1 / 2 N): smaller population, more likely fix; larger population more likely lost • Founder effect: new colony starts from few members (small N) of initial population 8

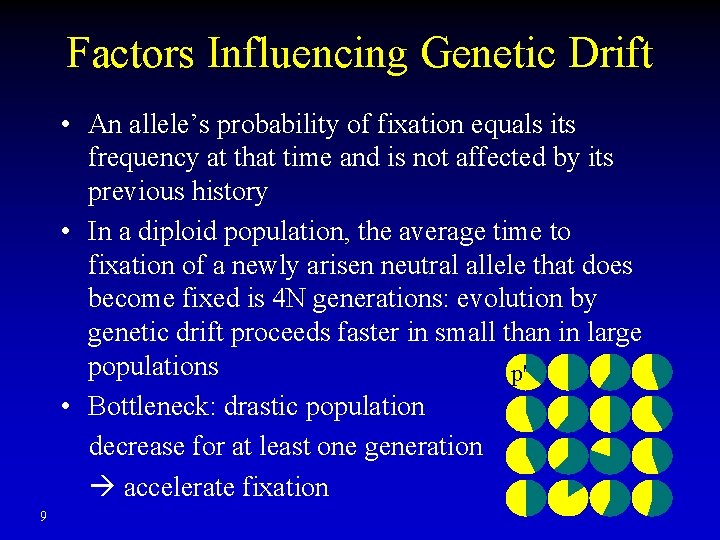
Factors Influencing Genetic Drift • An allele’s probability of fixation equals its frequency at that time and is not affected by its previous history • In a diploid population, the average time to fixation of a newly arisen neutral allele that does become fixed is 4 N generations: evolution by genetic drift proceeds faster in small than in large populations p' • Bottleneck: drastic population decrease for at least one generation accelerate fixation 9

Factors Influencing Genetic Drift • Initially genetically identical demes can evolve by chance to have different genetic constitutions • Pb (mutation X will fix) = allele frequency • Among genetically identical demes in a metapopulation, average allele frequency does not change but heterogeneity in each declines to 0 Metapopulation Deme p q Neutral alleles p' pt … 10

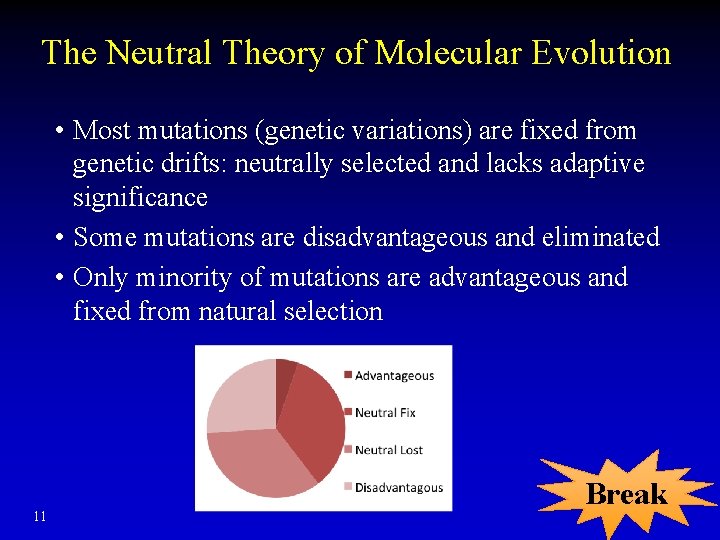
The Neutral Theory of Molecular Evolution • Most mutations (genetic variations) are fixed from genetic drifts: neutrally selected and lacks adaptive significance • Some mutations are disadvantageous and eliminated • Only minority of mutations are advantageous and fixed from natural selection 11 Break

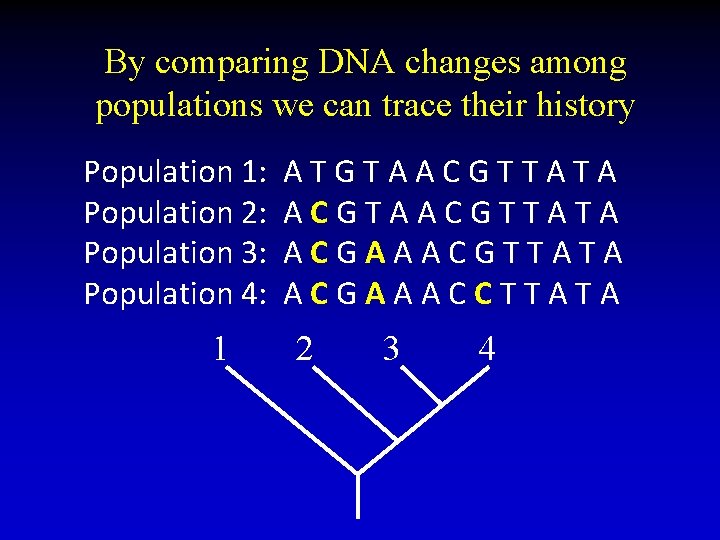
By comparing DNA changes among populations we can trace their history Population 1: Population 2: Population 3: Population 4: 1 ATGTAACGTTATA ACGAAACGTTATA ACGAAACCTTATA 2 3 4

From Phylogeny to Selection • The protein-coding portion of DNA has synonymous and nonsynonymous substitutions. Thus, some DNA changes do not have corresponding protein changes. • If the synonymous substitution rate (d. S) is greater than the nonsynonymous substitution rate (d. N), the DNA sequence is under negative (purifying) selection. • If d. S < d. N, positive selection occurs. E. g. a duplicated gene may evolve rapidly to assume new functions. 13

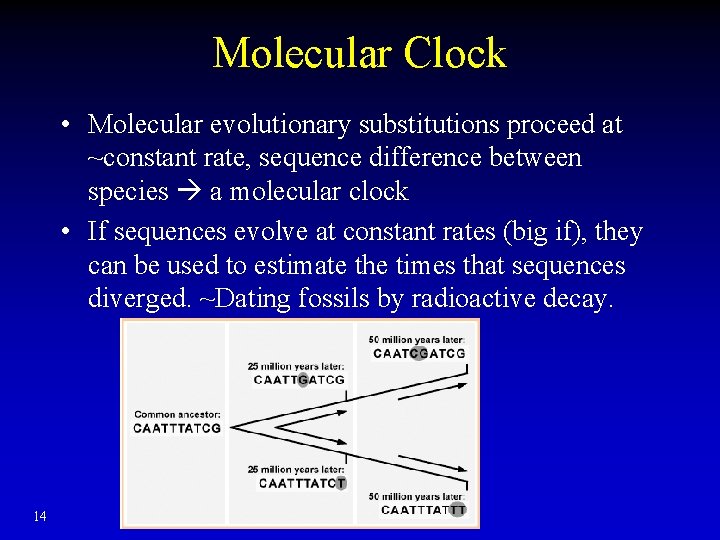
Molecular Clock • Molecular evolutionary substitutions proceed at ~constant rate, sequence difference between species a molecular clock • If sequences evolve at constant rates (big if), they can be used to estimate the times that sequences diverged. ~Dating fossils by radioactive decay. 14

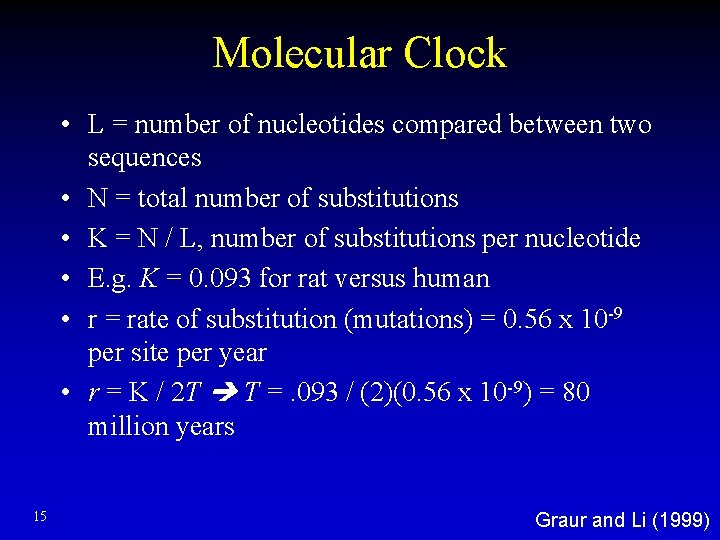
Molecular Clock • L = number of nucleotides compared between two sequences • N = total number of substitutions • K = N / L, number of substitutions per nucleotide • E. g. K = 0. 093 for rat versus human • r = rate of substitution (mutations) = 0. 56 x 10 -9 per site per year • r = K / 2 T T =. 093 / (2)(0. 56 x 10 -9) = 80 million years 15 Graur and Li (1999)

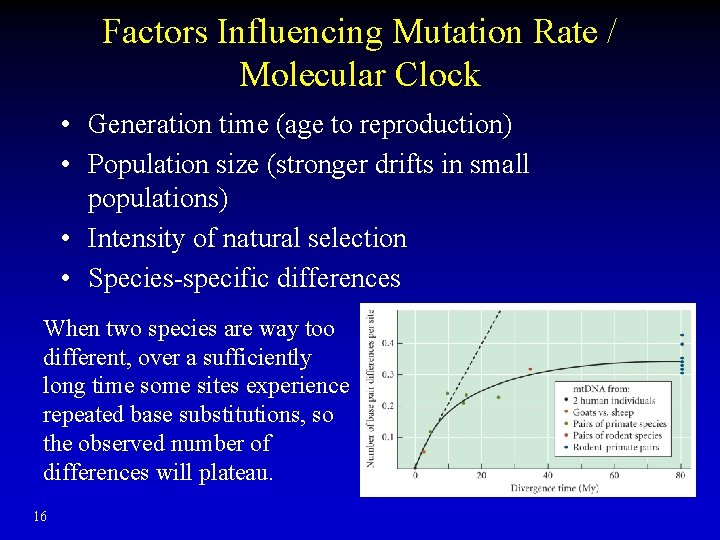
Factors Influencing Mutation Rate / Molecular Clock • Generation time (age to reproduction) • Population size (stronger drifts in small populations) • Intensity of natural selection • Species-specific differences When two species are way too different, over a sufficiently long time some sites experience repeated base substitutions, so the observed number of differences will plateau. 16

Factors Influencing Mutation Rate / Molecular Clock • Generation time (age to reproduction) • Population size (stronger drifts in small populations) • Intensity of natural selection • Species-specific differences • Change in protein function 17

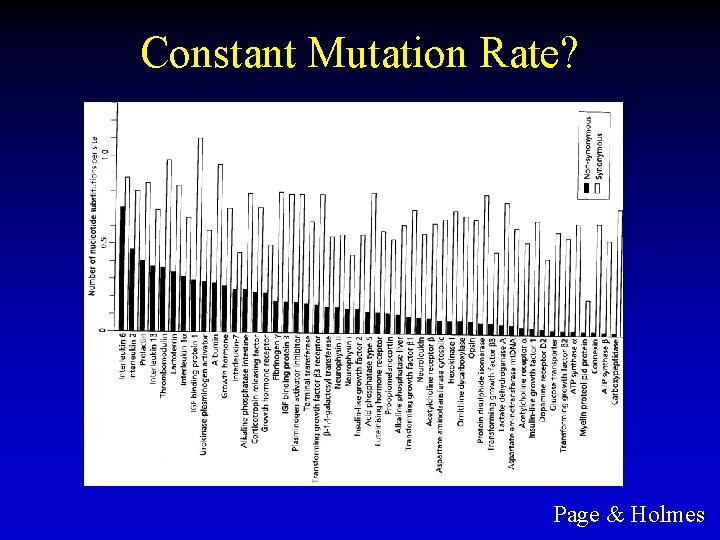
Constant Mutation Rate? Page & Holmes

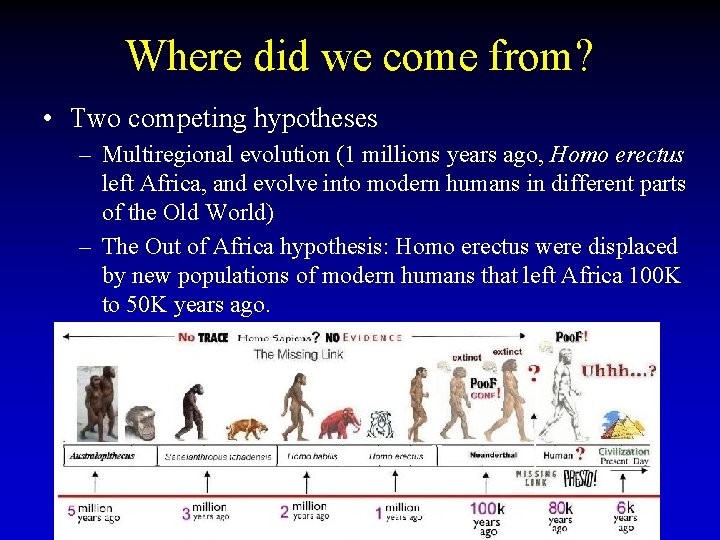
Where did we come from? • Two competing hypotheses – Multiregional evolution (1 millions years ago, Homo erectus left Africa, and evolve into modern humans in different parts of the Old World) – The Out of Africa hypothesis: Homo erectus were displaced by new populations of modern humans that left Africa 100 K to 50 K years ago.

• National Geographic Story Jan 2014 • If a fragment of DNA is shared by Neanderthals and non-Africans, but not Africans or other primates, it is likely to be a Neanderthal heirloom. • People living outside Africa carries 1 -4% of Neanderthal DNA (skin, hair, etc). 20 Break

Polymorphism • Polymorphism: sites/genes with “common” variation, less common allele frequency >= 1%, otherwise called rare variant and not polymorphic • Single Nucleotide Polymorphism – Come from DNA-replication mistake individual germ line cell, then transmitted – ~90% of human genetic variation • Copy number variations – May or may not be genetic 21 STAT 115

Why Should We Care • Disease gene discovery – Association studies, e. g. certain SNPs are susceptible for diabetes – Chromosome aberrations, duplication / deletion might cause cancer • Personalized Medicine – Drug only effective if you have one allele 22 STAT 115

SNP Distribution • Most common, 1 SNP / 100 -300 bp – Balance between mutation introduction rate and polymorphism lost rate – Most mutations lost within a few generations • 2/3 are CT differences • In non-coding regions, often less SNPs at more conserved regions • In coding regions, often more synonymous than non-synonymous SNPs 23 STAT 115

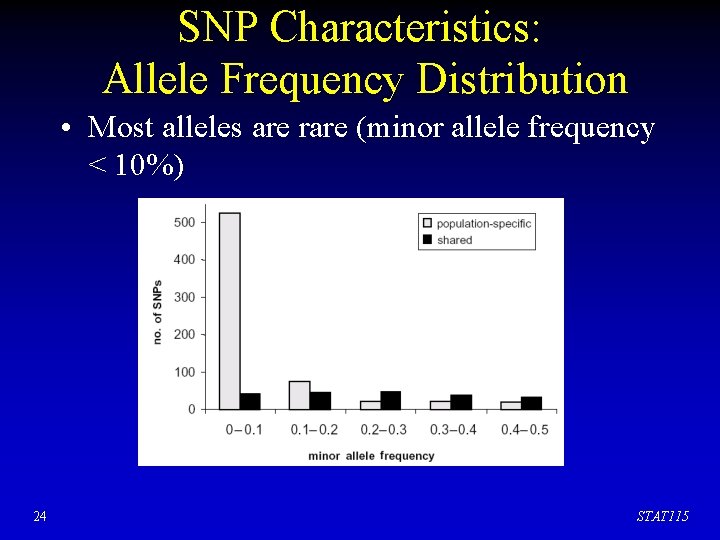
SNP Characteristics: Allele Frequency Distribution • Most alleles are rare (minor allele frequency < 10%) 24 STAT 115

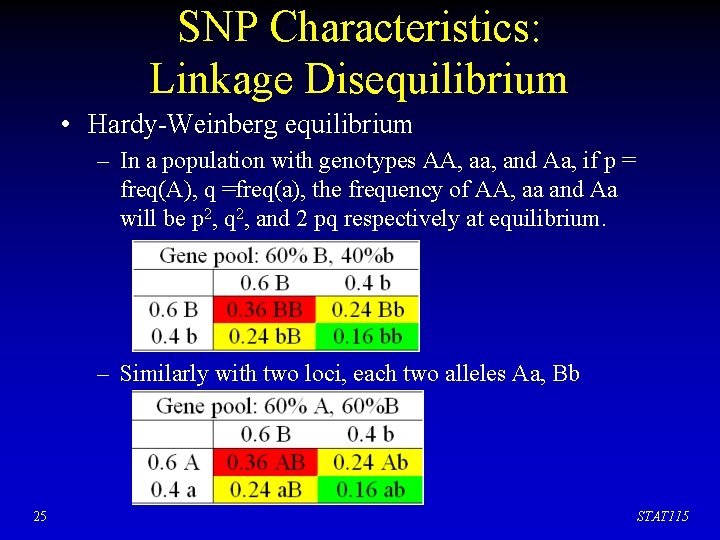
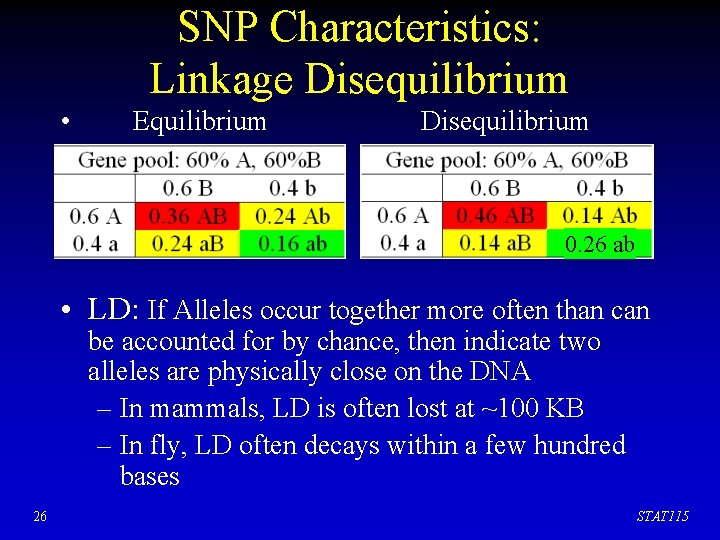
SNP Characteristics: Linkage Disequilibrium • Hardy-Weinberg equilibrium – In a population with genotypes AA, aa, and Aa, if p = freq(A), q =freq(a), the frequency of AA, aa and Aa will be p 2, q 2, and 2 pq respectively at equilibrium. – Similarly with two loci, each two alleles Aa, Bb 25 STAT 115

SNP Characteristics: Linkage Disequilibrium • Equilibrium Disequilibrium 0. 26 ab • LD: If Alleles occur together more often than can be accounted for by chance, then indicate two alleles are physically close on the DNA – In mammals, LD is often lost at ~100 KB – In fly, LD often decays within a few hundred bases 26 STAT 115

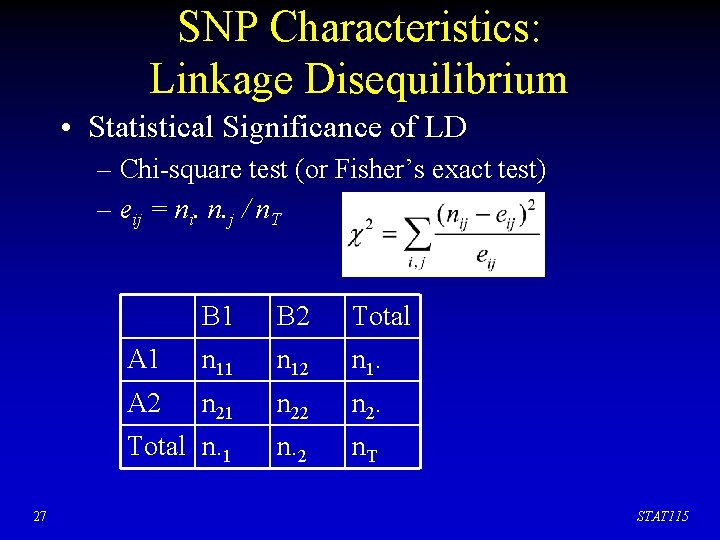
SNP Characteristics: Linkage Disequilibrium • Statistical Significance of LD – Chi-square test (or Fisher’s exact test) – eij = ni. n. j / n. T 27 B 1 B 2 Total A 1 n 12 n 1. A 2 n 21 n 22 n 2. Total n. 1 n. 2 n. T STAT 115

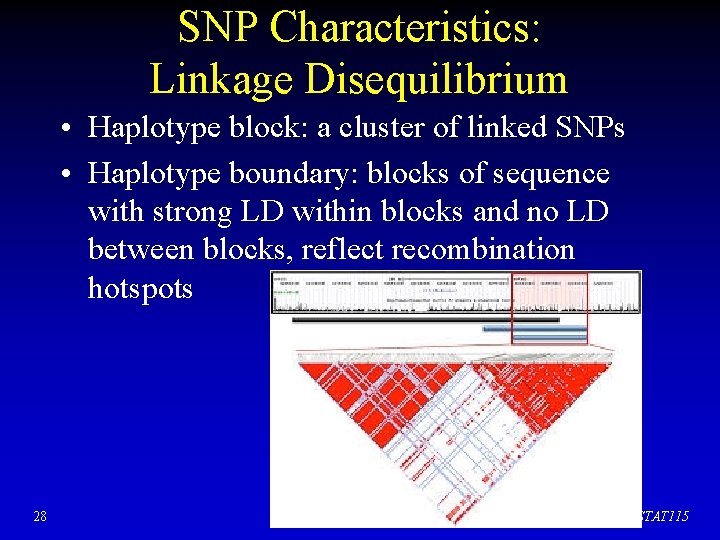
SNP Characteristics: Linkage Disequilibrium • Haplotype block: a cluster of linked SNPs • Haplotype boundary: blocks of sequence with strong LD within blocks and no LD between blocks, reflect recombination hotspots 28 STAT 115

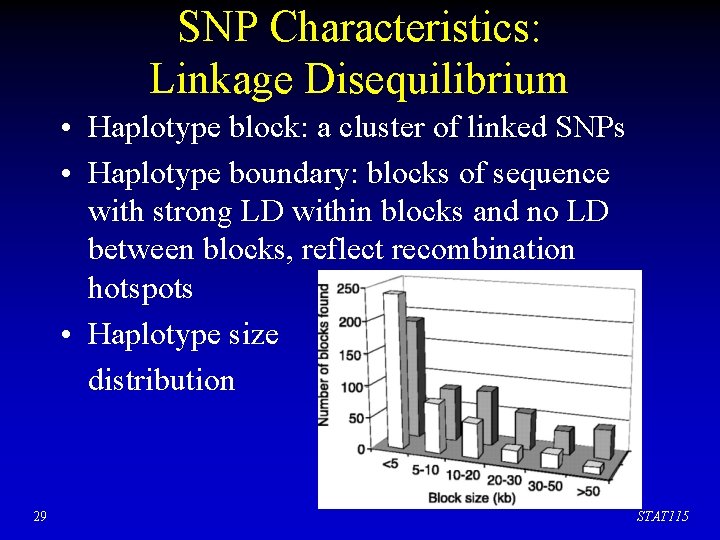
SNP Characteristics: Linkage Disequilibrium • Haplotype block: a cluster of linked SNPs • Haplotype boundary: blocks of sequence with strong LD within blocks and no LD between blocks, reflect recombination hotspots • Haplotype size distribution 29 STAT 115

Summary • Phenotype evolution (natural selection) vs molecular evolution (neutral theory) • Decrease of genetic variation over time • Fixation: population size, probability • Positive and negative selection (d. N / d. S ratio) • Molecular clock and migration patterns • Genome variations: SNP and CNV • Linkage disequilibrium from recombination 30

Acknowledgement • Francisco Ubeda • Jun Liu 31