Differential Gene Expression Xiaole Shirley Liu STAT 115




- Slides: 23

Differential Gene Expression Xiaole Shirley Liu STAT 115 / STAT 215

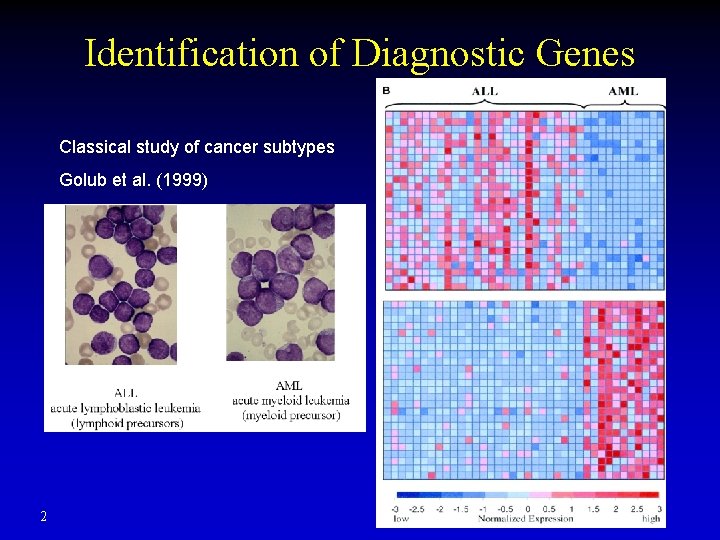
Identification of Diagnostic Genes Classical study of cancer subtypes Golub et al. (1999) 2

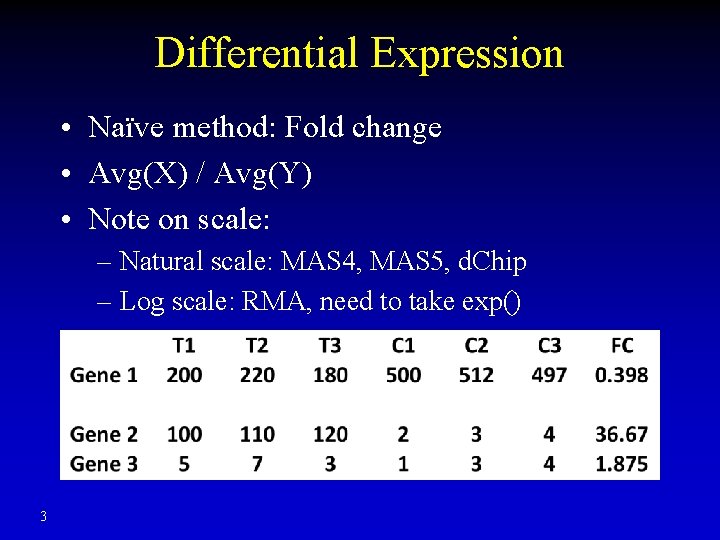
Differential Expression • Naïve method: Fold change • Avg(X) / Avg(Y) • Note on scale: – Natural scale: MAS 4, MAS 5, d. Chip – Log scale: RMA, need to take exp() 3

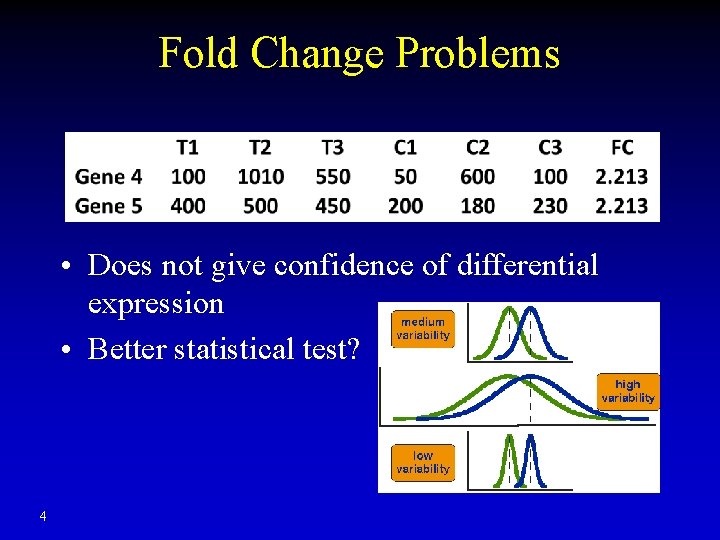
Fold Change Problems • Does not give confidence of differential expression • Better statistical test? 4

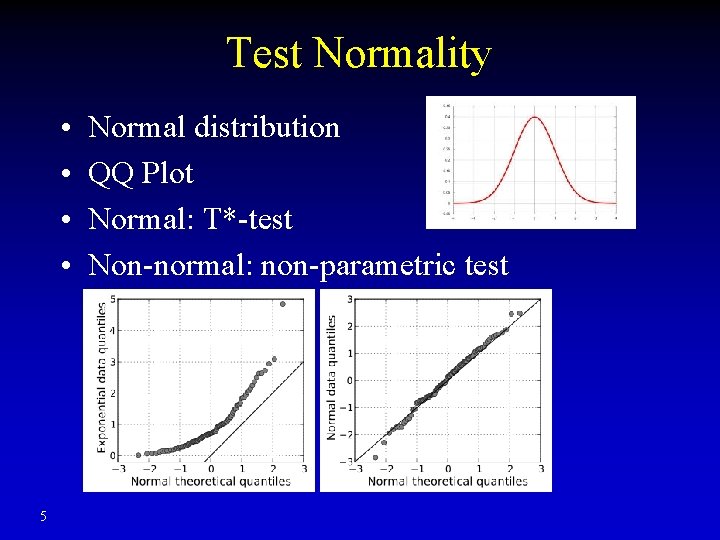
Test Normality • • 5 Normal distribution QQ Plot Normal: T*-test Non-normal: non-parametric test

Wilcoxon Rank Sum Test • Rank all data in row, count sum of ranks TT or TC • Significance calculated from permutation as well • E. g. 10 normal and 10 cancer – Min(T) = 55 – Max(T) = 155 – Significance(T=150) • Check U table (transformation of T) for stat significance • Non-parametric, less power with fewer samples 6 Break

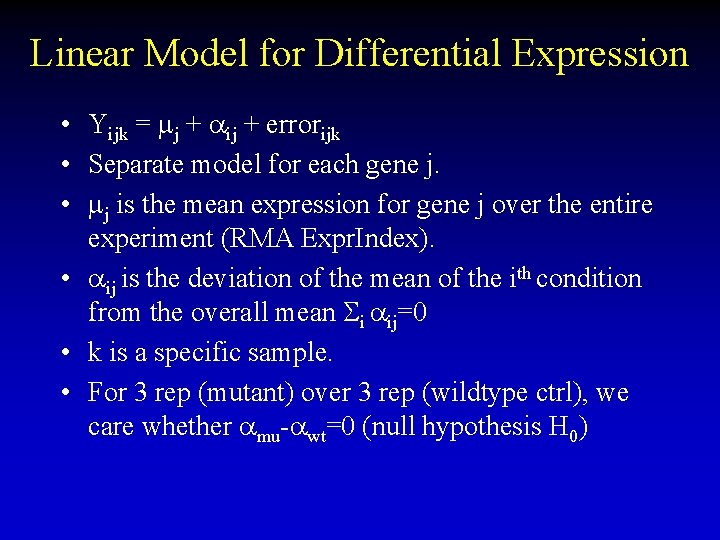
Linear Model for Differential Expression • Yijk = mj + ij + errorijk • Separate model for each gene j. • mj is the mean expression for gene j over the entire experiment (RMA Expr. Index). • ij is the deviation of the mean of the ith condition from the overall mean Si ij=0 • k is a specific sample. • For 3 rep (mutant) over 3 rep (wildtype ctrl), we care whether mu- wt=0 (null hypothesis H 0)

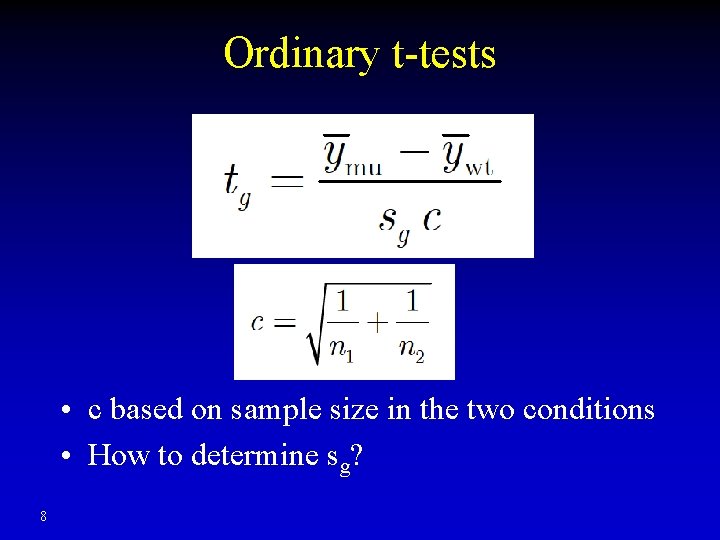
Ordinary t-tests • c based on sample size in the two conditions • How to determine sg? 8

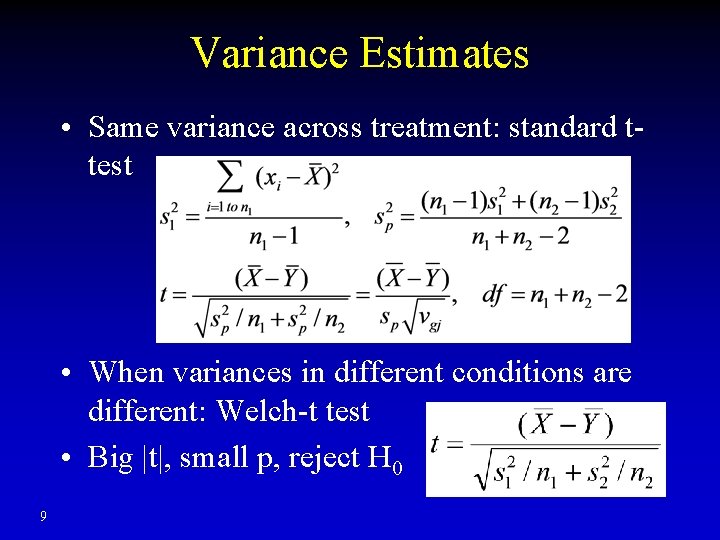
Variance Estimates • Same variance across treatment: standard ttest • When variances in different conditions are different: Welch-t test • Big |t|, small p, reject H 0 9

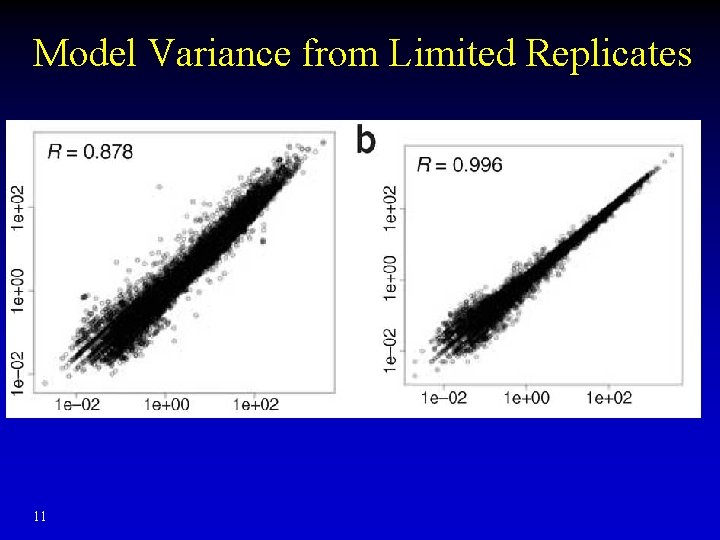
Variance Stabilization • Problem with estimating variance when the sample size is small (e. g. 2 -3 replicates in each condition) • Statistical Analysis of Microarrays (SAM) – Modified t*, increase sg based on sg of other genes on the array (i. e. lowest 5 percentile of sg) • LIMMA: Smyth 2004 – Empirical Bayes: borrow info from all genes 10

Model Variance from Limited Replicates 11

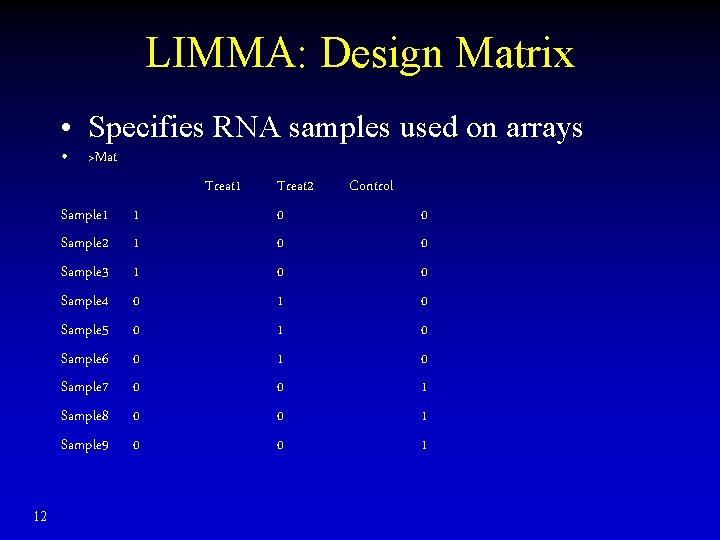
LIMMA: Design Matrix • Specifies RNA samples used on arrays • >Mat Treat 1 Sample 2 Sample 3 Sample 4 Sample 5 Sample 6 Sample 7 Sample 8 Sample 9 12 1 1 1 0 0 0 Treat 2 0 0 0 1 1 1 0 0 0 Control 0 0 0 1 1 1

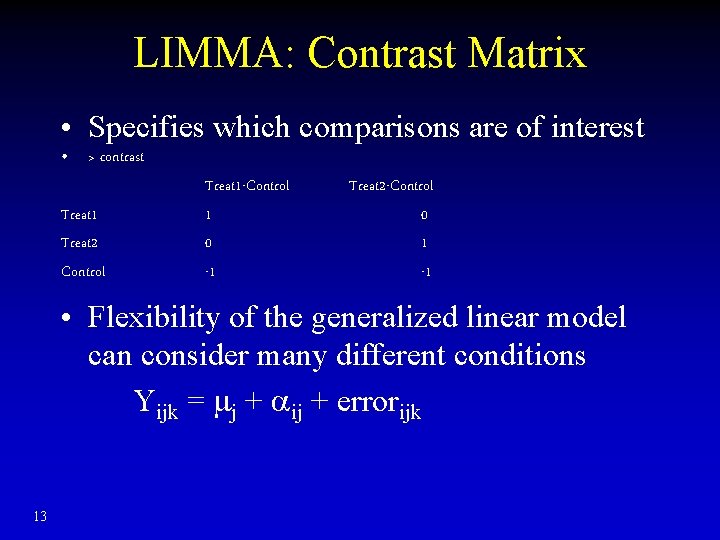
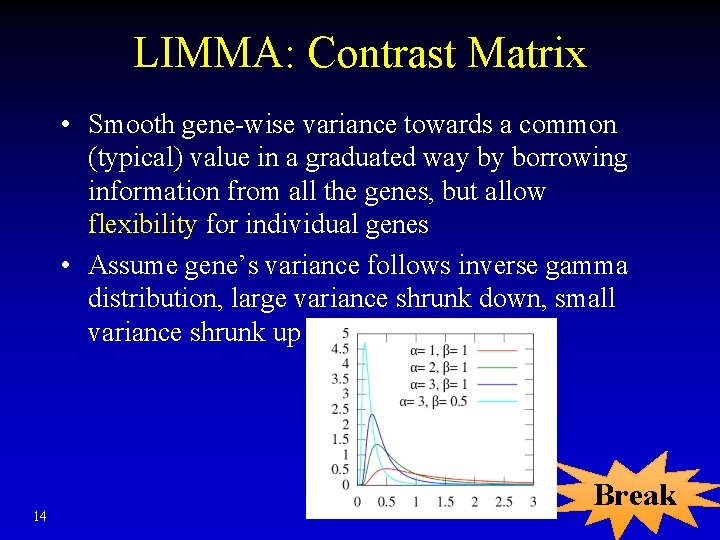
LIMMA: Contrast Matrix • Specifies which comparisons are of interest • > contrast Treat 1 Treat 2 Control Treat 1 -Control 1 0 -1 Treat 2 -Control 0 1 -1 • Flexibility of the generalized linear model can consider many different conditions Yijk = mj + ij + errorijk 13

LIMMA: Contrast Matrix • Smooth gene-wise variance towards a common (typical) value in a graduated way by borrowing information from all the genes, but allow flexibility for individual genes • Assume gene’s variance follows inverse gamma distribution, large variance shrunk down, small variance shrunk up 14 Break

Multiple Hypotheses Testing How many differential genes to report?

Multiple Hypotheses Testing • We test differential expression for every gene with p-value, e. g. 0. 01 • For ~20 K genes on the array, potentially 0. 01 x 20 K = 200 genes wrongly called • H 0: no diff expr; H 1: diff expr – Reject H 0: call something to be differential expressed • Should control family-wise error rate or false discovery rate 16

Family-Wise Error Rate • P(false rejection at most one hypothesis) < α P(no false rejection ) > 1 - α • Bonferroni correction: to control the familywise error rate for testing m hypotheses at level α, we need to control the false rejection rate for each individual test at α/m • If α is 0. 05, for 20 K gene prediction, pvalue cutoff is 0. 05/20 K = 2. 5 E-6 • Too conservative for differential expressed gene selection 17

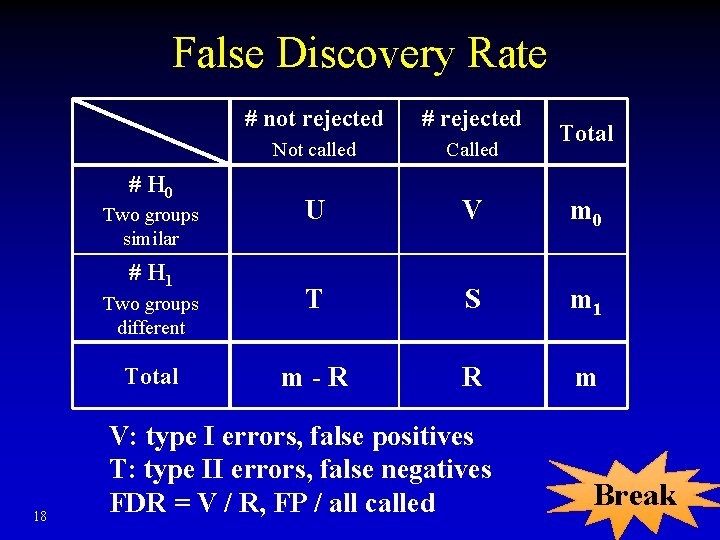
False Discovery Rate # not rejected # rejected Not called Called U V m 0 Two groups different T S m 1 Total m-R R m # H 0 Two groups similar # H 1 18 V: type I errors, false positives T: type II errors, false negatives FDR = V / R, FP / all called Total Break

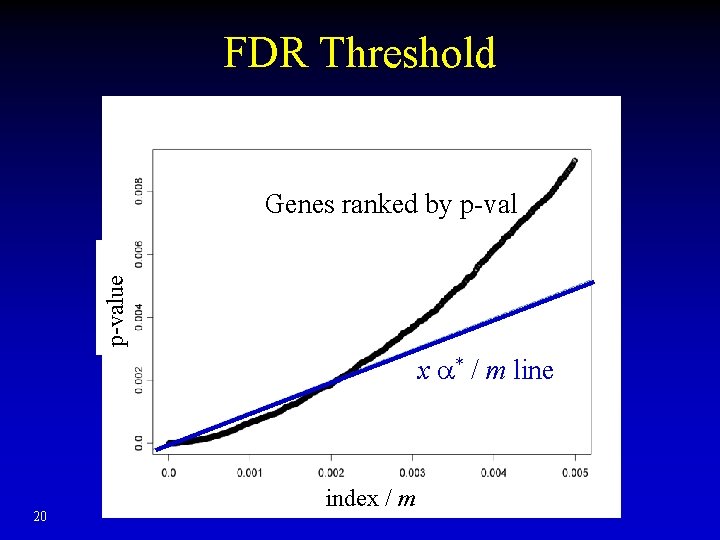
False Discovery Rate • Less conservative than family-wise error rate • Benjamini and Hochberg (1995) method for FDR control, e. g. FDR ≤ * – Assume all the p-val from different tests are independent – Draw all m genes (x), ranked by p-val (y) – Draw line y = x * / m, x = 1…m – Call the genes below the line 19

FDR Threshold p-value Genes ranked by p-val x * / m line 20 index / m

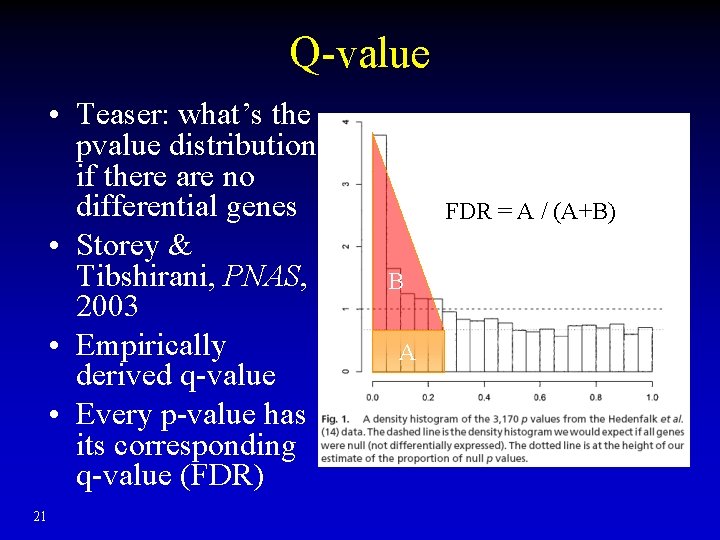
Q-value • Teaser: what’s the pvalue distribution if there are no differential genes • Storey & Tibshirani, PNAS, 2003 • Empirically derived q-value • Every p-value has its corresponding q-value (FDR) 21 FDR = A / (A+B) B A

Practical Use of FDR • Very useful concepts in most of genomics or high throughput studies • Pvalue and FDR are monotonic • Common FDR: 1%, 5%, 10%, also filter by fold change • Give rough estimate of signal / noise and experimental quality • For expression, most people are comfortable with ~500 -2000 differentially expressed genes 22

Summary • Differential Expression – Fold change – T* test on normally distributed data – LIMMA uses hierarchical model to stabilize gene-wise variance • Adjust for multiple hypotheses testing – FWER: conservative – FDR: Benjamini-Hochberg, qvalue 23