SNPs and GWAS Xiaole Shirley Liu STAT 115215

![SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – 24 possible haplotype, SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – 24 possible haplotype,](https://slidetodoc.com/presentation_image/ca1b41eda2ecc0dcdd5a1678695a2e58/image-6.jpg)






- Slides: 28

SNPs and GWAS Xiaole Shirley Liu STAT 115/215, BIO/BST 282

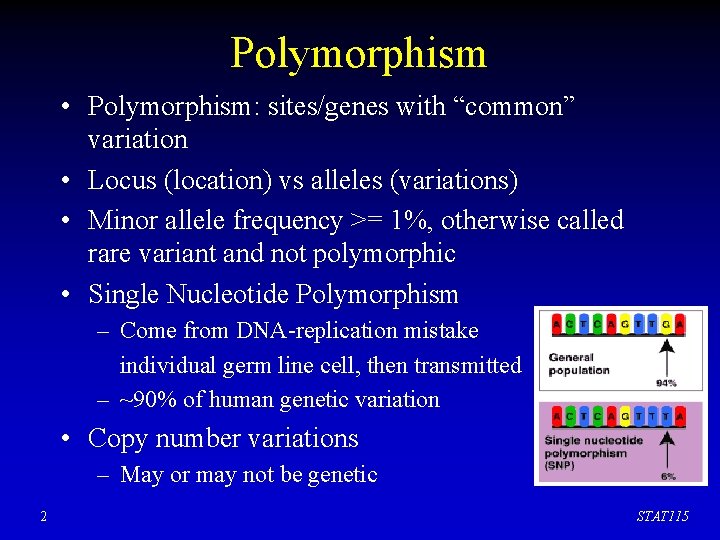
Polymorphism • Polymorphism: sites/genes with “common” variation • Locus (location) vs alleles (variations) • Minor allele frequency >= 1%, otherwise called rare variant and not polymorphic • Single Nucleotide Polymorphism – Come from DNA-replication mistake individual germ line cell, then transmitted – ~90% of human genetic variation • Copy number variations – May or may not be genetic 2 STAT 115

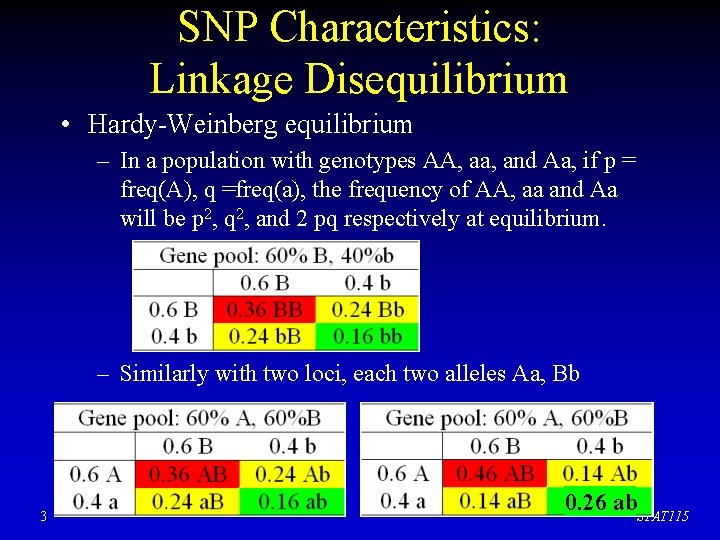
SNP Characteristics: Linkage Disequilibrium • Hardy-Weinberg equilibrium – In a population with genotypes AA, aa, and Aa, if p = freq(A), q =freq(a), the frequency of AA, aa and Aa will be p 2, q 2, and 2 pq respectively at equilibrium. – Similarly with two loci, each two alleles Aa, Bb 3 0. 26 ab. STAT 115

SNP Characteristics: Linkage Disequilibrium • LD: If Alleles occur together more often than can be accounted for by chance, then indicate two alleles are physically close on the DNA • Haplotype block: a cluster of linked SNPs • Haplotype boundary: blocks of sequence with strong LD within blocks and no LD between blocks, reflect recombination hotspots 4 STAT 115

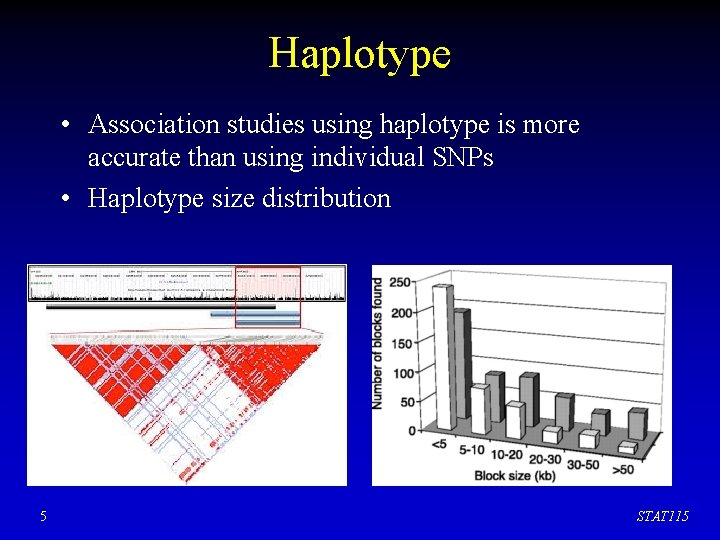
Haplotype • Association studies using haplotype is more accurate than using individual SNPs • Haplotype size distribution 5 STAT 115
![SNP Profiling CT AG T X C AC TA 24 possible haplotype SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – 24 possible haplotype,](https://slidetodoc.com/presentation_image/ca1b41eda2ecc0dcdd5a1678695a2e58/image-6.jpg)
SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – 24 possible haplotype, although often a few common ones explain 90% variations • Tagging (non-redundant) SNPs that capture most variations in haplotypes – reference SNP ID number: rs 12345678 • SNP arrays covering whole genome • Now WES or WGS • Geno-type 2 alleles 6 STAT 115

Association Studies • Association between genetic markers and phenotype – E. g. Cystic Fibrosis ~70% of Cystic Fibrosis patients have a deletion of 3 base pairs resulting in the loss of a phenylalanine amino acid at position 508 of the CFTR gene • Especially, find disease genes, SNP / haplotype markers, for susceptibility prediction and diagnosis 7

Influences individual decisions on life styles, prevention, screening, and treatment 8

Warfarin and CYP 2 C 9: SNPs in Pharmacogenomics • Warfarin anticoagulant drug; CYP 2 C 9 gene metabolizes warfarin. • A patient requiring low dosage warfarin compared to normal population, has an odd ratio of 6. 21 for having 1 variant allele • Subgroup of patients who are poor metabolisers of warfarin are potentially at higher risk of bleeding Break Aithal et al. , 1999, Lancet.

Genome-Wide Association Studies • Quality Control – Unusual similarity between individual – Wrong sex – Trio has non-Mendelian inheritance – Genotyping quality • Two strategies: – Family-based association studies – Population-based case-control association studies 10

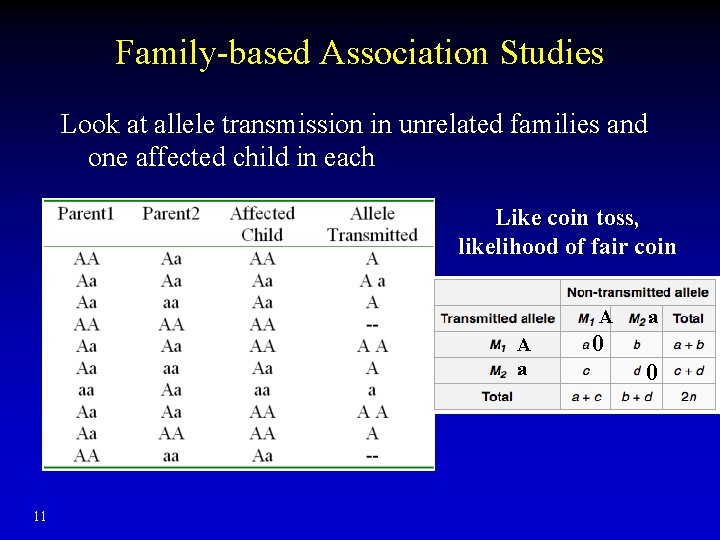
Family-based Association Studies Look at allele transmission in unrelated families and one affected child in each Like coin toss, likelihood of fair coin A A a 11 a 0 0

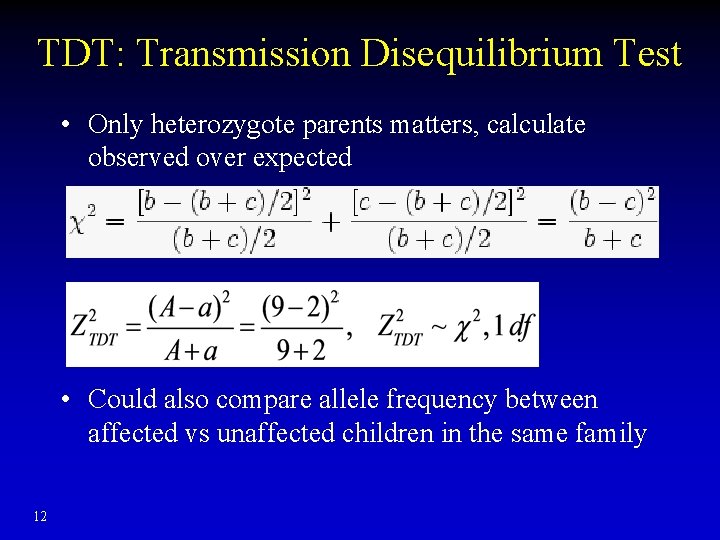
TDT: Transmission Disequilibrium Test • Only heterozygote parents matters, calculate observed over expected • Could also compare allele frequency between affected vs unaffected children in the same family 12

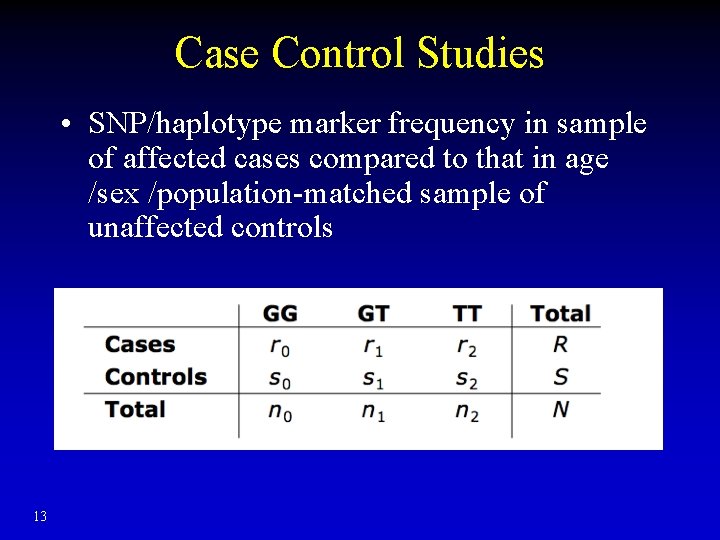
Case Control Studies • SNP/haplotype marker frequency in sample of affected cases compared to that in age /sex /population-matched sample of unaffected controls 13

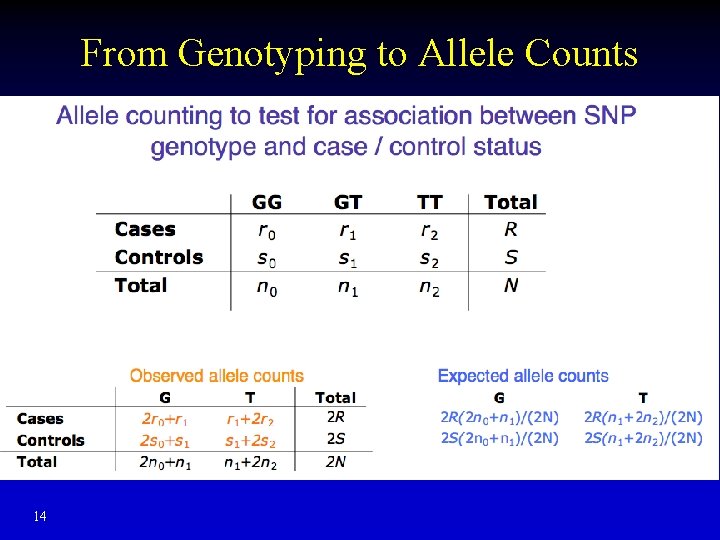
From Genotyping to Allele Counts 14

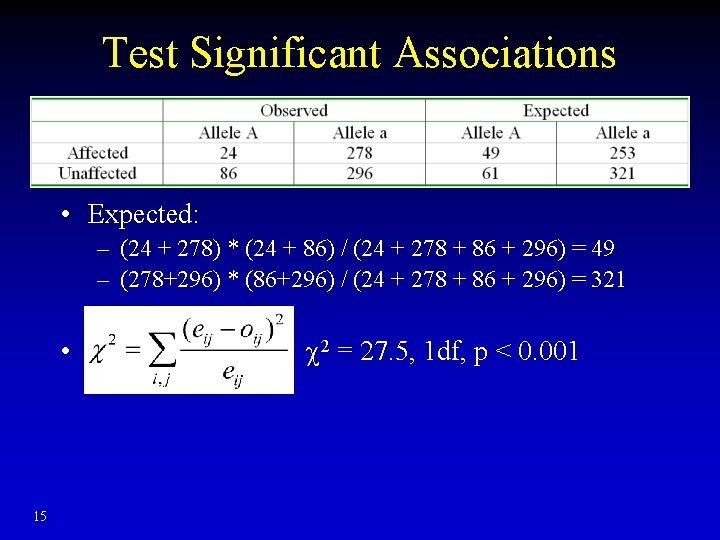
Test Significant Associations • Expected: – (24 + 278) * (24 + 86) / (24 + 278 + 86 + 296) = 49 – (278+296) * (86+296) / (24 + 278 + 86 + 296) = 321 • 2 = 27. 5, 1 df, p < 0. 001 15

16

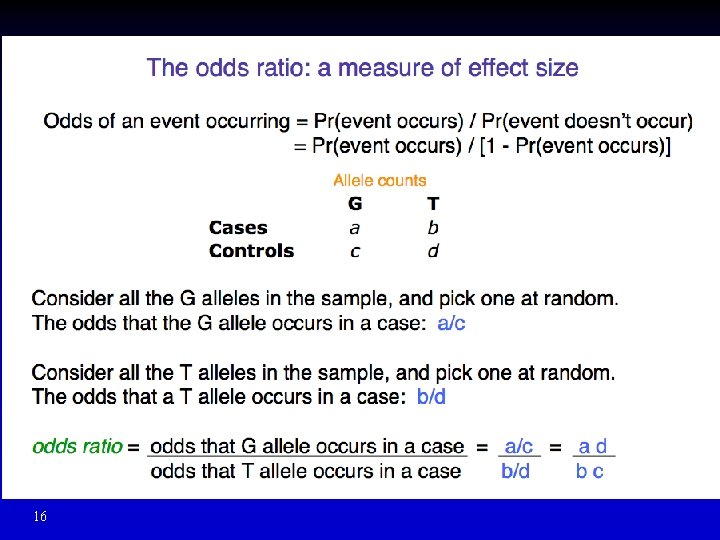
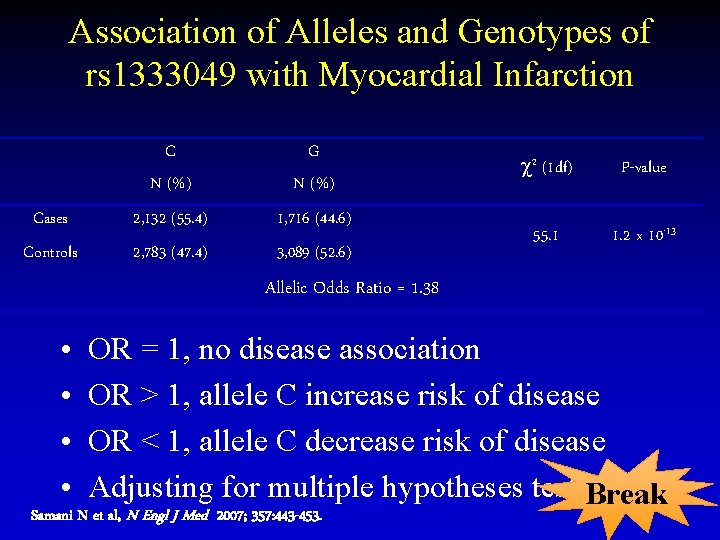
Association of Alleles and Genotypes of rs 1333049 with Myocardial Infarction C N (%) G N (%) Cases 2, 132 (55. 4) 1, 716 (44. 6) Controls 2, 783 (47. 4) 3, 089 (52. 6) 2 (1 df) P-value 55. 1 1. 2 x 10 -13 Allelic Odds Ratio = 1. 38 • • OR = 1, no disease association OR > 1, allele C increase risk of disease OR < 1, allele C decrease risk of disease Adjusting for multiple hypotheses testing? Break Samani N et al, N Engl J Med 2007; 357: 443 -453.

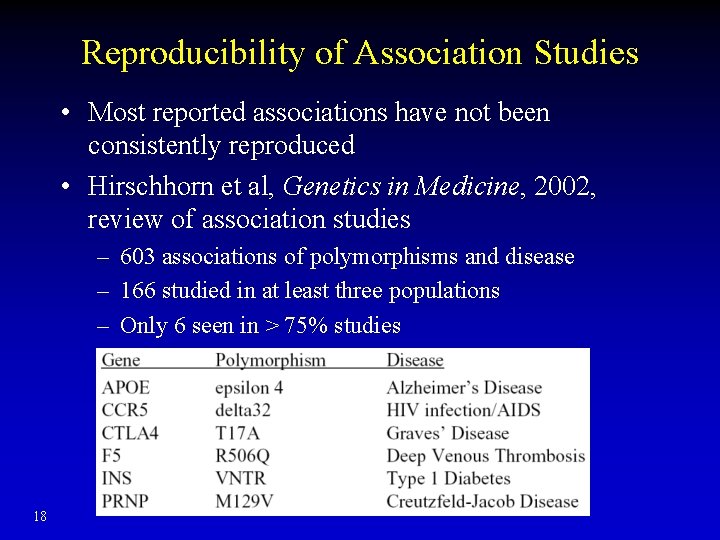
Reproducibility of Association Studies • Most reported associations have not been consistently reproduced • Hirschhorn et al, Genetics in Medicine, 2002, review of association studies – 603 associations of polymorphisms and disease – 166 studied in at least three populations – Only 6 seen in > 75% studies 18

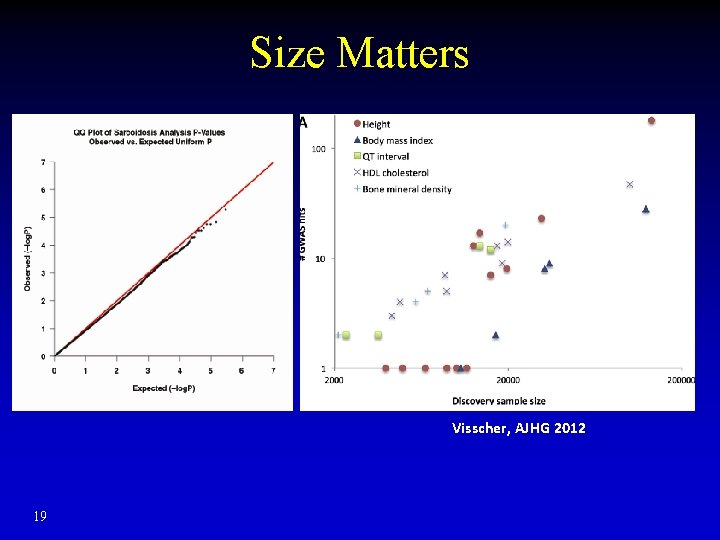
Size Matters Visscher, AJHG 2012 19

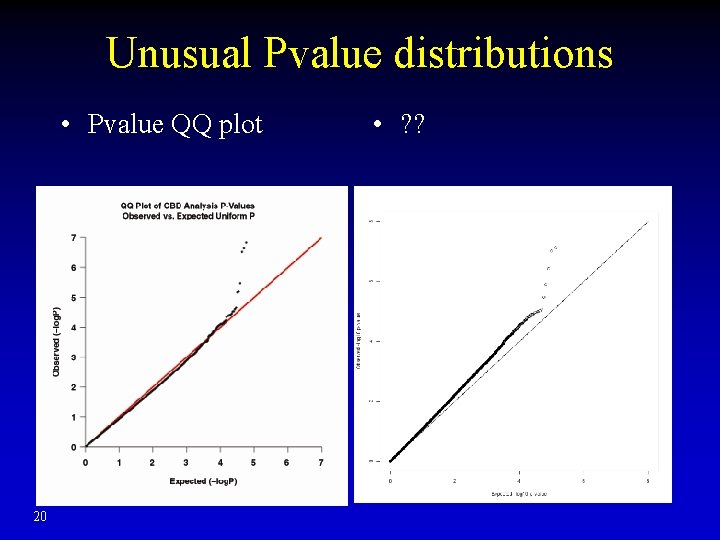
Unusual Pvalue distributions • Pvalue QQ plot 20 • ? ?

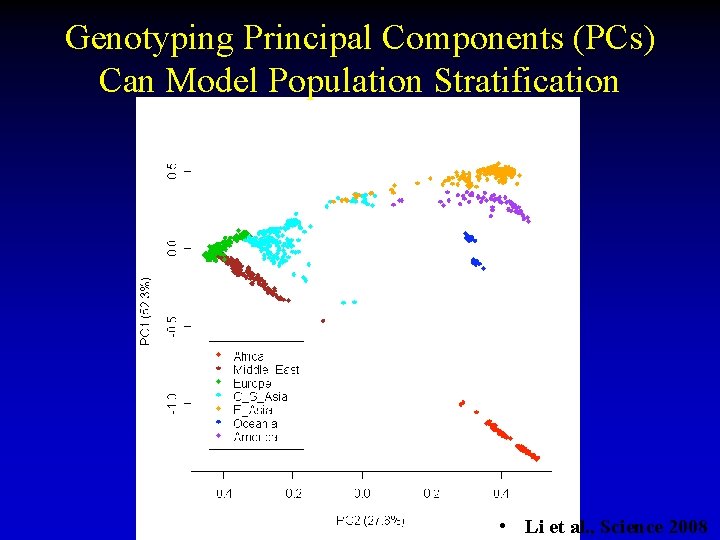
Population Stratification • Population stratification – e. g. some SNP unique to ethnic group – Need to make sure sample groups match – Hidden environmental structure ● ● 21 Two populations have different disease frequency, and different allele frequency. Association picks up the fact they are different populations!

Genotyping Principal Components (PCs) Can Model Population Stratification • Li et al. , Science 2008

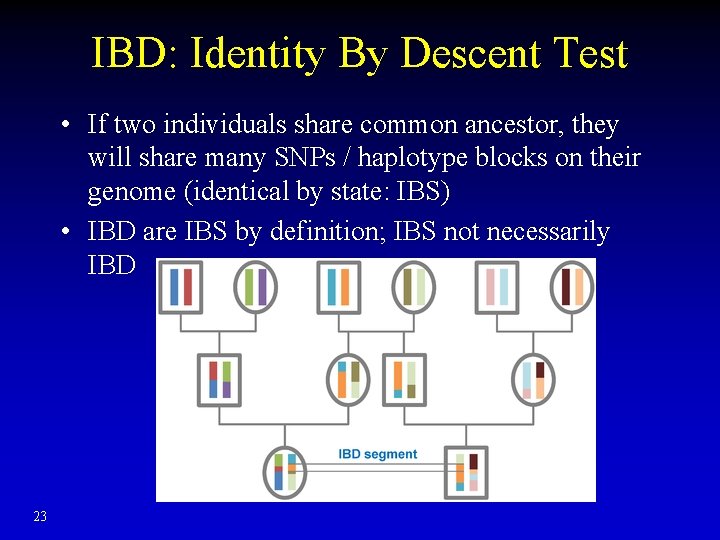
IBD: Identity By Descent Test • If two individuals share common ancestor, they will share many SNPs / haplotype blocks on their genome (identical by state: IBS) • IBD are IBS by definition; IBS not necessarily IBD 23

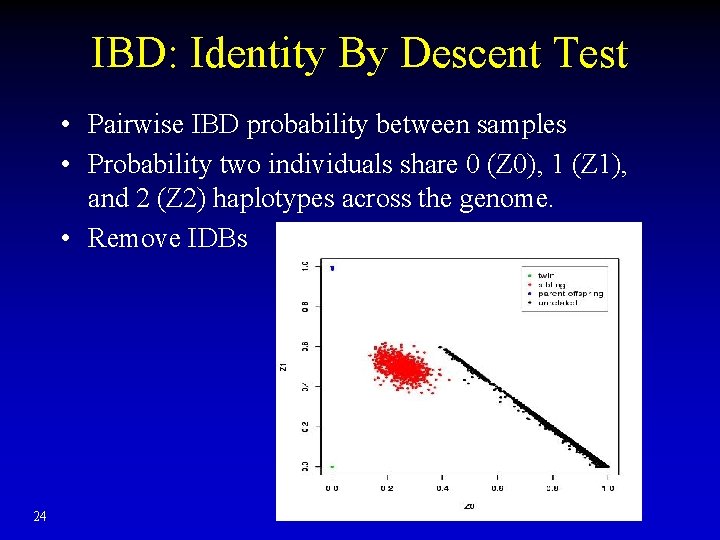
IBD: Identity By Descent Test • Pairwise IBD probability between samples • Probability two individuals share 0 (Z 0), 1 (Z 1), and 2 (Z 2) haplotypes across the genome. • Remove IDBs 24

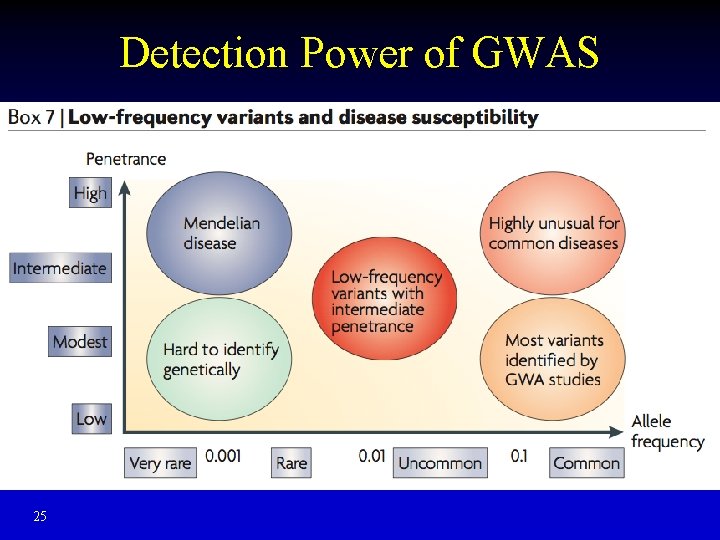
Detection Power of GWAS 25

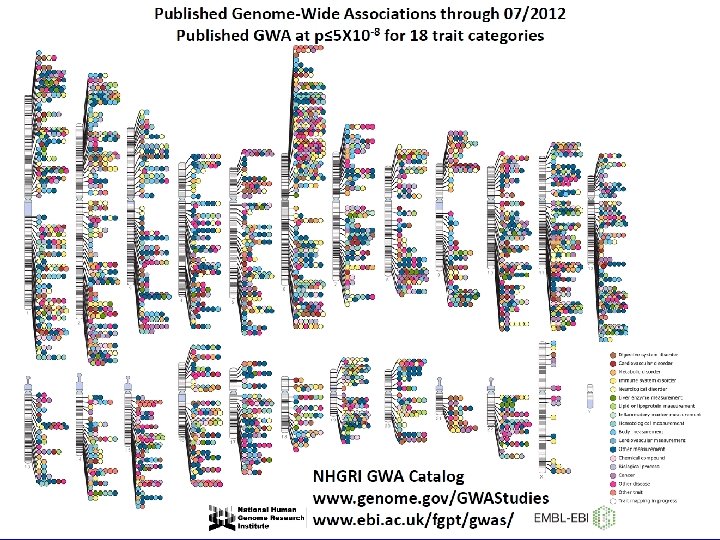
Manolio et al. , Clin Invest 2008

Summary • SNP, LD, haplotypes and tagging SNPs • GWAS: – Family based association studies: TDT transmitted allele to affected child – Case control studies: X-sq (allele frequency difference in case and controls) and OR • Increase reproducibility by size and reduce population stratification and IBD 27 STAT 115

Acknowledgement • • • 28 Francisco Ubeda Jun Liu Tim Niu Bo Li Cheng Li Jim Stankovich Teri Manolio David Evans Guodong Wu Stefano Mont Wei Wang Soumya Raychaudhuri • • • Kenneth Kidd Judith Kidd Glenys Thomson Joel Hirschhorn Greg Gibson Spencer Muse Jim Stankovich Teri Manolio Benjamin Neale Enrico Petretto
 Stat115 harvard
Stat115 harvard Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Gwas power calculation
Gwas power calculation Gwas method
Gwas method Gwas method
Gwas method Alex liu cecilia liu
Alex liu cecilia liu Líu líu lo lo ta ca hát say sưa
Líu líu lo lo ta ca hát say sưa Cornelia m. ruland
Cornelia m. ruland Boy or girl slinger
Boy or girl slinger Sentiment analysis and opinion mining bing liu
Sentiment analysis and opinion mining bing liu The lottery shirley jackson questions
The lottery shirley jackson questions Shirley cvc
Shirley cvc Why would miss strangeworth be interested in whether linda
Why would miss strangeworth be interested in whether linda The lottery by shirley jackson discussion questions
The lottery by shirley jackson discussion questions Shirley ardell mason
Shirley ardell mason Shirley ardell mason
Shirley ardell mason Seven types of ambiguity shirley jackson
Seven types of ambiguity shirley jackson Jak ania nazwała miejsca
Jak ania nazwała miejsca Shirley d'sa
Shirley d'sa Shirley hills primary
Shirley hills primary Shirley dyke
Shirley dyke Charles by shirley jackson vocabulary
Charles by shirley jackson vocabulary Charles by shirley jackson literary elements
Charles by shirley jackson literary elements Shirley toulson about the poet
Shirley toulson about the poet Shirley kavanagh
Shirley kavanagh The lottery by shirley jackson theme
The lottery by shirley jackson theme The lottery shirley jackson symbols
The lottery shirley jackson symbols Shirley c. heim middle school
Shirley c. heim middle school