Haplotypes and GWAS Xiaole Shirley Liu STAT 115STAT

![SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – Possible haplotype: 24 SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – Possible haplotype: 24](https://slidetodoc.com/presentation_image/173bbd7371263442a4e957c34cfe7f29/image-3.jpg)









- Slides: 35

Haplotypes and GWAS Xiaole Shirley Liu STAT 115/STAT 215/

Haplotype • Haplotype block: a cluster of linked SNPs • Haplotype boundary: blocks of sequence with strong LD within blocks and no LD between blocks, reflect recombination hotspots • Association studies using haplotype is more accurate than using individual SNPs • Haplotype size distribution 2 STAT 115
![SNP Profiling CT AG T X C AC TA Possible haplotype 24 SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – Possible haplotype: 24](https://slidetodoc.com/presentation_image/173bbd7371263442a4e957c34cfe7f29/image-3.jpg)
SNP Profiling • [C/T] [A/G] T X C [A/C] [T/A] – Possible haplotype: 24 – In reality, a few common haplotypes explain 90% variations • Tagging SNPs: Redundant – SNPs that capture most variations in haplotypes – removes redundancy 3 STAT 115

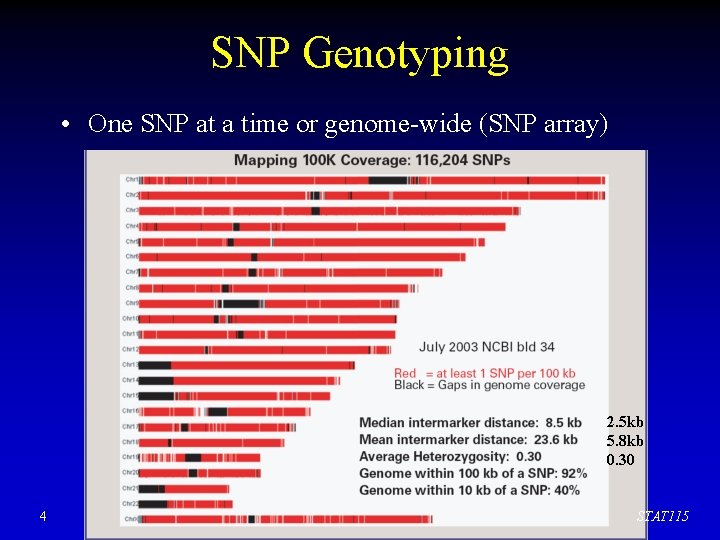
SNP Genotyping • One SNP at a time or genome-wide (SNP array) 2. 5 kb 5. 8 kb 0. 30 4 STAT 115

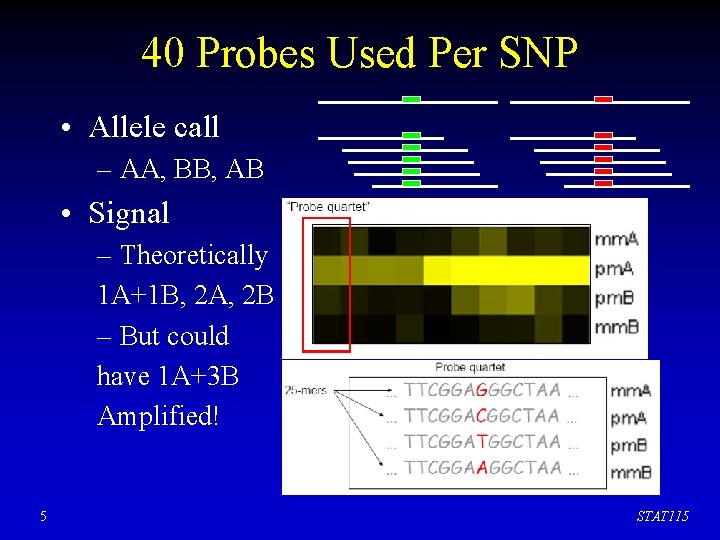
40 Probes Used Per SNP • Allele call – AA, BB, AB • Signal – Theoretically 1 A+1 B, 2 A, 2 B – But could have 1 A+3 B Amplified! 5 STAT 115

Haplotype Inference • Genotyping only tells an individual is e. g. Aa BB Cc, but it doesn’t tell whether haplotype is: ABC + a. Bc, or ABc + a. BC • Haplotype can often be inferred if parental genotype is known – Similar to blood typing, e. g. F: A, M: AB, C: B F: , M: , C: • Otherwise, look at the population genotypes, infer common haplotypes 6 STAT 115

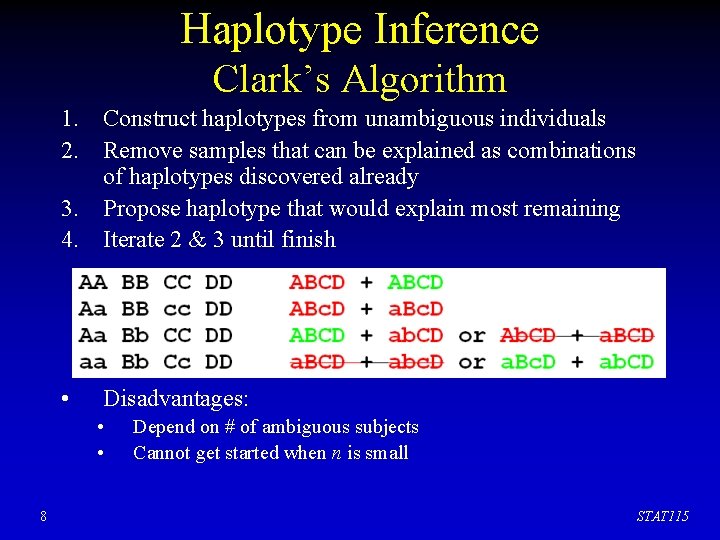
Haplotype Inference Clark’s Algorithm 1. Construct haplotypes from unambiguous individuals 2. Remove samples that can be explained as combinations of haplotypes discovered already 3. Propose haplotype that would explain most remaining 4. Iterate 2 & 3 until finish 7 STAT 115

Haplotype Inference Clark’s Algorithm 1. Construct haplotypes from unambiguous individuals 2. Remove samples that can be explained as combinations of haplotypes discovered already 3. Propose haplotype that would explain most remaining 4. Iterate 2 & 3 until finish • Disadvantages: • • 8 Depend on # of ambiguous subjects Cannot get started when n is small STAT 115

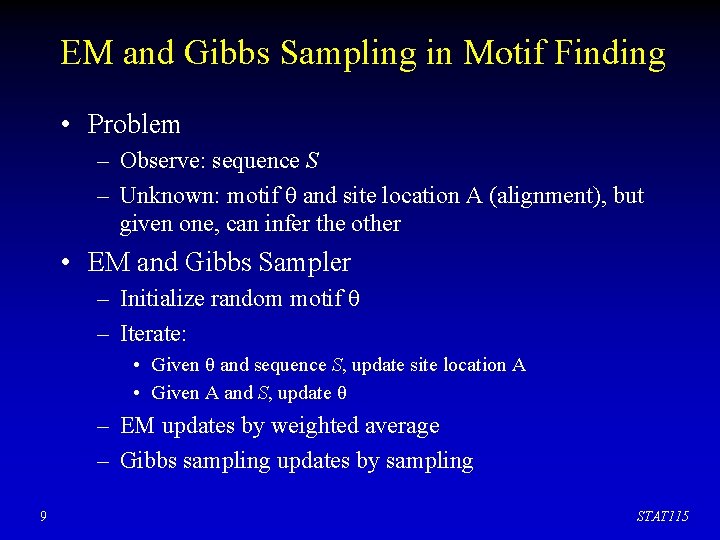
EM and Gibbs Sampling in Motif Finding • Problem – Observe: sequence S – Unknown: motif θ and site location A (alignment), but given one, can infer the other • EM and Gibbs Sampler – Initialize random motif θ – Iterate: • Given θ and sequence S, update site location A • Given A and S, update θ – EM updates by weighted average – Gibbs sampling updates by sampling 9 STAT 115

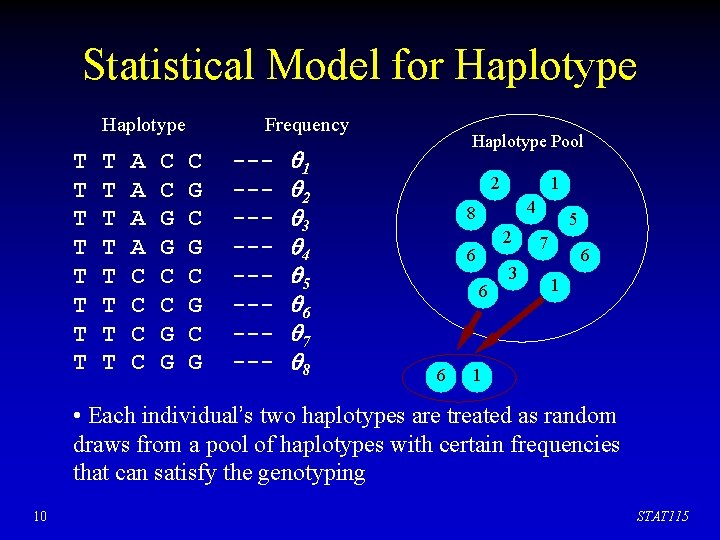
Statistical Model for Haplotype T T T T A A C C C G G Frequency C G C G --------- 1 2 3 4 5 6 7 8 Haplotype Pool 2 1 4 8 2 6 6 6 3 5 7 6 1 1 • Each individual’s two haplotypes are treated as random draws from a pool of haplotypes with certain frequencies that can satisfy the genotyping 10 STAT 115

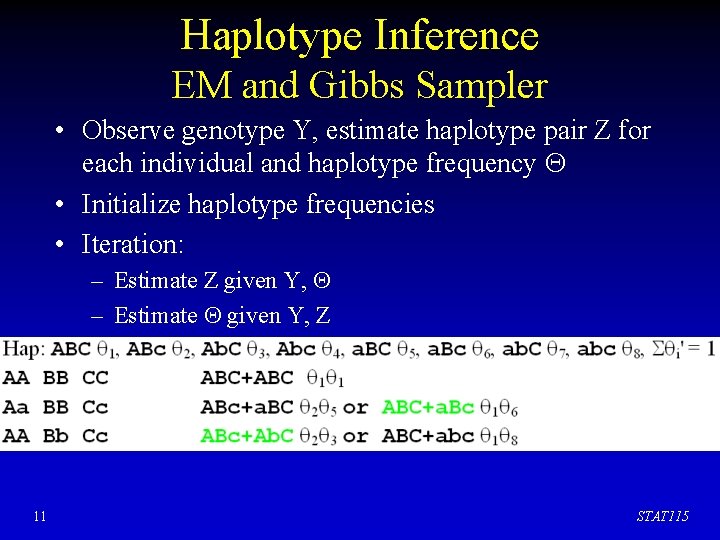
Haplotype Inference EM and Gibbs Sampler • Observe genotype Y, estimate haplotype pair Z for each individual and haplotype frequency • Initialize haplotype frequencies • Iteration: – Estimate Z given Y, – Estimate given Y, Z 11 STAT 115

Haplotype Inference EM and Gibbs Sampler • Observe genotype Y, estimate haplotype pair Z for each individual and haplotype frequency • Initialize haplotype frequencies • Iteration: – Estimate Z given Y, – Estimate given Y, Z 12 STAT 115

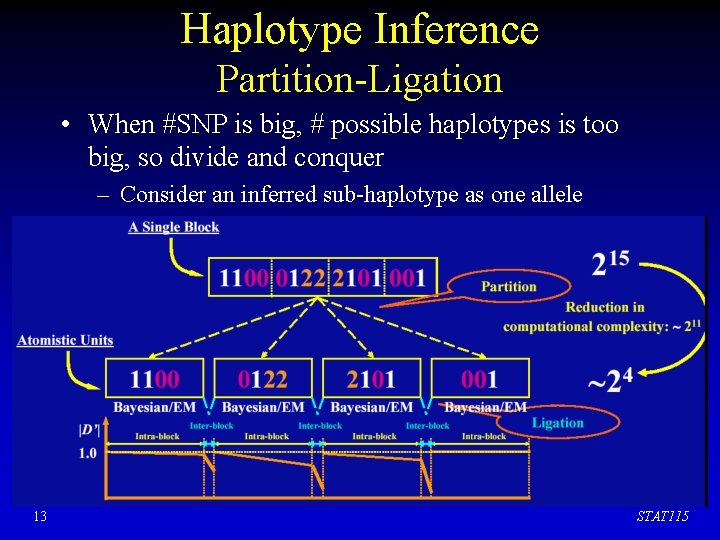
Haplotype Inference Partition-Ligation • When #SNP is big, # possible haplotypes is too big, so divide and conquer – Consider an inferred sub-haplotype as one allele 13 STAT 115

Hapmap of Human Genome • Hap. Map: catalog of common genetic variants in human – What are these variants – Where do they occur in our DNA – How are they distributed within populations and between populations around the world • Goals: – Define haplotype “blocks” across the genome – Enable unbiased, genome-wide association studies 14 STAT 115

1000 Genomes Projects • Characterization of human genome sequence variation • Foundation for investigating the relationship between genotype and phenotype 15 Break STAT 115

Association Studies • Association between genetic markers and phenotype – E. g. Cystic Fibrosis ~70% of Cystic Fibrosis patients have a deletion of 3 base pairs resulting in the loss of a phenylalanine amino acid at position 508 of the CFTR gene • Especially, find disease genes, SNP / haplotype markers, for susceptibility prediction and diagnosis 16

Warfarin and CYP 2 C 9: SNPs in Pharmacogenomics • Warfarin anticoagulant drug; CYP 2 C 9 gene metabolizes warfarin. • A patient requiring low dosage warfarin compared to normal population, has an odd ratio of 6. 21 for having 1 variant allele • Subgroup of patients who are poor metabolisers of warfarin are potentially at higher risk of bleeding Aithal et al. , 1999, Lancet.

Influences individual decisions on life styles, prevention, screening, and treatment 18

Genome-Wide Association Studies • Quality Control – Unusual similarity between individual – Wrong sex – Trio has non-Mendelian inheritance – Genotyping quality • Two strategies: – Family-based association studies – Population-based case-control association studies 19

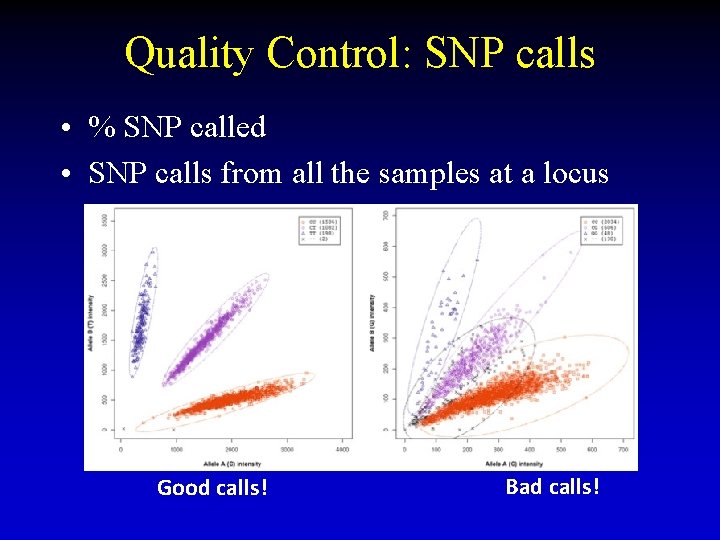
Quality Control: SNP calls • % SNP called • SNP calls from all the samples at a locus Good calls! Bad calls!

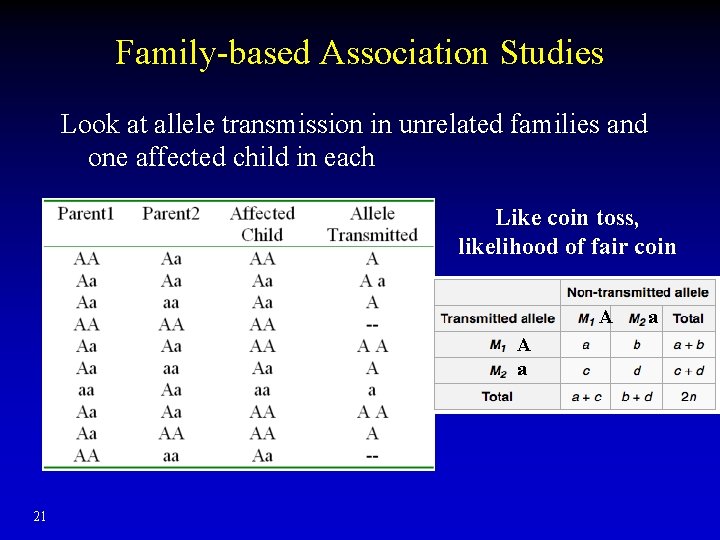
Family-based Association Studies Look at allele transmission in unrelated families and one affected child in each Like coin toss, likelihood of fair coin A A a 21 a

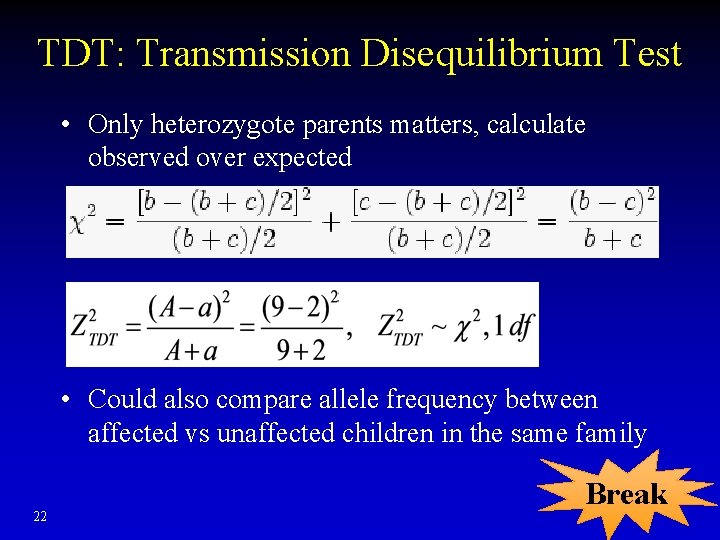
TDT: Transmission Disequilibrium Test • Only heterozygote parents matters, calculate observed over expected • Could also compare allele frequency between affected vs unaffected children in the same family 22 Break

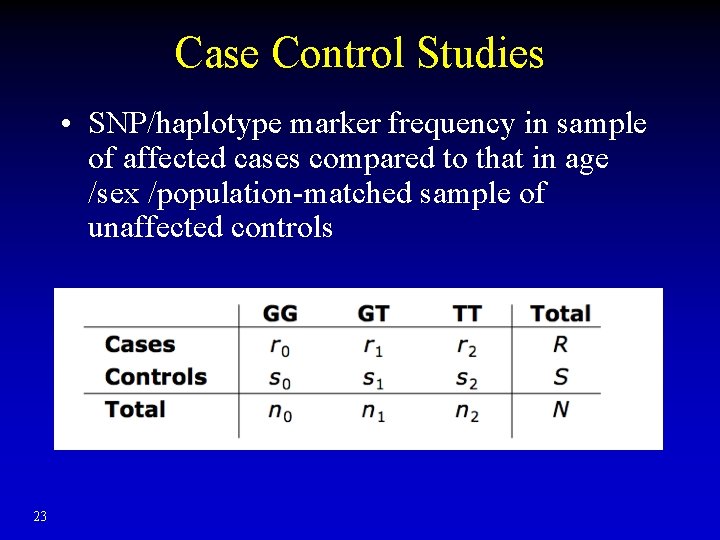
Case Control Studies • SNP/haplotype marker frequency in sample of affected cases compared to that in age /sex /population-matched sample of unaffected controls 23

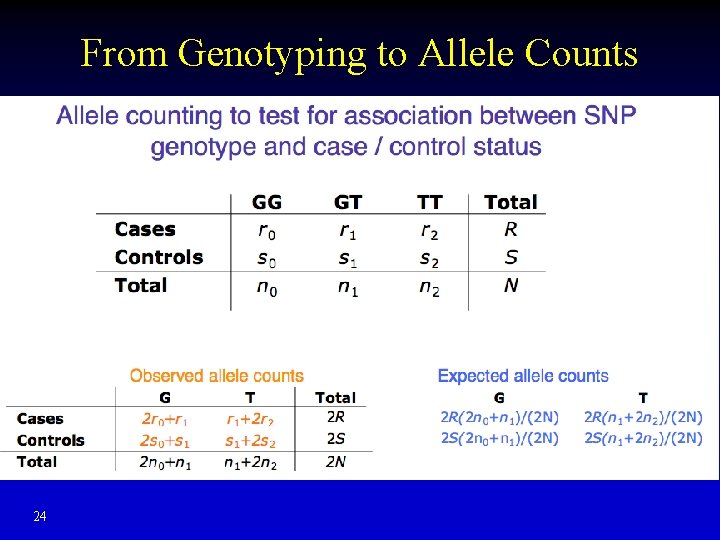
From Genotyping to Allele Counts 24

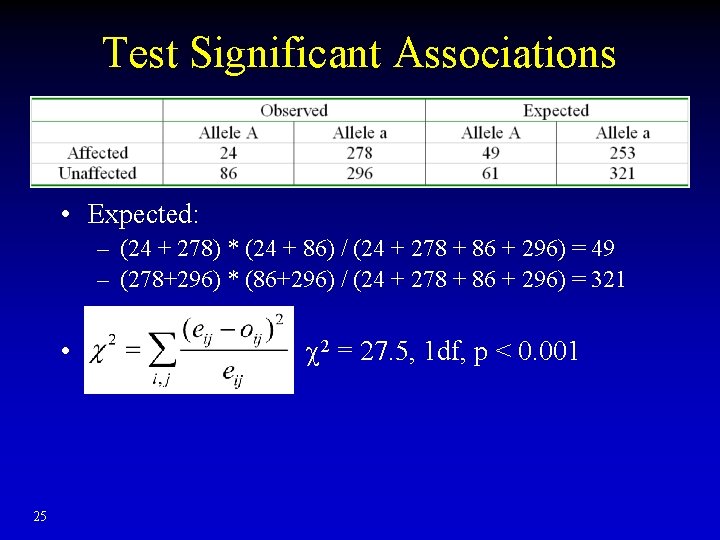
Test Significant Associations • Expected: – (24 + 278) * (24 + 86) / (24 + 278 + 86 + 296) = 49 – (278+296) * (86+296) / (24 + 278 + 86 + 296) = 321 • 25 2 = 27. 5, 1 df, p < 0. 001

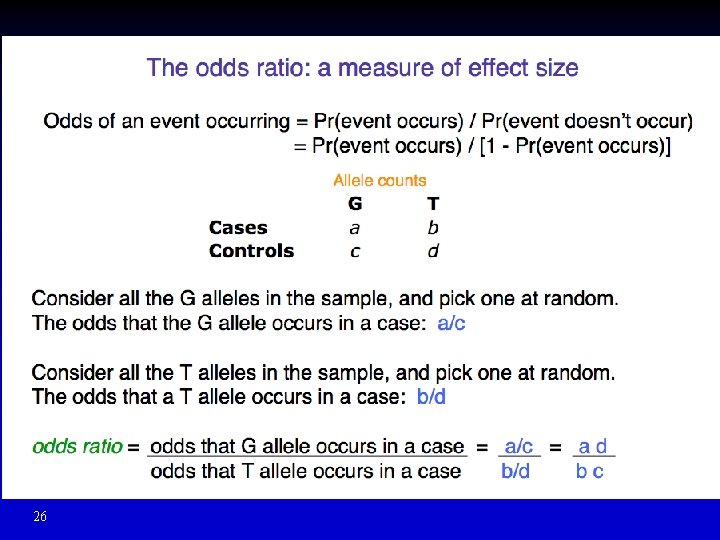
26

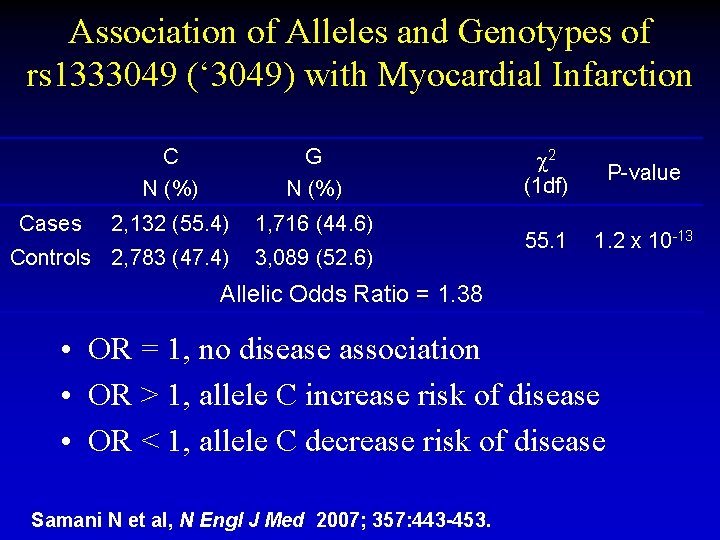
Association of Alleles and Genotypes of rs 1333049 (‘ 3049) with Myocardial Infarction C N (%) G N (%) 2, 132 (55. 4) 1, 716 (44. 6) Controls 2, 783 (47. 4) 3, 089 (52. 6) Cases 2 (1 df) P-value 55. 1 1. 2 x 10 -13 Allelic Odds Ratio = 1. 38 • OR = 1, no disease association • OR > 1, allele C increase risk of disease • OR < 1, allele C decrease risk of disease Samani N et al, N Engl J Med 2007; 357: 443 -453.

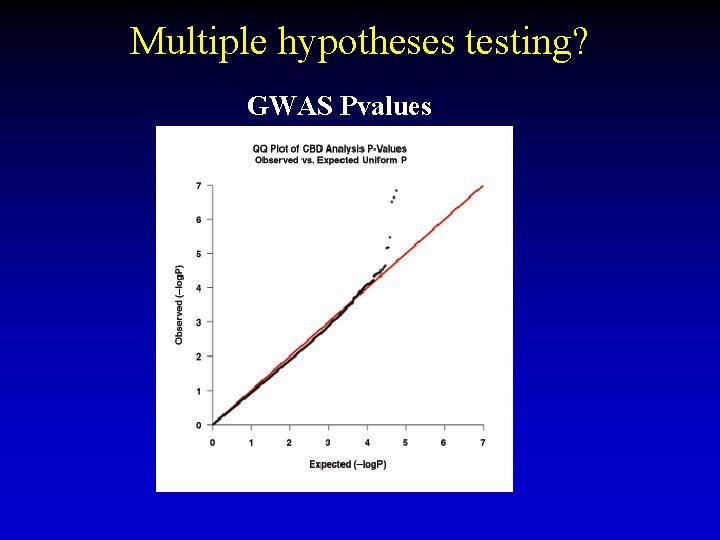
Multiple hypotheses testing? GWAS Pvalues

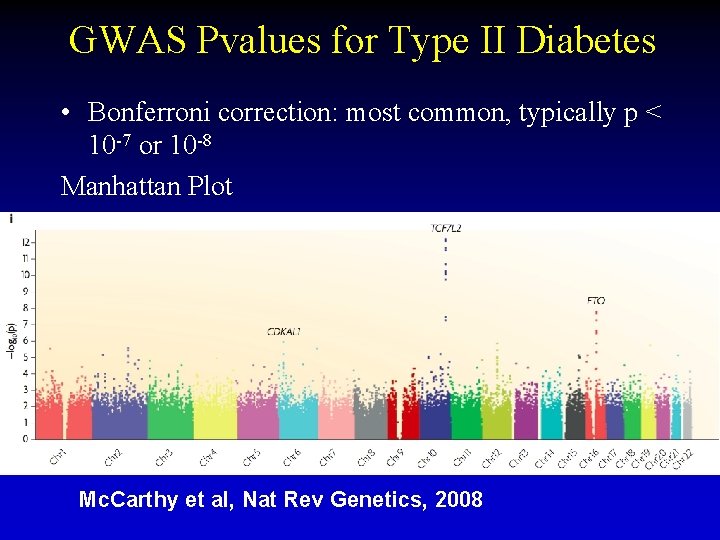
GWAS Pvalues for Type II Diabetes • Bonferroni correction: most common, typically p < 10 -7 or 10 -8 Manhattan Plot Mc. Carthy et al, Nat Rev Genetics, 2008

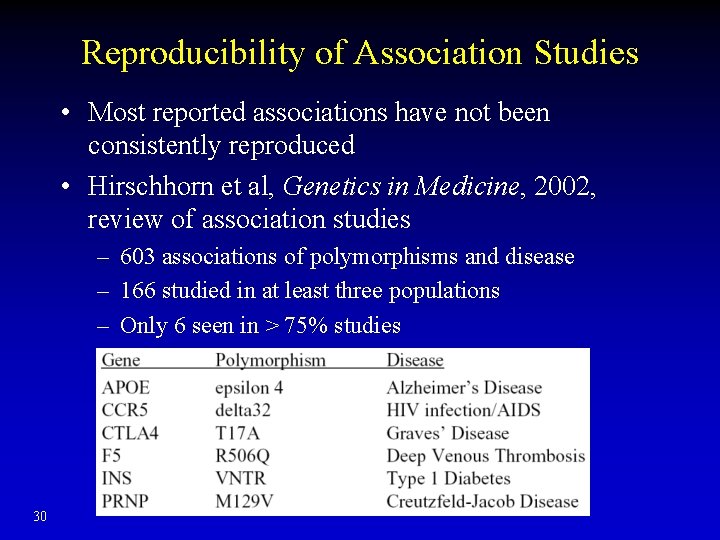
Reproducibility of Association Studies • Most reported associations have not been consistently reproduced • Hirschhorn et al, Genetics in Medicine, 2002, review of association studies – 603 associations of polymorphisms and disease – 166 studied in at least three populations – Only 6 seen in > 75% studies 30

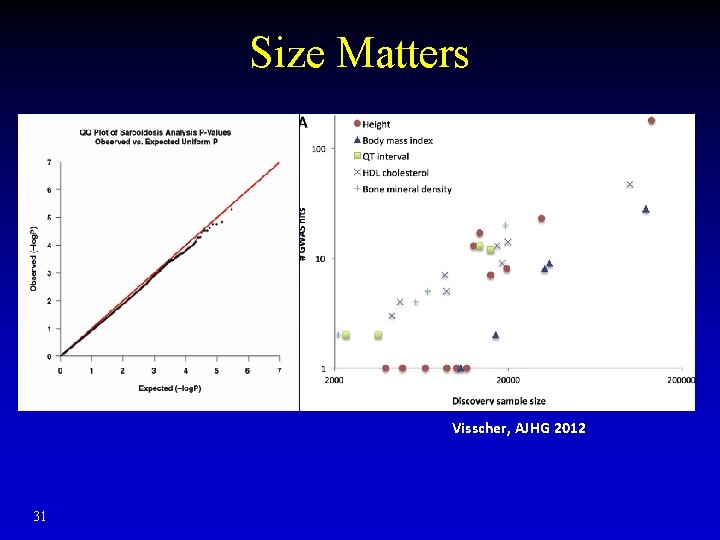
Size Matters Visscher, AJHG 2012 31

How to Improve Statistical Power? • Without increasing samples? • Test association of disease with haplotypes instead of individual SNPs – Also reduce genotyping errors • Split samples: – First half narrow down promising SNPs / haplotypes – Second half refining hits (much fewer multiple hypotheses) • Increase sample size: precision medicine initiative cohort ~ 1 million volunteers 32

Manolio et al. , Clin Invest 2008

Summary • Haplotype inference – Clarks: resolve unambiguous first, propose new haplotypes to maximize explanation – EM & Gibbs: iteratively infer haplotype frequency and individuals’ haplotypes • Tagging SNPs and GWAS • Family based association studies: TDT transmitted allele to affected child • Case control studies: X-sq (allele frequency difference in case and controls) and OR 34 STAT 115

Acknowledgement • • • 35 Jun Liu & Tim Niu Cheng Li & Yuhyun Park Kenneth Kidd, Judith Kidd and Glenys Thomson Joel Hirschhorn Greg Gibson & Spencer Muse Jim Stankovich Teri Manolio David Evans Guodong Wu Stefano Monti Bo Li
 12345678
12345678 Xiaole liu
Xiaole liu Xiaole liu
Xiaole liu Haplotype
Haplotype Gwas power calculation
Gwas power calculation Gwas method
Gwas method Nature review genetics
Nature review genetics Alex liu cecilia liu
Alex liu cecilia liu Líu líu lo lo ta ca hát say sưa
Líu líu lo lo ta ca hát say sưa Cornelia m ruland
Cornelia m ruland Lucy liu my girl drew
Lucy liu my girl drew Bing images
Bing images The lottery shirley jackson questions
The lottery shirley jackson questions Shirleycvc
Shirleycvc Miss strangeworth character traits
Miss strangeworth character traits The lottery by shirley jackson discussion questions
The lottery by shirley jackson discussion questions Shirley ardell mason
Shirley ardell mason Shirley ardell mason central park in spring
Shirley ardell mason central park in spring Shirley jackson seven types of ambiguity
Shirley jackson seven types of ambiguity Zacisze słowika ania z zielonego wzgórza
Zacisze słowika ania z zielonego wzgórza Dr shirley d'sa
Dr shirley d'sa Shirley hills primary
Shirley hills primary Neeshub
Neeshub Michael jackson vocabulary
Michael jackson vocabulary What is the climax of the story charles
What is the climax of the story charles Biography of shirley toulson
Biography of shirley toulson Shirley kavanagh
Shirley kavanagh The lottery theme
The lottery theme The lottery by shirley jackson symbolism
The lottery by shirley jackson symbolism Shirley c. heim middle school
Shirley c. heim middle school Shirley c heim middle school calendar
Shirley c heim middle school calendar The lottery shirley jackson protagonist
The lottery shirley jackson protagonist Shirley lerner
Shirley lerner Grundy shirley
Grundy shirley Dr shirley wray
Dr shirley wray Shirley heim middle school lunch menu
Shirley heim middle school lunch menu