SEQUENCE RETRIEVAL SYSTEM SRS Ashwin Sivakumar 021203 Hands














![Some examples /^glu/ /ase$/ /c. t/ /c. *t/ /sm[iy]th/ /rho[1 -9]/ /mue? ller/ will Some examples /^glu/ /ase$/ /c. t/ /c. *t/ /sm[iy]th/ /rho[1 -9]/ /mue? ller/ will](https://slidetodoc.com/presentation_image_h/1177d8a851c95db324626b3f67f9e528/image-21.jpg)











- Slides: 62

SEQUENCE RETRIEVAL SYSTEM SRS Ashwin Sivakumar, 02/12/03 Hands on Workshop on Protein Analysis (HOW)

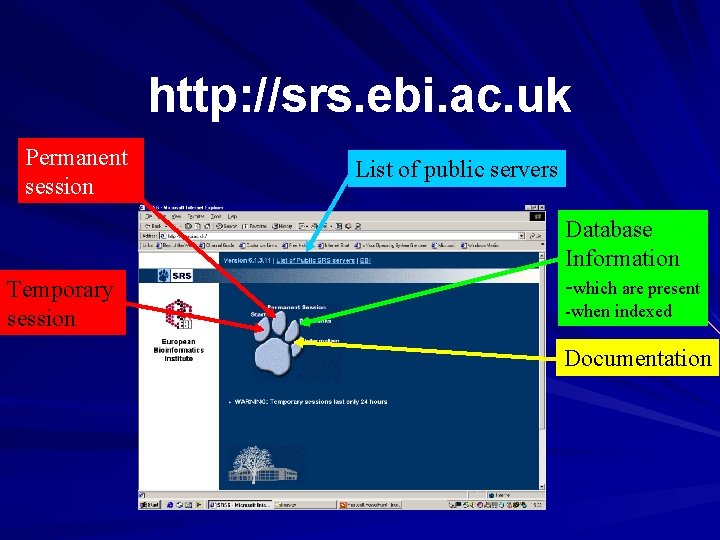
http: //srs. ebi. ac. uk Permanent session Temporary session List of public servers Database Information -which are present -when indexed Documentation

What is SRS? Central resource for molecular biology data Data retrieval system - more than 250 databanks have been indexed. More than 35 SRS servers over the WWW Data analysis applications server - 11 protein applications - 6 nucleic acid applications - Uniform query interface on the web

Data Jungle Sequencing information genetics Structural biology molecular biology medicine physiology gene expression toxicology

History of SRS 1990 - Main author Dr. Thure Etzold – Development started in EMBL, Heidelberg 1997 – Moved to EBI in Cambridge. Development work was supported by various grants amongst others from the EMBnet. 1998 – Etzold and his group join Lion. Biosciences

Why SRS? Information retrieval – Easy way to retrieve information from sequence and sequence-related databases – Possibility to search for multiple words/other criteria Linkage between different databases – E. g. Find all primary structures with known three-dimensional structure . . . and much more

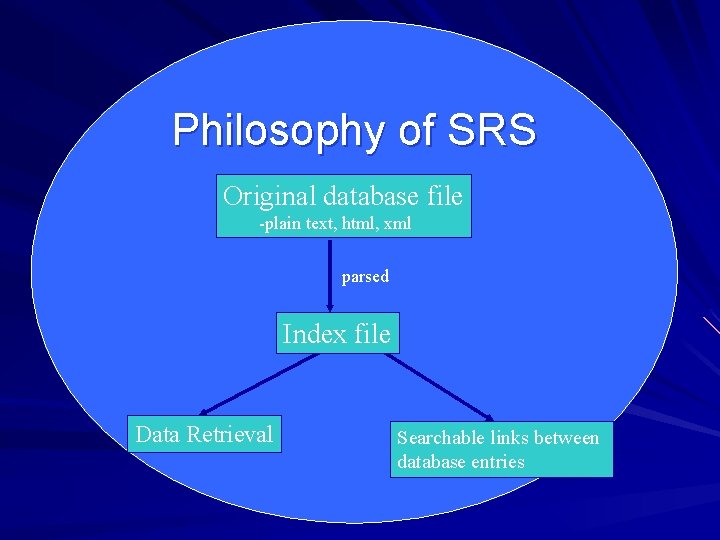
Philosophy of SRS Original database file -plain text, html, xml parsed Index file Data Retrieval Searchable links between database entries

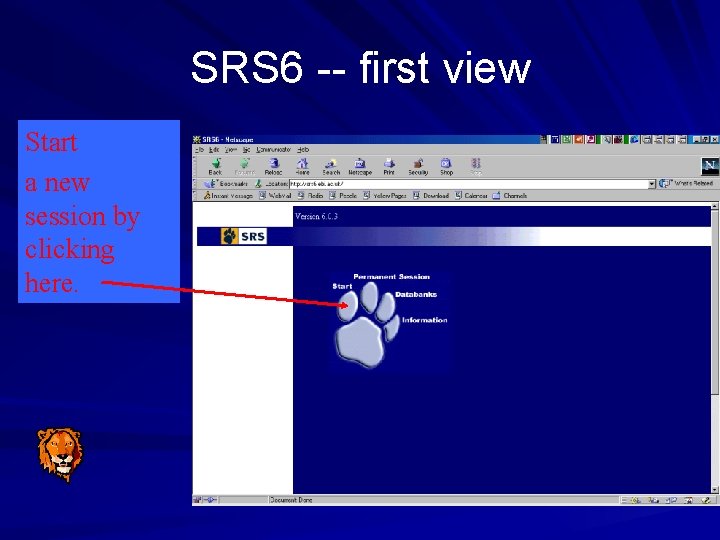
Temporary Projects Queries and views are stored by the project manager temporarily Temporary sessions last 24 hours Useful when you: – Do not need to keep your results – look something up quickly – Run an occasional application Click on ‘Start’ paw on SRS start page

Permanent Projects Queries and views are stored by the project manager in a single location They are available for use in the future Useful when: – You want to return to a session – Want to have many projects in the same session Begin by clicking ‘Permanent session’ paw on SRS start page – Just need to enter an SRS user name and re-enter this to return to same session again later

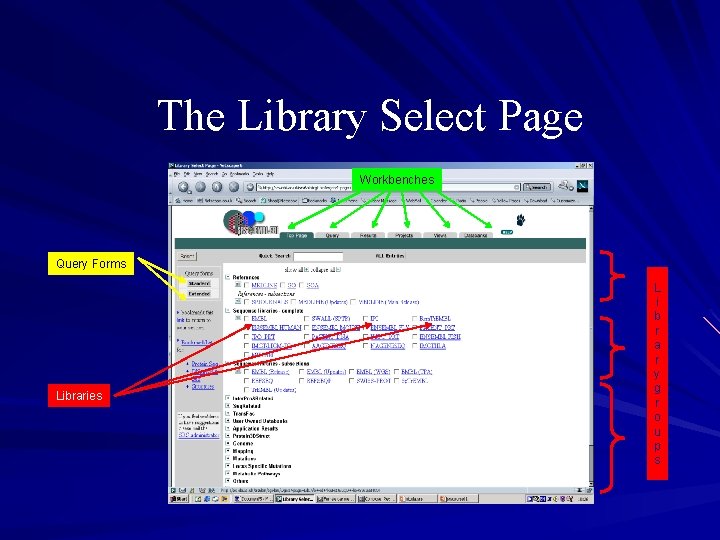
The Library Select Page Workbenches Query Forms Libraries L i b r a r y g r o u p s

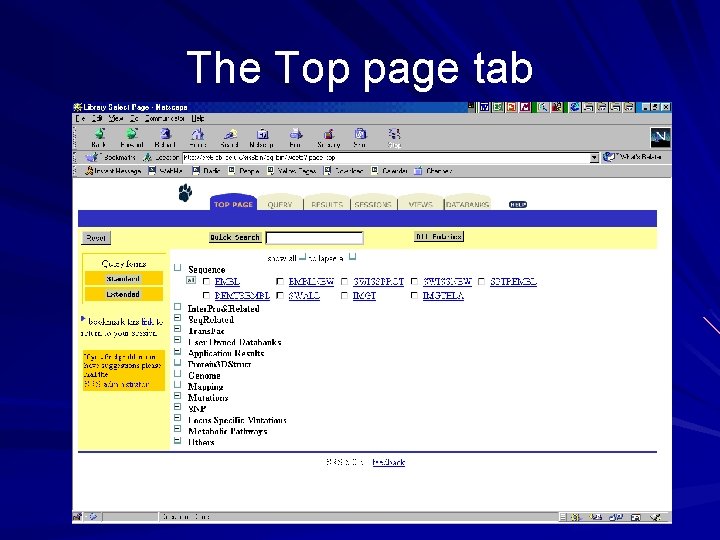
SRS main toolbar tabs Top Page: displays databases in different database groups Query: displays either the standard or extended query form Results or “the query manager”: maintains a history of all the results obtained during a session Projects or “the project manager”: maintains a history of all queries and views used during a session Views: allows a user to define a user specific view for one or more databases Databanks: contains a list and some facts about the databases available in the system

Search terms in SRS indexed fields can be searched using any of the following: – Single word search – Multiple word phrases – Numbers and dates – Regular expressions – Wildcards

Search methods Quick search button: – Works by searching all datafields of type text – The quickest way to generate query results – For very general/broad searches Example: get all mouse and mouse related proteins in SWISS-PROT All Entries button: – Returns all entries in the database selected Search forms : allow you to specify your area of interest in more detail – Standard query form – Extended query form

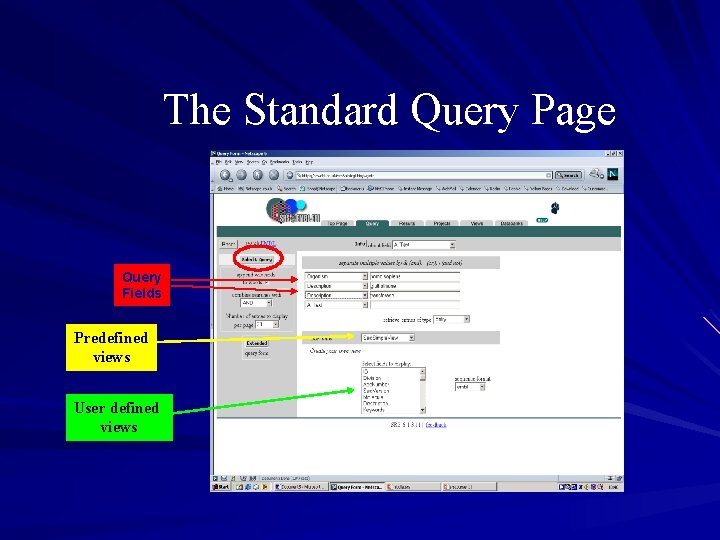
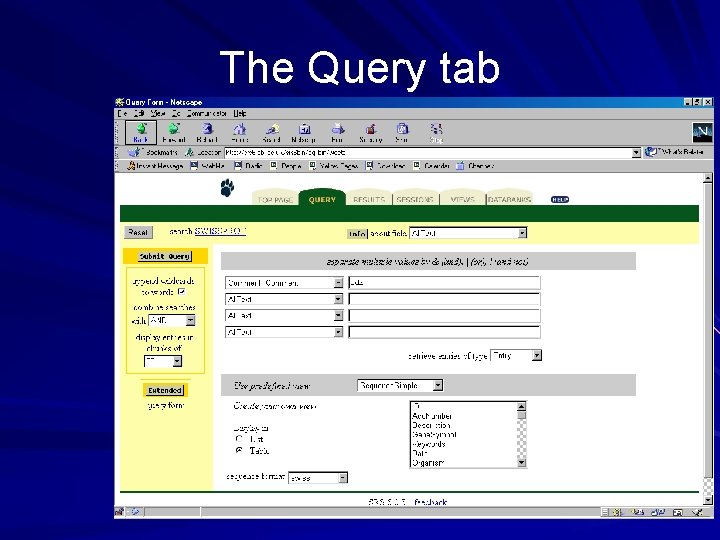
Standard query form Enter up to 4 separate search terms against up to 4 datafields simultaneously Combine entries with logical operators ( and & or | butnot ! ) Choose the number of entries to display per page Retrieve entries of type (entry or subentry(name)) Choose a view – use an SRS predefined view – create one of your own by selecting specific fields from a dropdown menu (and choose whether to view a list or table in SRS 7)

The Standard Query Page Query Fields Predefined views User defined views

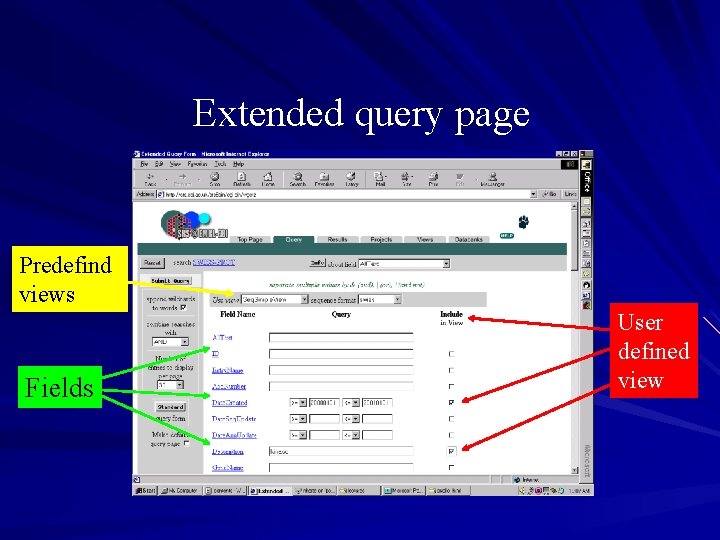
Extended query form Can enter search terms for as many fields as you want Combine searches with logical operators ( and & or | butnot ! ) Choose how many results to display per page Choose view and sequence format to use – Can choose an SRS predefined view – Define your own view by clicking the boxes next to the fields that you want to have displayed (list or table option in SRS 7) Each field name has a hyperlink to the description page for that field Form provides less than ‘<‘ and greater than ‘>’ for numerical fields Choose what type of entries to retrieve (entry, subentry (name)) – on extended form if you query a subentry field, it defaults to returning results of type subentry

Extended query page Predefind views Fields User defined view

Differences in these 2 forms Ranges – standard must use ‘: ’ – extended provides ‘<‘ and ‘>’ Type retrieval – standard defaults to retrieving entries of type ‘entry’ – extended defaults to retrieving entries of type entry unless you query a subentry field in which case the default is the subentry type Controlled vocabulary fields – standard does not provide you with a list for these fields – extended provides a drop down menu for these fields allowing you to select an option

Wildcards These are useful when: – Searching for a group of words (eg. Words starting ‘cell’ and ending ‘ase’ : cell*ase) – If unclear about how a word is spelt in a database Two types: – * one or more characters of any value – ? Single character of any value Any number of wildcards can be placed anywhere in a search word Placing a wildcard at the start of a word or string may increase response time because all words in the index have to be checked against the string

Regular expressions NB: Must appear within forward slashes (/) Some operators: ^ marks the start of a string /^glu/ begins with ‘glu’ $ marks the end of a string /ase$/ ends with ‘ase’. dot is any single character […] characters in square brackets are regarded as a set, any of which can be matched [0 -9] specifies a range of 1 to 9 * the preceding group may be repeated zero or more times + the preceding group may be repeated one or more times ? The preceding character/group occurs one or zero times
![Some examples glu ase c t c t smiyth rho1 9 mue ller will Some examples /^glu/ /ase$/ /c. t/ /c. *t/ /sm[iy]th/ /rho[1 -9]/ /mue? ller/ will](https://slidetodoc.com/presentation_image_h/1177d8a851c95db324626b3f67f9e528/image-21.jpg)
Some examples /^glu/ /ase$/ /c. t/ /c. *t/ /sm[iy]th/ /rho[1 -9]/ /mue? ller/ will find terms beginning with ‘glu’ will find terms ending with ‘ase’ will find the words cat, cot, cut……. will find terms beginning with ‘c’ and then any number of characters and ending with ‘t’ will find the words ‘smith’ or ‘smyth’ will find the word ‘rho’ followed by a number from 1 -9 will find ‘muller’ or ‘mueller’ NB. The ‘*’ symbol has two meanings: -within forward slashes ‘/’ it means the preceding group may be repeated zero or more times - outside forward slashes it means any character

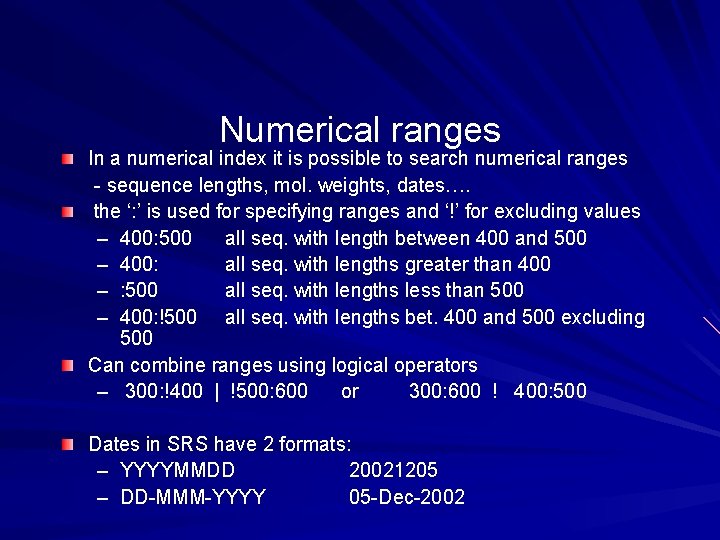
Numerical ranges In a numerical index it is possible to search numerical ranges - sequence lengths, mol. weights, dates…. the ‘: ’ is used for specifying ranges and ‘!’ for excluding values – 400: 500 all seq. with length between 400 and 500 – 400: all seq. with lengths greater than 400 – : 500 all seq. with lengths less than 500 – 400: !500 all seq. with lengths bet. 400 and 500 excluding 500 Can combine ranges using logical operators – 300: !400 | !500: 600 or 300: 600 ! 400: 500 Dates in SRS have 2 formats: – YYYYMMDD 20021205 – DD-MMM-YYYY 05 -Dec-2002

Some examples – Find entries with sequences having length betwwen 300 and 400 excluding 400 and between 500 and 600 excluding 500: 300: !400 | !500: 600 or 300: 600 ! 400: 500 – Find entries that were created in the first half of 2001: 01 -jan-2001: 30 -jun-2001 or 20010101: 20010630 – Find all entries updated since May this year: 01 -may-2002: or 20020501:

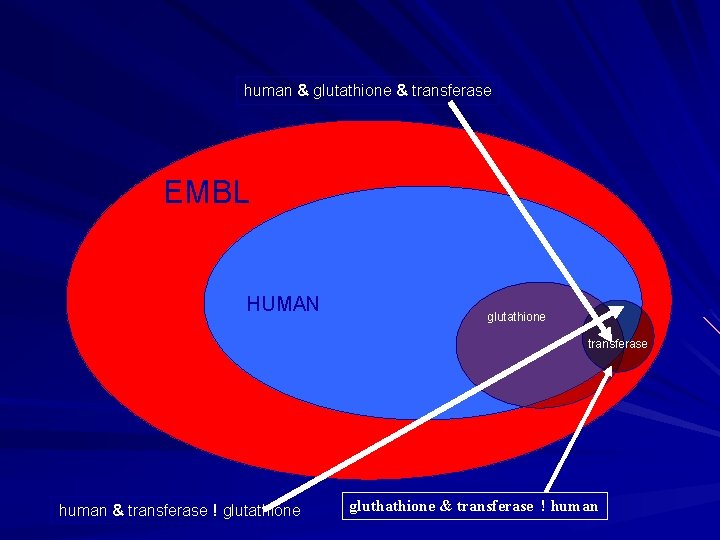
SRS Indexing SRS indexes database records using a ‘word by word’ approach. - DE Human glutathione transferase - The SRS description index will contain terms ‘human’, ‘glutathione’ and ‘transferase’. (&) AND : ‘human & glutathione & transferase’ (|) OR : ‘human | glutathione | transferase’ (!) BUTNOT : ‘human ! glutathione ! transferase’

human & glutathione & transferase EMBL HUMAN glutathione transferase human & transferase ! glutathione gluthathione & transferase ! human

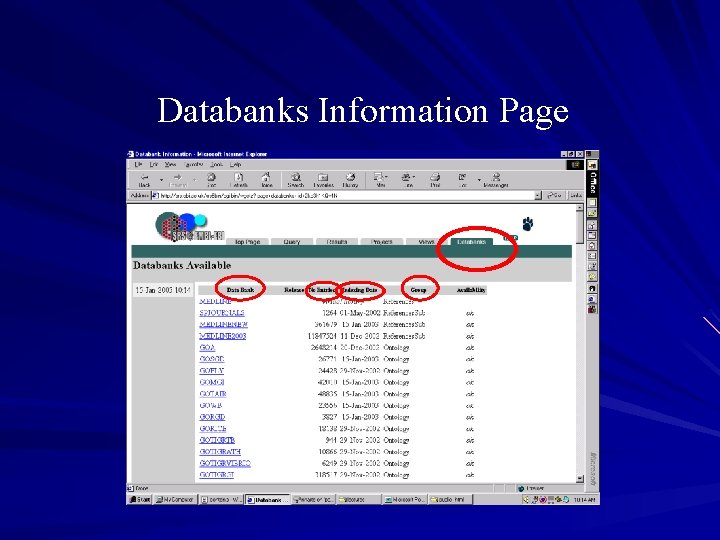
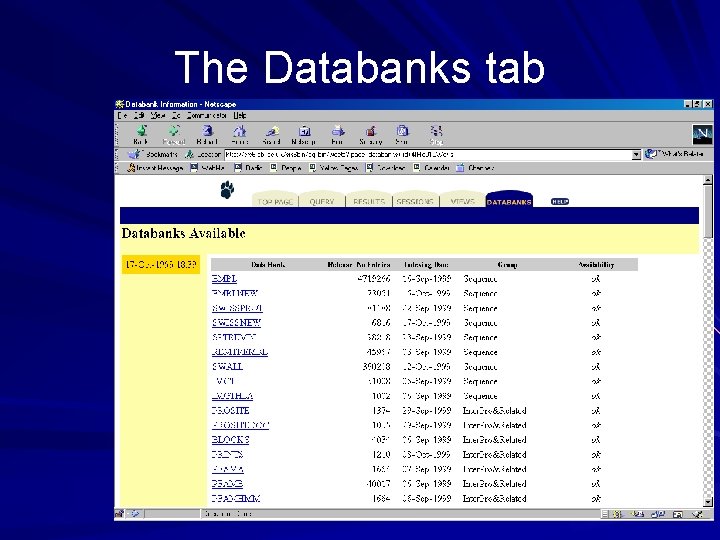
Databanks information page Lists the databases available in the system and a summary about them: – Number of entries in the database – Date it was indexed – Group it belongs to – Its availability status Hyperlinks to information page specific to each database

Databanks Information Page

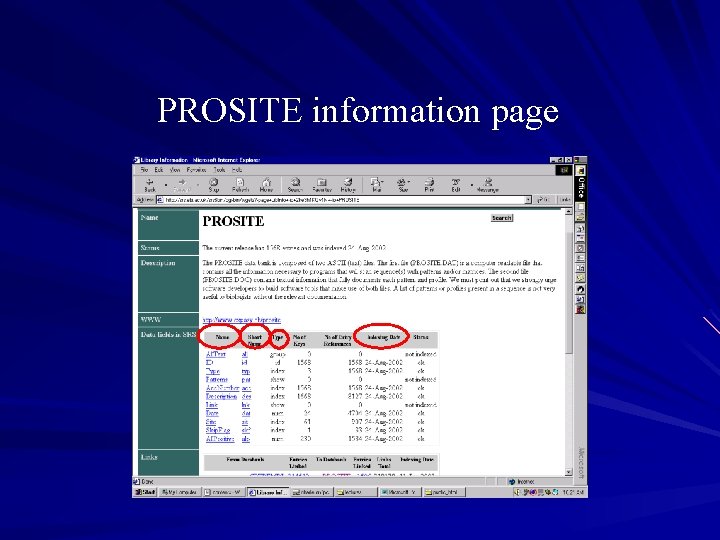
Database information page Provides a detailed description about the database contents, source, ftp site, literature… Lists information about the fields that are present in the database including: – Name of field – Short name for field – Type of field index : it is indexed num : indexed and a numerical field id: unique field show: not indexed, just for display – Number of keys for that field – Date it was indexed Lists databases that it is linked to and how many entries are linked respectively

PROSITE information page

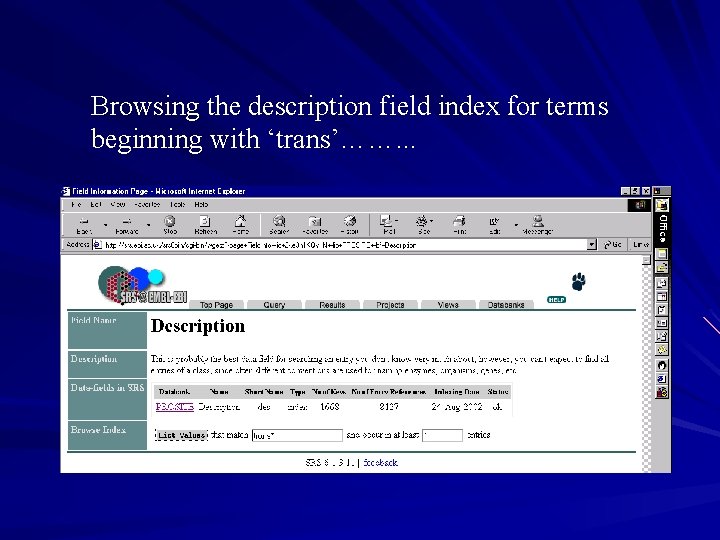
Browsing indices This gives information on what is being indexed for a particular field – Single words, multiple words, controlled vocabulary…. . To browse an index go to the information page for a particular field from a certain database – If you want to look at all indexed terms use ‘*’ – If you want all terms beginning with trans use ‘trans*’ – If you want all terms containing the string trans use ‘*trans*’

Browsing the description field index for terms beginning with ‘trans’……. . .

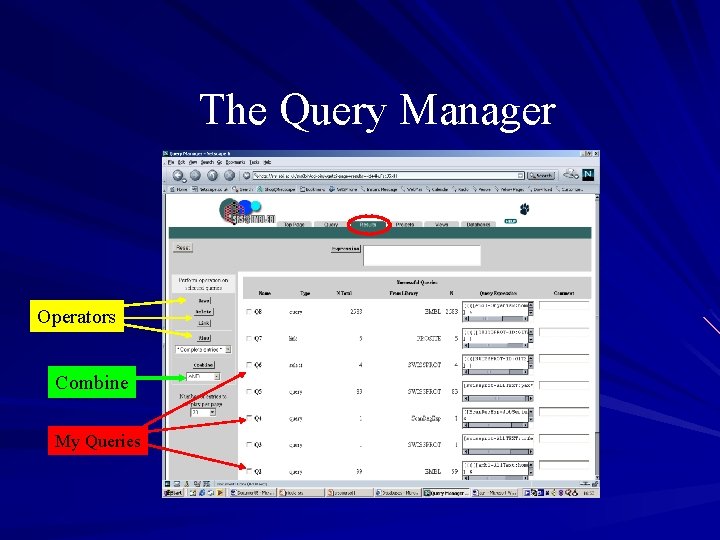
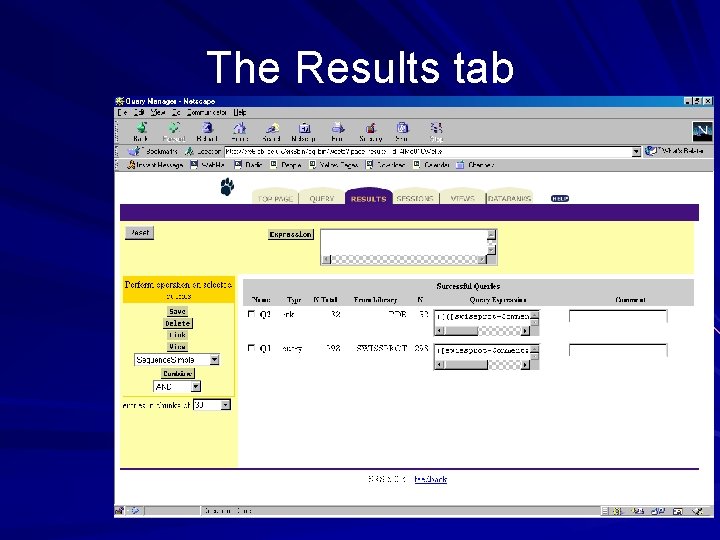
Query manager Found under the results tab Saves a history of results obtained in the session Page allows you to return to previous results and: – Combine them using logical operators – thus allowing you to perform a multi-step query – Use a different view to display them – Perform further actions link, save, delete

The Query Manager Operators Combine My Queries

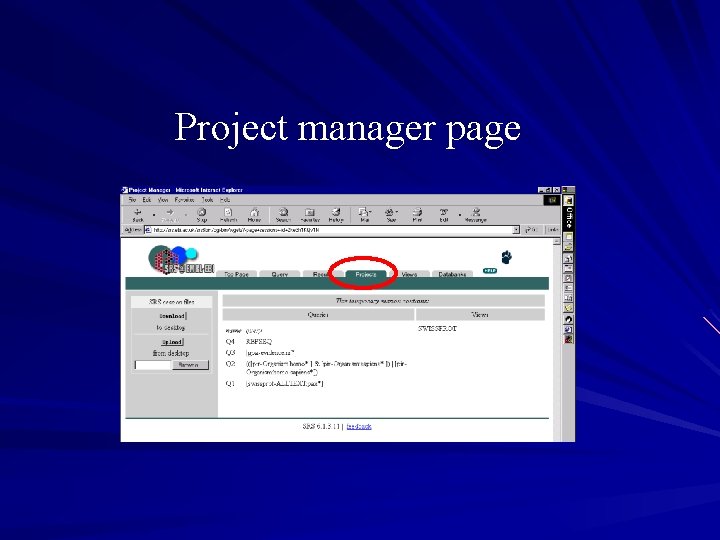
Project manager Found under the projects tab Saves a history of queries performed in the session Can upload/download SRS session files from a desktop In a permanent session, the project manager can also: – Manage numerous SRS projects at the same time – Move queries/views between projects – Upload/download projects to desktop – Delete projects

Project manager page

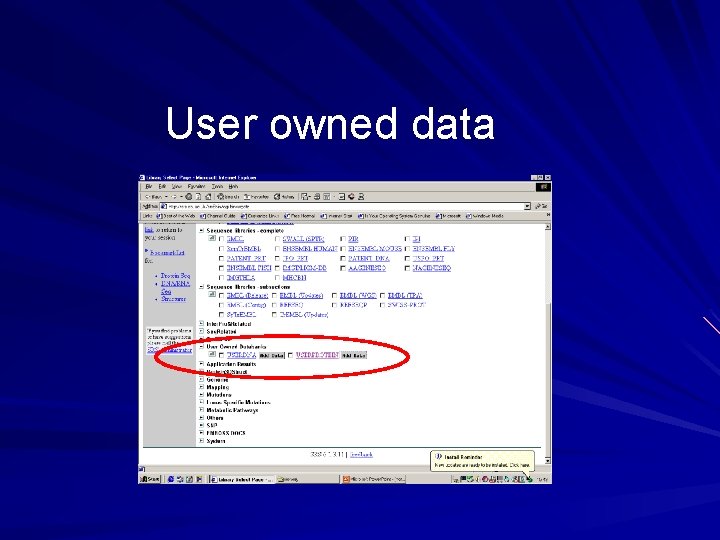
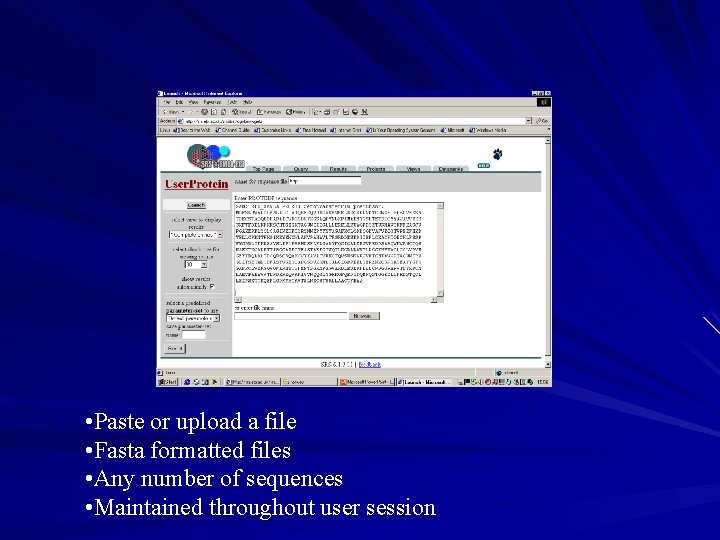
User owned databanks Found in the category ‘user owned databanks’ on top page User can upload their own nucleotide or protein sequence data into a user owned database – sequences must be in fasta format – any number of sequences can be uploaded – database is specific to the individual and to the session Can launch applications on database sequences

User owned data

• Paste or upload a file • Fasta formatted files • Any number of sequences • Maintained throughout user session

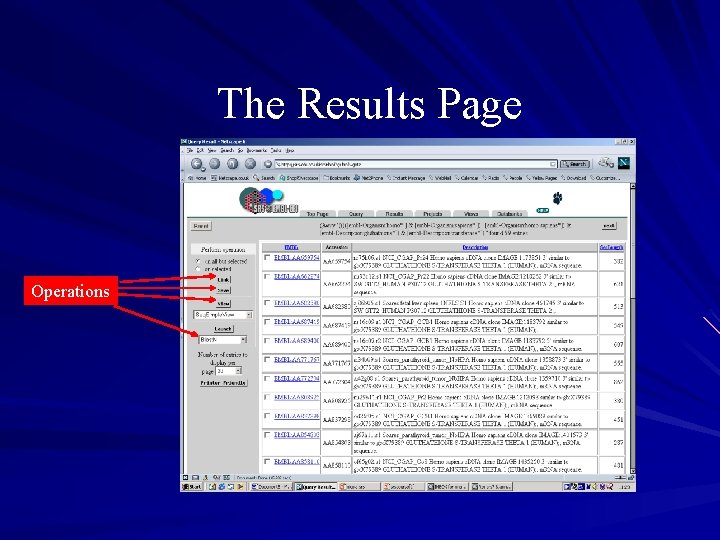
Operations on results Linking : link results to other databases Saving: save results in different formats to the browser or a file Viewing: view results using different formats Sequence analysis: launch applications on the results SRS 6 – 11 protein applications, 6 nucleic acid apps. SRS 7 – more than 100 applications available

The Results Page Operations

SRS 6 versus SRS 7 provides over 100 applications while SRS 6 provides 17 You can retrieve results in either list or table format in SRS 7 In SRS 6 only the table format is available Current EBI version 7. 1. 1

SRS 6 -- first view Start a new session by clicking here.

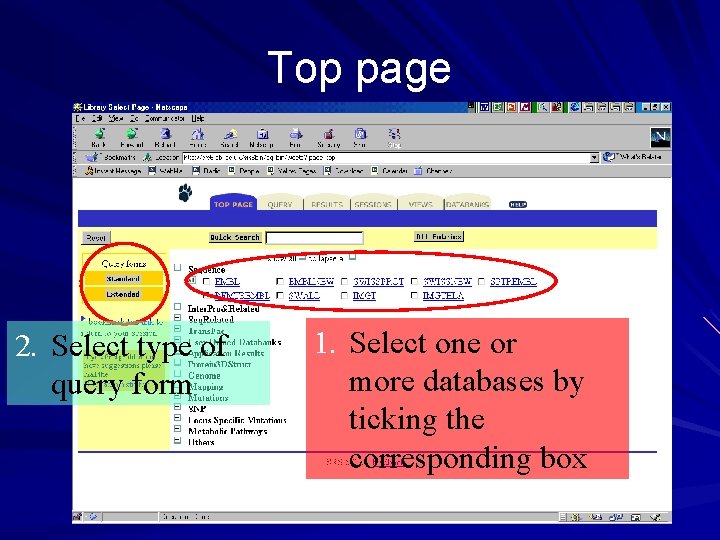
Top page 2. Select type of query form 1. Select one or more databases by ticking the corresponding box

Different types of database in SRS Sequence & structure – DNA, protein, three-dimensional structures Sequence-related Gene-related – Genome, mapping, mutations, transcription factors – SNP Bibliographic – Medline, enzyme User-defined

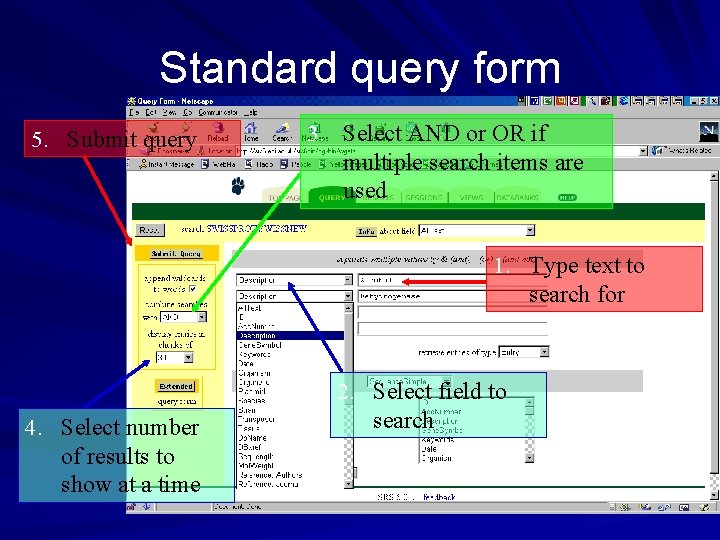
Standard query form 5. Submit query 3. Select AND or OR if multiple search items are used 1. Type text to search for 4. Select number of results to show at a time 2. Select field to search

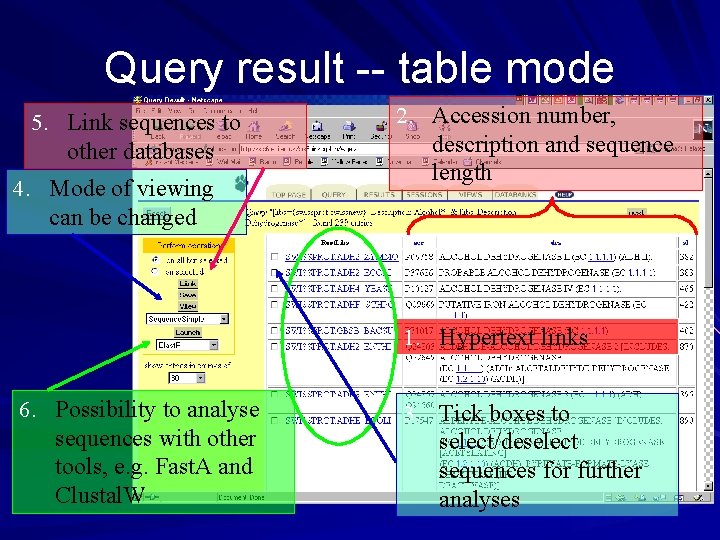
Query result -- table mode 5. Link sequences to other databases 4. Mode of viewing can be changed 2. Accession number, description and sequence length 1. Hypertext links 6. Possibility to analyse sequences with other tools, e. g. Fast. A and Clustal. W 3. Tick boxes to select/deselect sequences for further analyses

Example query Use SRS to answer the following question: For which short-chain dehydrogenases/ reductases (SDR) are threedimensional structure known in PDB?

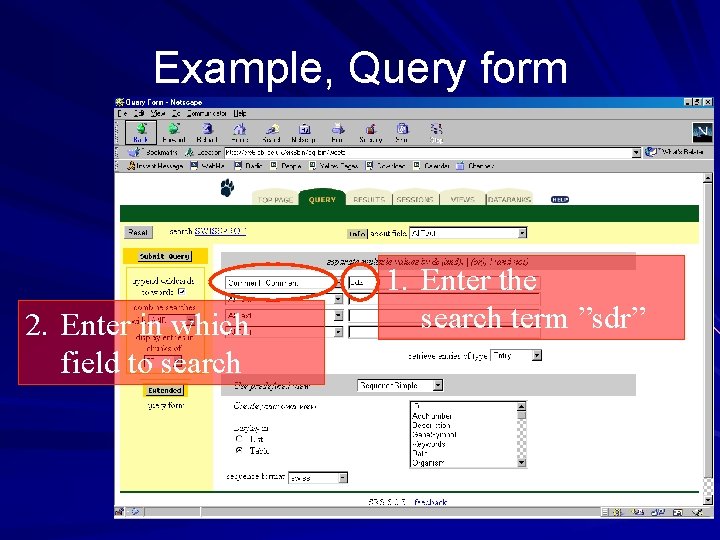
Example, Query form 2. Enter in which field to search 1. Enter the search term ”sdr”

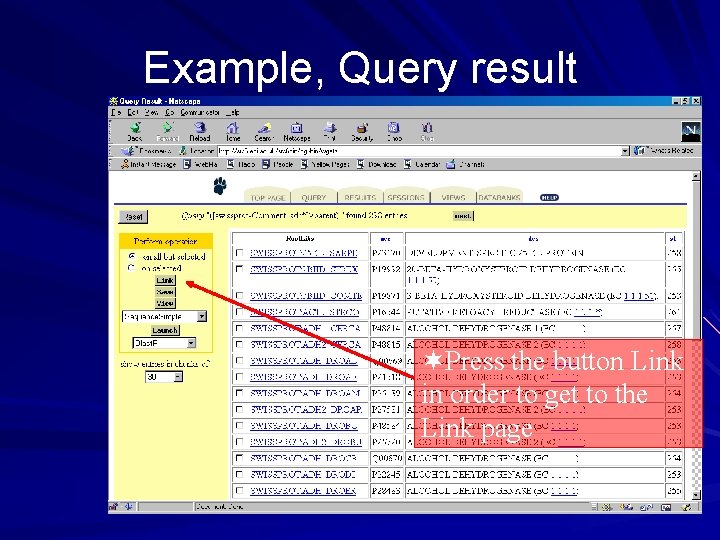
Example, Query result ¬Press the button Link in order to get to the Link page

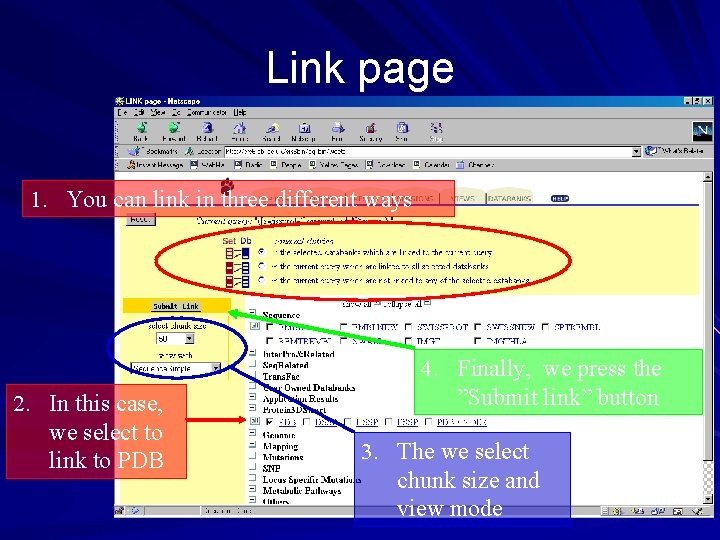
Link page 1. You can link in three different ways 2. In this case, we select to link to PDB 4. Finally, we press the ”Submit link” button 3. The we select chunk size and view mode

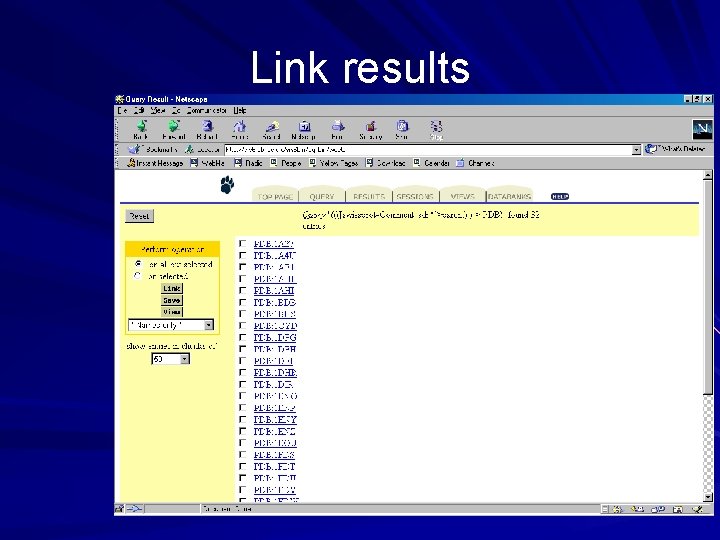
Link results

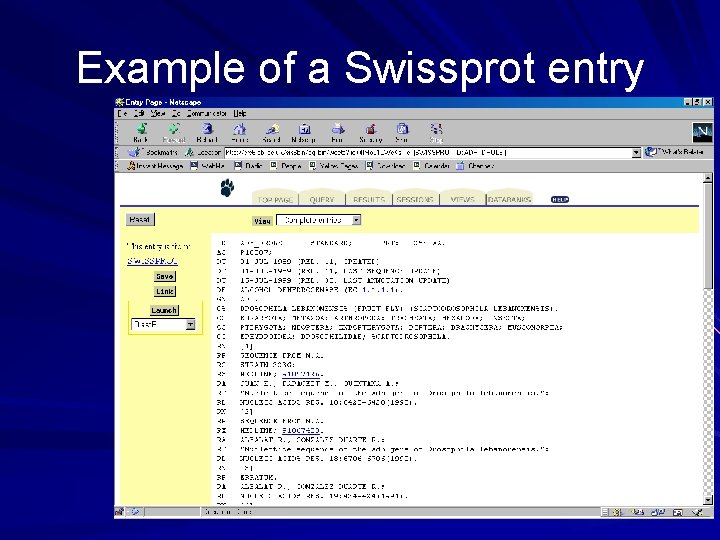
Example of a Swissprot entry

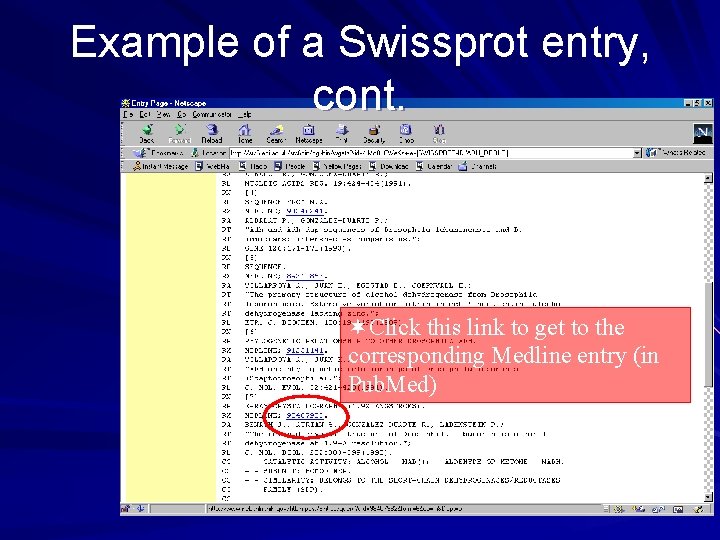
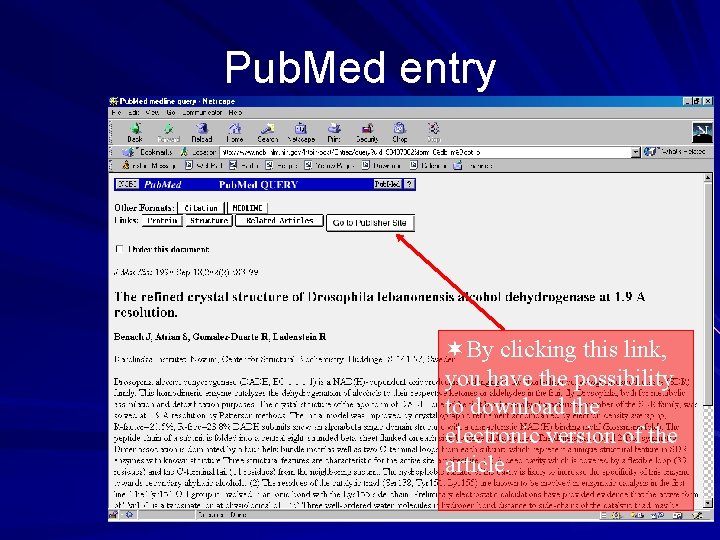
Example of a Swissprot entry, cont. ¬Click this link to get to the corresponding Medline entry (in Pub. Med)

Pub. Med entry ¬By clicking this link, you have the possibility to download the electronic version of the article.

The Top page tab

The Query tab

The Results tab

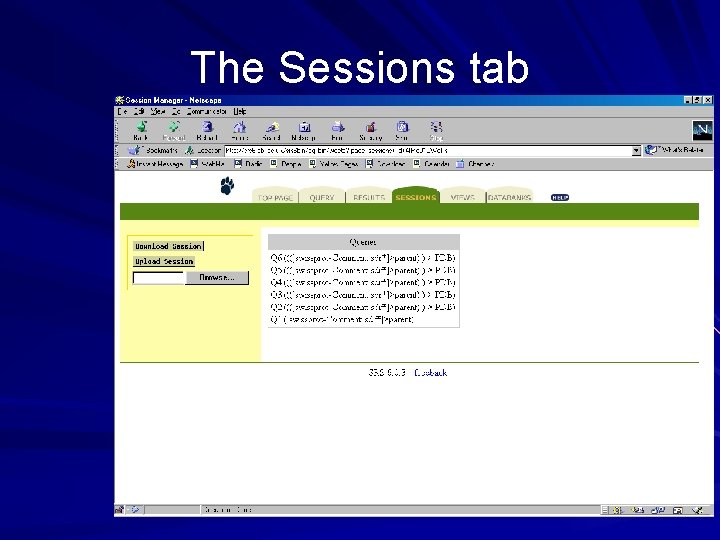
The Sessions tab

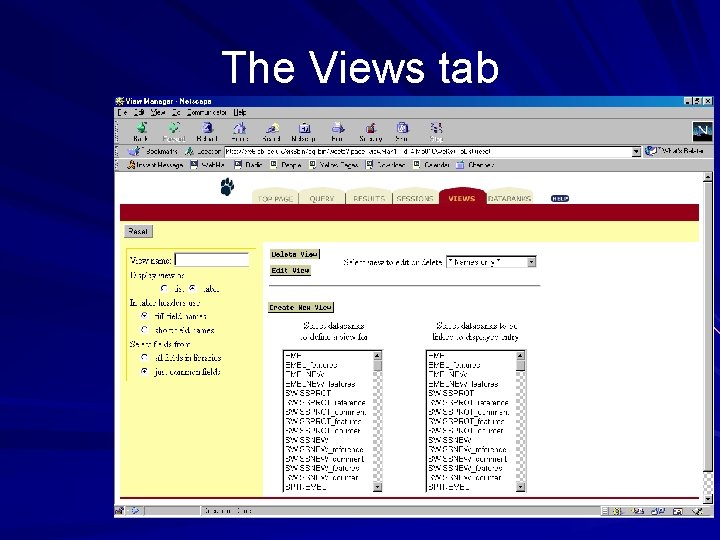
The Views tab

The Databanks tab

Acknowledgements ¤ Bengt Persson MBB, Karolinska institutet (demos) ¤ 2 can tutorial on SRS at EBI http: //downloads. lionbio. co. uk/publi csrs. html (The latest SRS server list)

server breakup srs. sanger. ac. uk (5) srs. ebi. ac. uk (5) srs. csc. fi (5) titanic. thep. lu. se/srs 71/ (5) If you think the load on a server is slowing your query, chose an alternative server to practice on.