NAMIC National Alliance for Medical Image Computing http







- Slides: 49

NA-MIC National Alliance for Medical Image Computing http: //na-mic. org Slicer 3 EMSegment Tutorial January 2008 NAMIC All-Hand’s Meeting Brad Davis, Yuman Yuan, Sebastien Barre, Will Schroeder, Polina Goland, Ron Kikinis, Kilian Pohl

Program • • EM Segmenter basics Algorithm “views” in Slicer 3 EM segment under the hood Tutorial – Data – Simple Segmentation Interface – Template Builder Interface • Questions (all week…) National Alliance for Medical Image Computing http: //na-mic. org

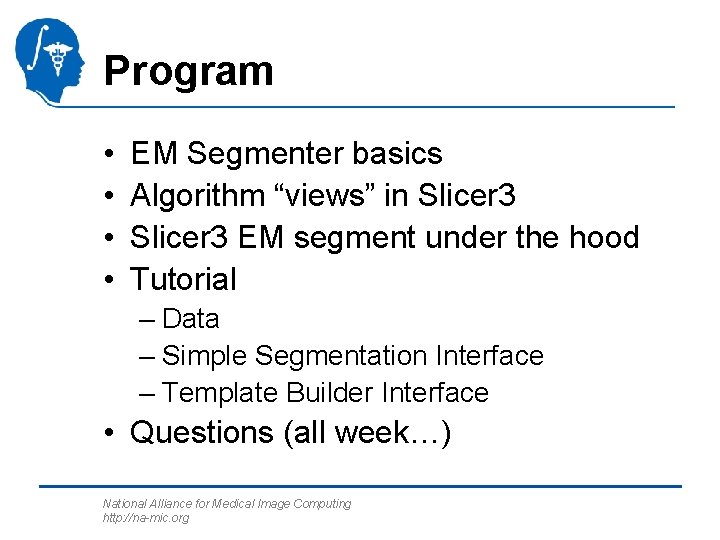
EM Segment Basics National Alliance for Medical Image Computing http: //na-mic. org Figure: Pohl et al.

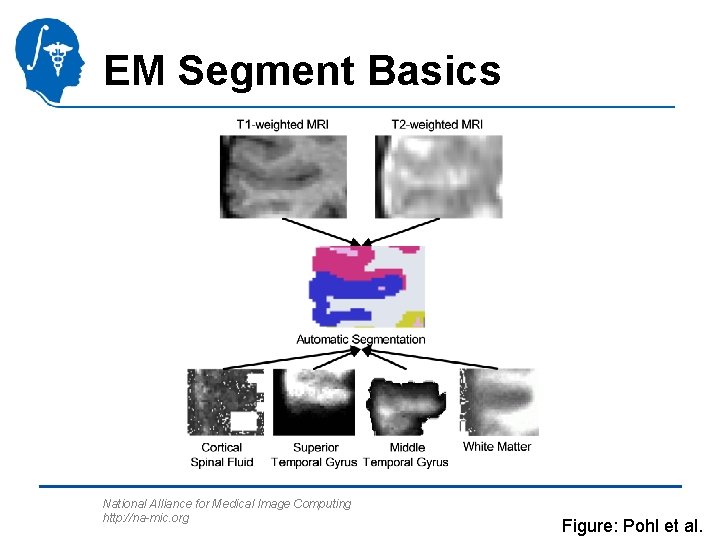
EM Segment Basics National Alliance for Medical Image Computing http: //na-mic. org Figures: Pohl et al.

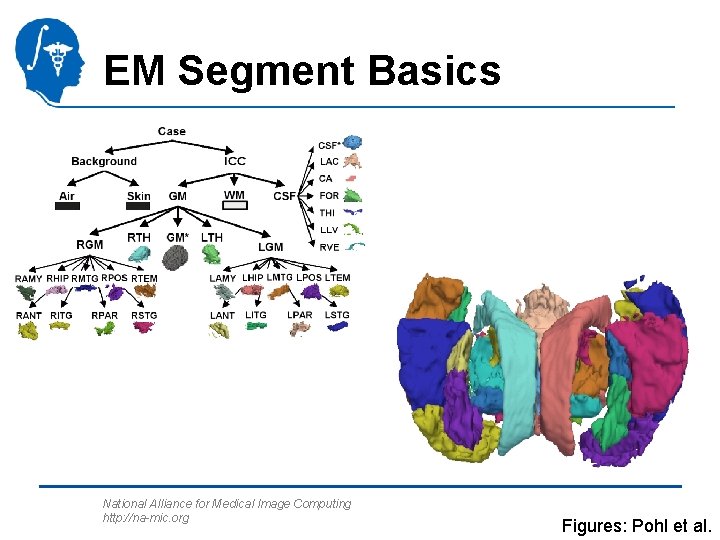
EM Segment Basics Parameters Target Images Segmentation Result Atlas Images WM GM CSF Air Skin/Skull National Alliance for Medical Image Computing http: //na-mic. org

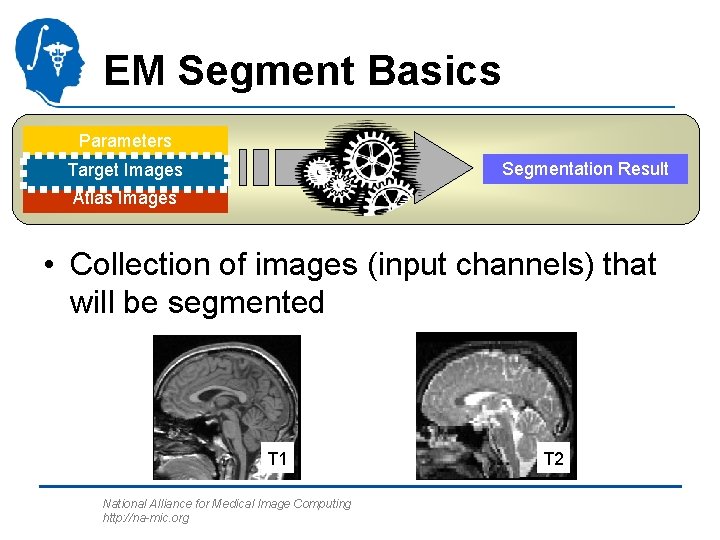
EM Segment Basics Parameters Segmentation Result Target Images Atlas Images • Collection of images (input channels) that will be segmented T 1 National Alliance for Medical Image Computing http: //na-mic. org T 2

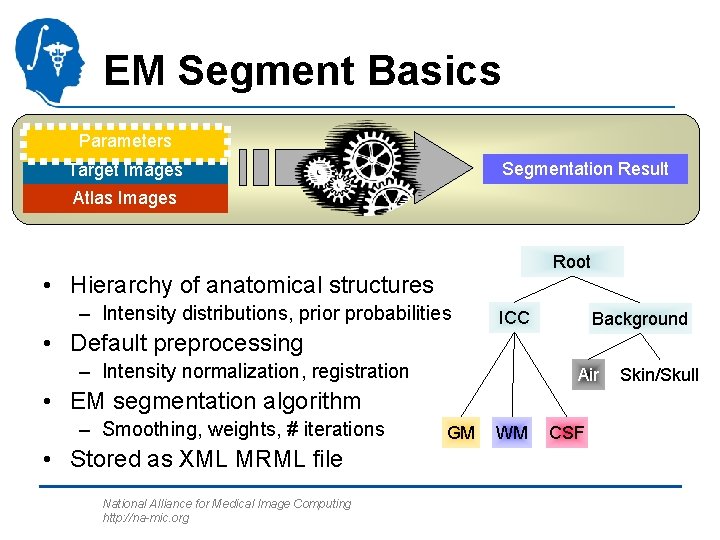
EM Segment Basics Parameters Segmentation Result Target Images Atlas Images Root • Hierarchy of anatomical structures – Intensity distributions, prior probabilities ICC Background • Default preprocessing – Intensity normalization, registration Air • EM segmentation algorithm – Smoothing, weights, # iterations • Stored as XML MRML file National Alliance for Medical Image Computing http: //na-mic. org GM WM CSF Skin/Skull

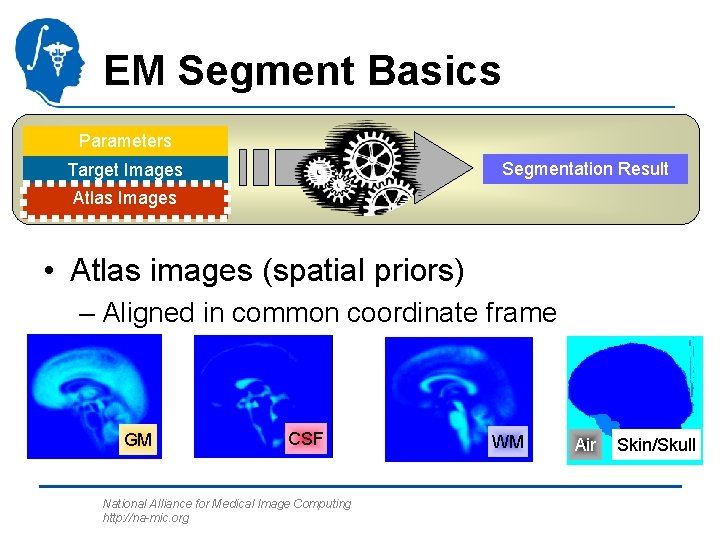
EM Segment Basics Parameters Segmentation Result Target Images Atlas Images • Atlas images (spatial priors) – Aligned in common coordinate frame GM CSF National Alliance for Medical Image Computing http: //na-mic. org WM Air Skin/Skull

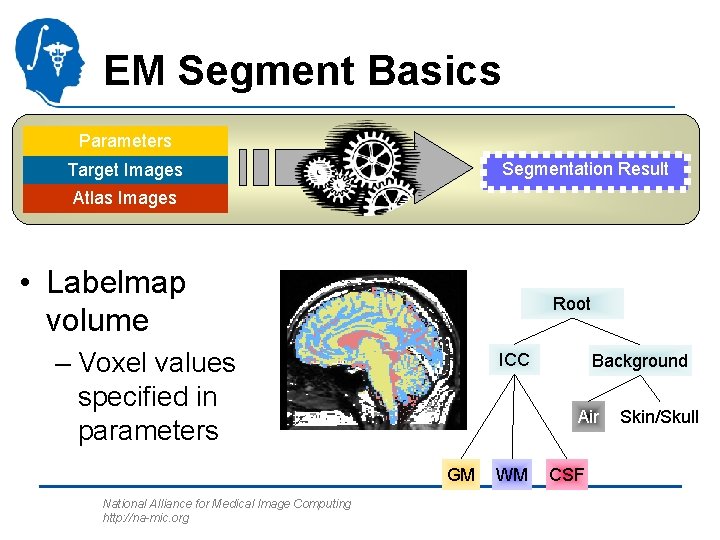
EM Segment Basics Parameters Segmentation Result Target Images Atlas Images • Labelmap volume Root – Voxel values specified in parameters ICC Air GM National Alliance for Medical Image Computing http: //na-mic. org Background WM CSF Skin/Skull

Algorithm Development • 12 -year history of algorithm development – Wells et al. 1996 (TMI): EM framework for simultaneous estimation of bias field and label map – Kapur et al. 1999 (Ph. D Thesis): Model noise via Markov Random Field – Van Leemput et al. 1999 (TMI): Non-spatial tissue priors – Pohl et al. • 2002 (MICCAI): Deformable registration to align atlas • 2004 (ISBI): Hierarchical framework to model anatomical dependencies • 2007 (TMI): Up-to-date, detailed description National Alliance for Medical Image Computing http: //na-mic. org

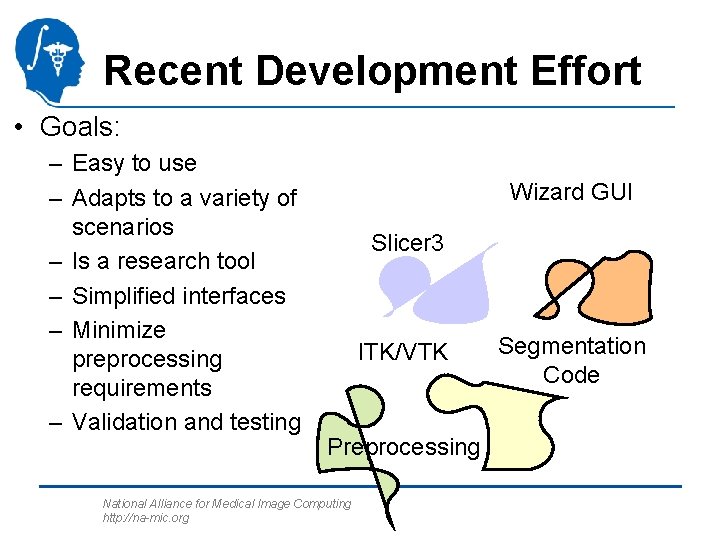
Recent Development Effort • Goals: – Easy to use – Adapts to a variety of scenarios – Is a research tool – Simplified interfaces – Minimize preprocessing requirements – Validation and testing Wizard GUI Slicer 3 ITK/VTK Preprocessing National Alliance for Medical Image Computing http: //na-mic. org Segmentation Code

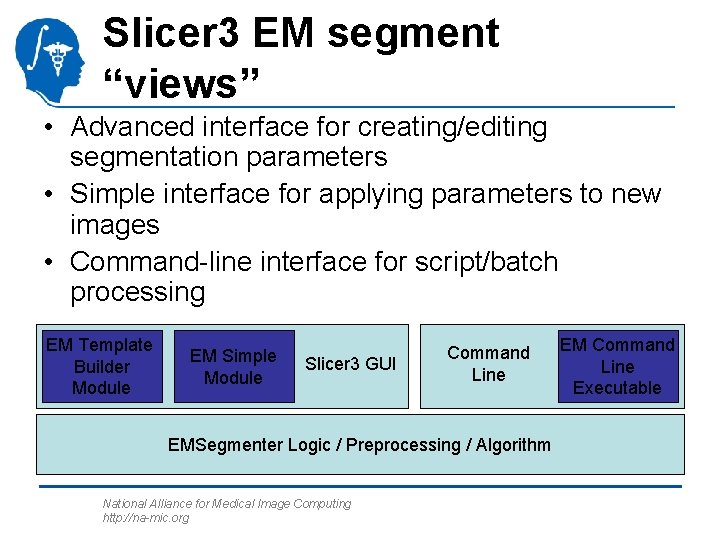
Slicer 3 EM segment “views” • Advanced interface for creating/editing segmentation parameters • Simple interface for applying parameters to new images • Command-line interface for script/batch processing EM Template Builder Module EM Simple Module Slicer 3 GUI Command Line EMSegmenter Logic / Preprocessing / Algorithm National Alliance for Medical Image Computing http: //na-mic. org EM Command Line Executable

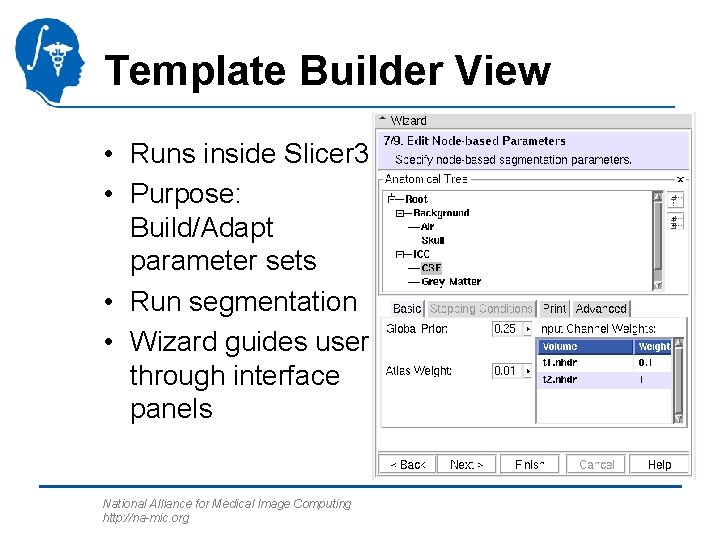
Template Builder View • Runs inside Slicer 3 • Purpose: Build/Adapt parameter sets • Run segmentation • Wizard guides user through interface panels National Alliance for Medical Image Computing http: //na-mic. org

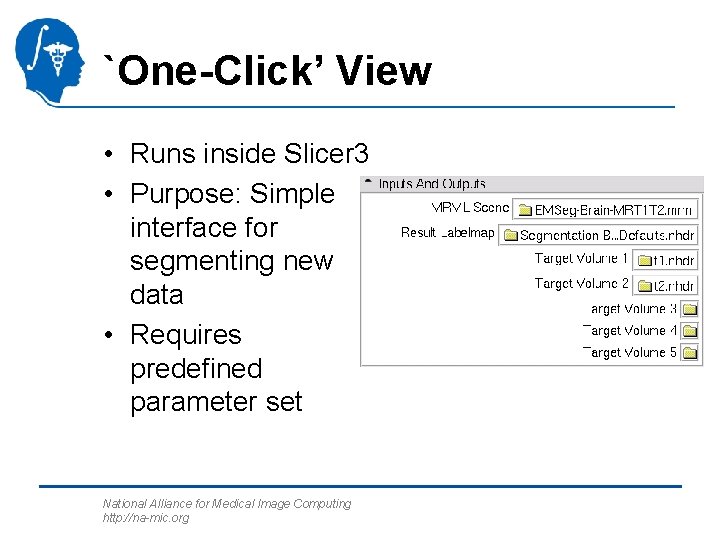
`One-Click’ View • Runs inside Slicer 3 • Purpose: Simple interface for segmenting new data • Requires predefined parameter set National Alliance for Medical Image Computing http: //na-mic. org

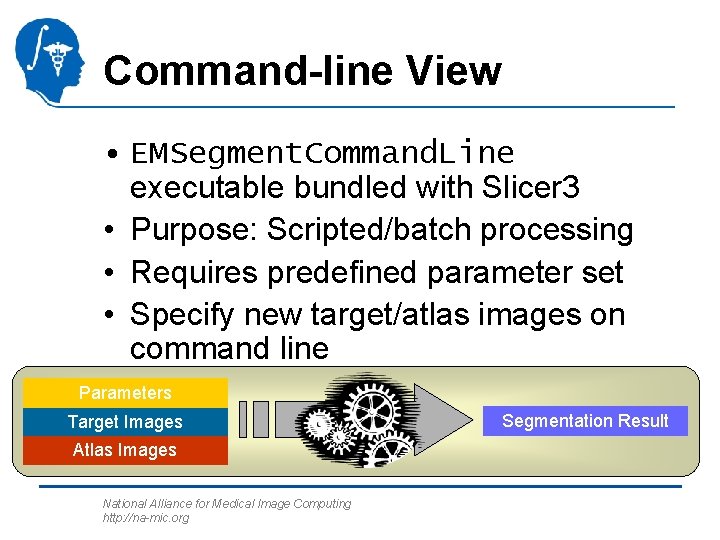
Command-line View • EMSegment. Command. Line executable bundled with Slicer 3 • Purpose: Scripted/batch processing • Requires predefined parameter set • Specify new target/atlas images on command line Parameters Target Images Atlas Images National Alliance for Medical Image Computing http: //na-mic. org Segmentation Result

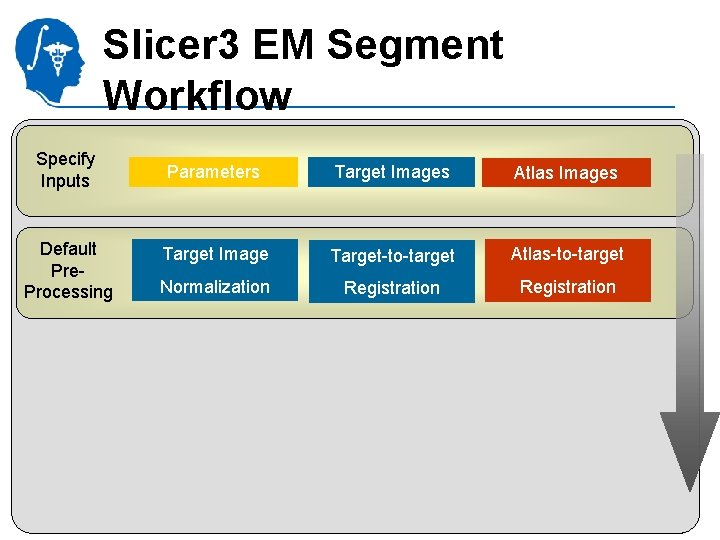
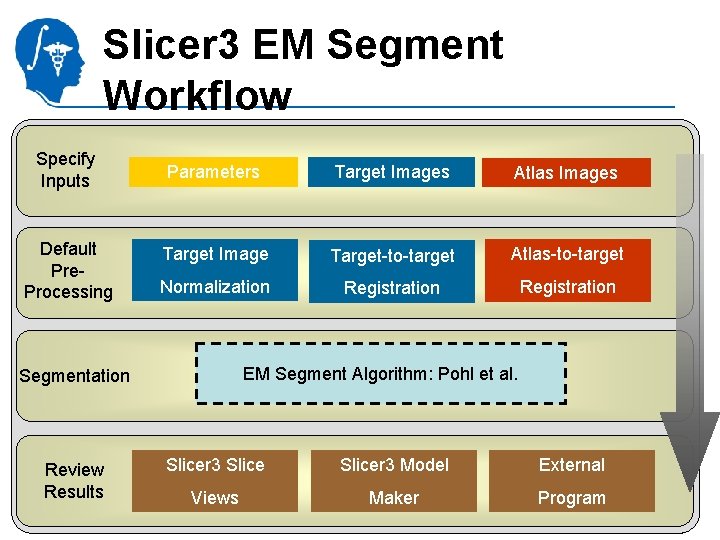
Slicer 3 EM Segment Workflow Specify Inputs Parameters Target Images National Alliance for Medical Image Computing http: //na-mic. org Atlas Images

Slicer 3 EM Segment Workflow Specify Inputs Default Pre. Processing Parameters Target Images Atlas Images Target Image Target-to-target Atlas-to-target Normalization Registration National Alliance for Medical Image Computing http: //na-mic. org

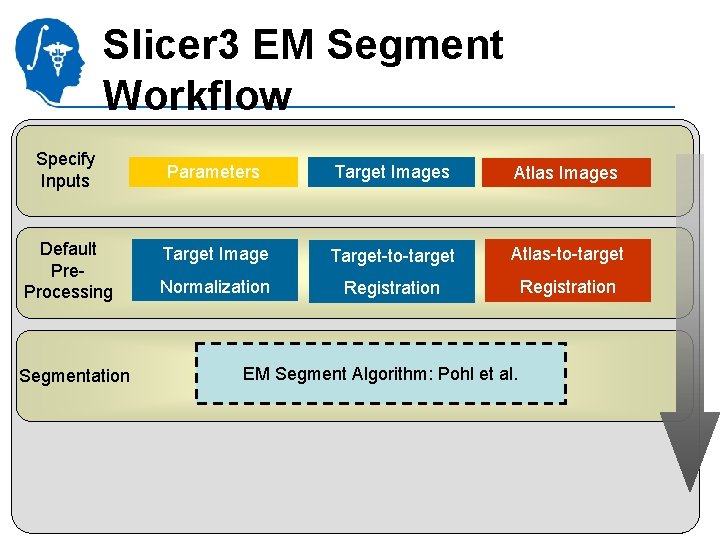
Slicer 3 EM Segment Workflow Specify Inputs Default Pre. Processing Segmentation Parameters Target Images Atlas Images Target Image Target-to-target Atlas-to-target Normalization Registration EM Segment Algorithm: Pohl et al. National Alliance for Medical Image Computing http: //na-mic. org

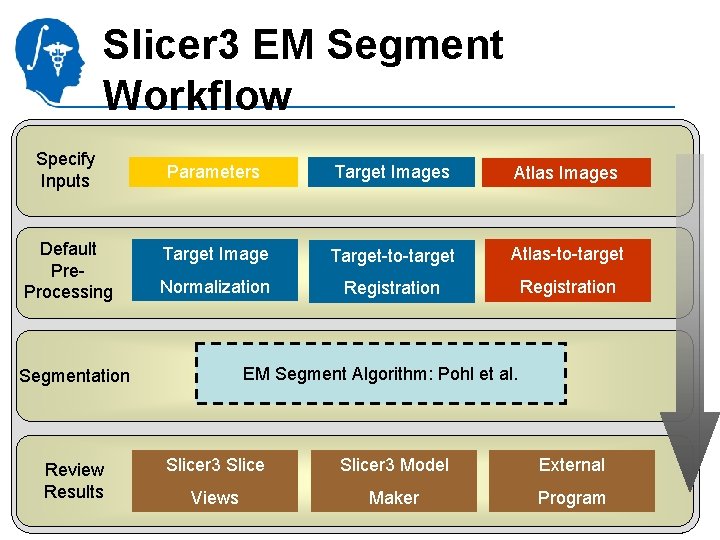
Slicer 3 EM Segment Workflow Specify Inputs Default Pre. Processing Segmentation Review Results Parameters Target Images Atlas Images Target Image Target-to-target Atlas-to-target Normalization Registration EM Segment Algorithm: Pohl et al. Slicer 3 Model National Alliance. Views for Medical Image Computing http: //na-mic. org Maker External Program

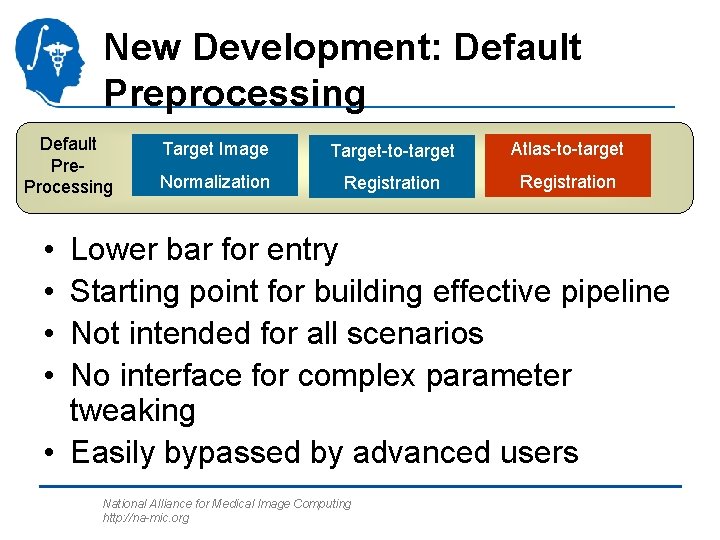
New Development: Default Preprocessing Default Pre. Processing Target Image Target-to-target Atlas-to-target Normalization Registration • • Lower bar for entry Starting point for building effective pipeline Not intended for all scenarios No interface for complex parameter tweaking • Easily bypassed by advanced users National Alliance for Medical Image Computing http: //na-mic. org

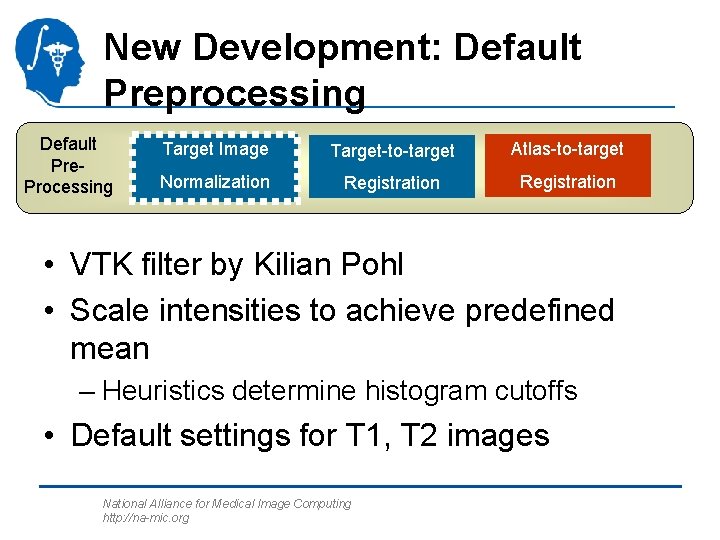
New Development: Default Preprocessing Default Pre. Processing Target Image Target-to-target Atlas-to-target Normalization Registration • VTK filter by Kilian Pohl • Scale intensities to achieve predefined mean – Heuristics determine histogram cutoffs • Default settings for T 1, T 2 images National Alliance for Medical Image Computing http: //na-mic. org

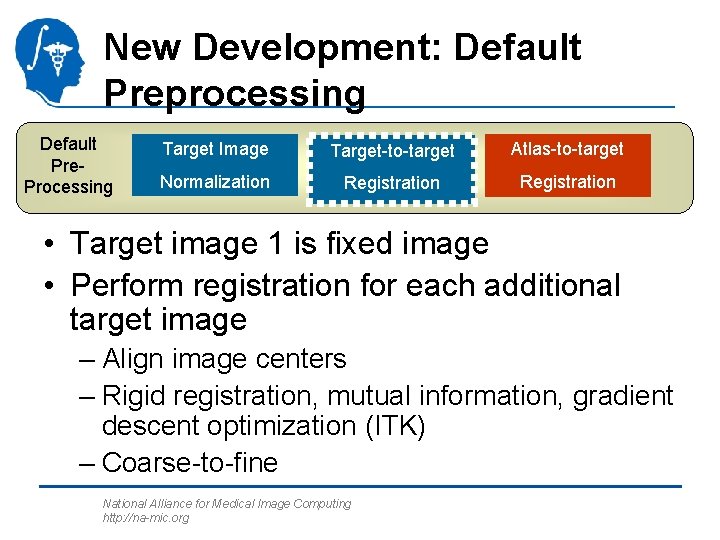
New Development: Default Preprocessing Default Pre. Processing Target Image Target-to-target Atlas-to-target Normalization Registration • Target image 1 is fixed image • Perform registration for each additional target image – Align image centers – Rigid registration, mutual information, gradient descent optimization (ITK) – Coarse-to-fine National Alliance for Medical Image Computing http: //na-mic. org

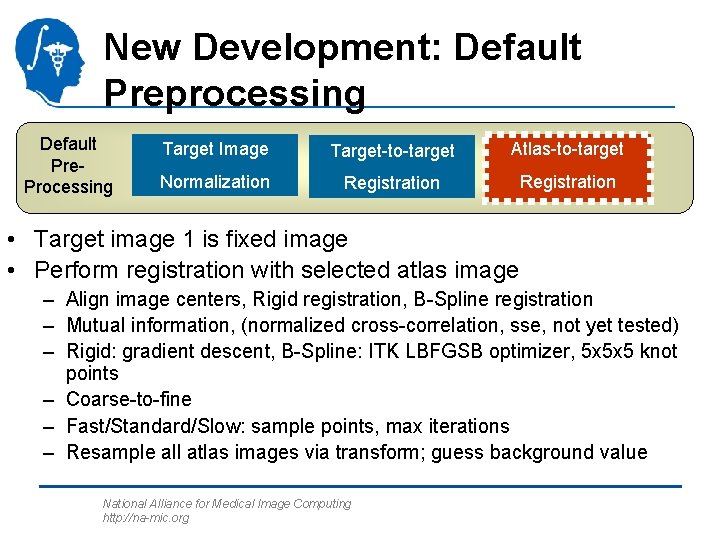
New Development: Default Preprocessing Default Pre. Processing Target Image Target-to-target Atlas-to-target Normalization Registration • Target image 1 is fixed image • Perform registration with selected atlas image – Align image centers, Rigid registration, B-Spline registration – Mutual information, (normalized cross-correlation, sse, not yet tested) – Rigid: gradient descent, B-Spline: ITK LBFGSB optimizer, 5 x 5 x 5 knot points – Coarse-to-fine – Fast/Standard/Slow: sample points, max iterations – Resample all atlas images via transform; guess background value National Alliance for Medical Image Computing http: //na-mic. org

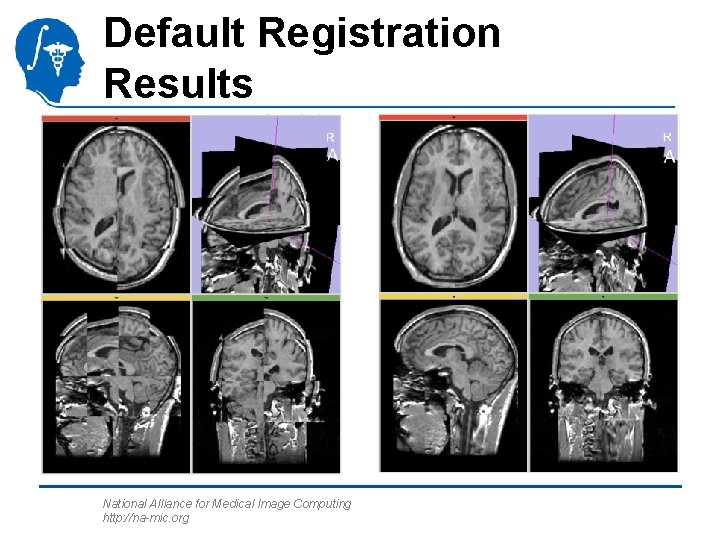
Default Registration Results National Alliance for Medical Image Computing http: //na-mic. org

Tutorial Data Walkthrough 1/3 • README: extra release notes • Predefined parameter file – MR T 1 & T 2 brain segmentation: White matter, gray matter, CSF • Target Images – Image. Data_Input sub directory – Target: T 1 & T 2 images used in parameter file – Target_New. Patient 1, Target_New. Patient 2: New data to try with the parameter file National Alliance for Medical Image Computing http: //na-mic. org

Tutorial Data Walkthrough 2/3 • Atlas Images – Image. Data_Input sub directory – Atlas: atlas images used in parameter file – Atlas_New. Atlas: Extra atlas data for demonstration purposes (currently a copy of first atlas) National Alliance for Medical Image Computing http: //na-mic. org

Tutorial Data Walkthrough 3/3 • Low resolution data for testing – Full sized data: 30 minutes – Low resolution data: 1 minute • Data, parameters have “_small” appended to filename National Alliance for Medical Image Computing http: //na-mic. org

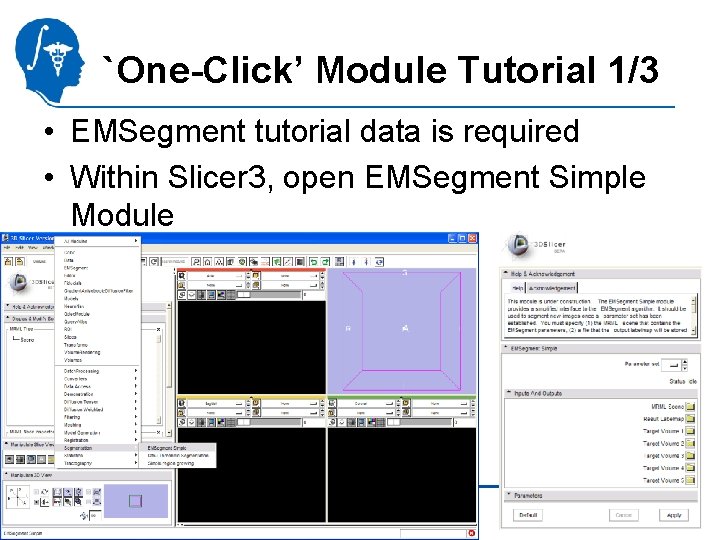
`One-Click’ Module Tutorial 1/3 • EMSegment tutorial data is required • Within Slicer 3, open EMSegment Simple Module National Alliance for Medical Image Computing http: //na-mic. org

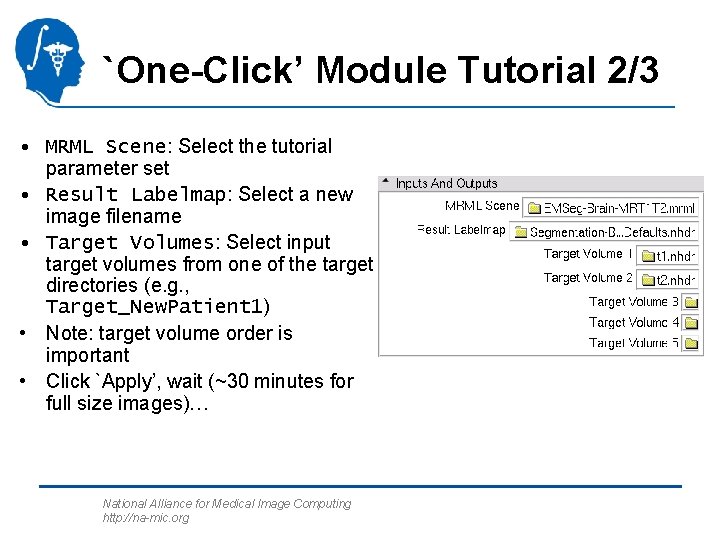
`One-Click’ Module Tutorial 2/3 • MRML Scene: Select the tutorial parameter set • Result Labelmap: Select a new image filename • Target Volumes: Select input target volumes from one of the target directories (e. g. , Target_New. Patient 1) • Note: target volume order is important • Click `Apply’, wait (~30 minutes for full size images)… National Alliance for Medical Image Computing http: //na-mic. org

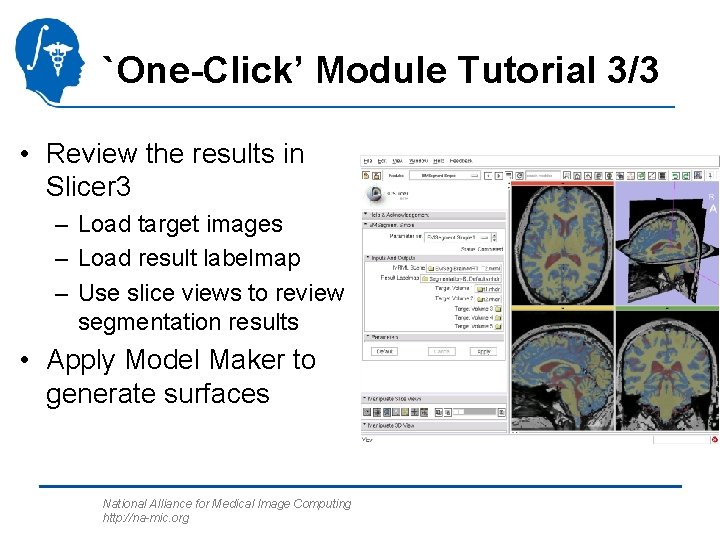
`One-Click’ Module Tutorial 3/3 • Review the results in Slicer 3 – Load target images – Load result labelmap – Use slice views to review segmentation results • Apply Model Maker to generate surfaces National Alliance for Medical Image Computing http: //na-mic. org

EMSegment Command-line Examples • Script examples available in tutorial directory – New images – New atlas – New image and atlas – See README National Alliance for Medical Image Computing http: //na-mic. org

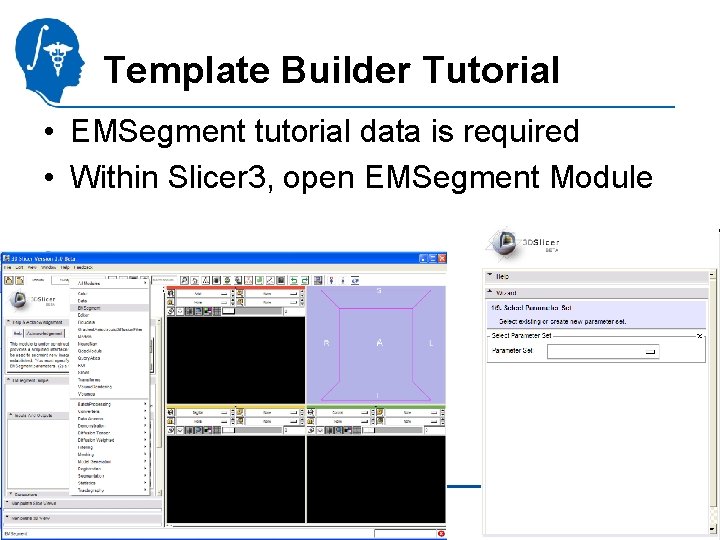
Template Builder Tutorial • EMSegment tutorial data is required • Within Slicer 3, open EMSegment Module National Alliance for Medical Image Computing http: //na-mic. org

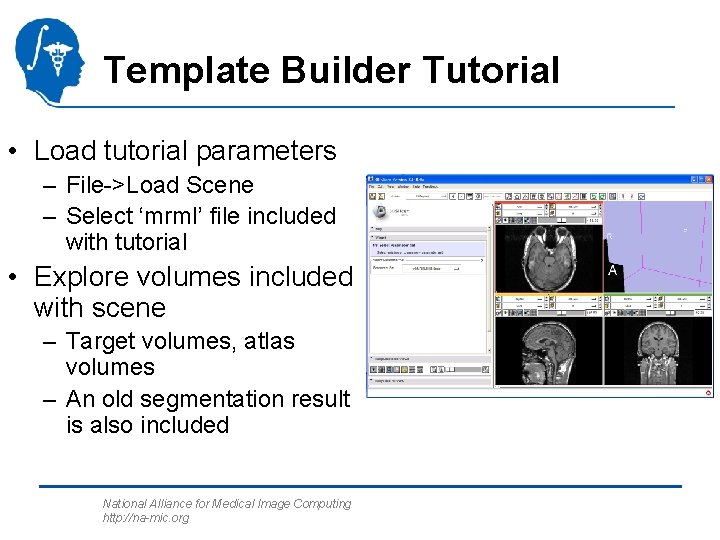
Template Builder Tutorial • Load tutorial parameters – File->Load Scene – Select ‘mrml’ file included with tutorial • Explore volumes included with scene – Target volumes, atlas volumes – An old segmentation result is also included National Alliance for Medical Image Computing http: //na-mic. org

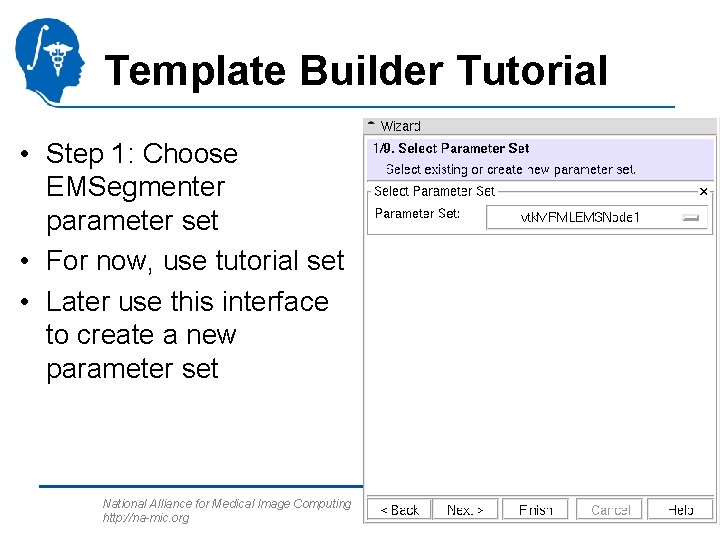
Template Builder Tutorial • Step 1: Choose EMSegmenter parameter set • For now, use tutorial set • Later use this interface to create a new parameter set National Alliance for Medical Image Computing http: //na-mic. org

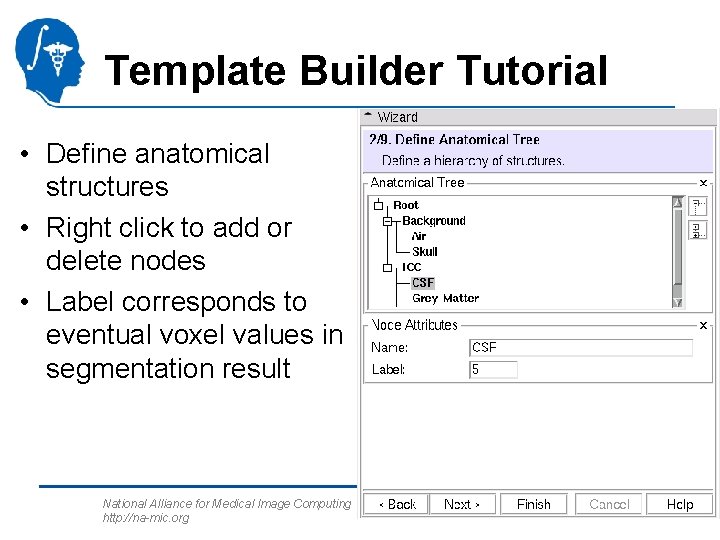
Template Builder Tutorial • Define anatomical structures • Right click to add or delete nodes • Label corresponds to eventual voxel values in segmentation result National Alliance for Medical Image Computing http: //na-mic. org

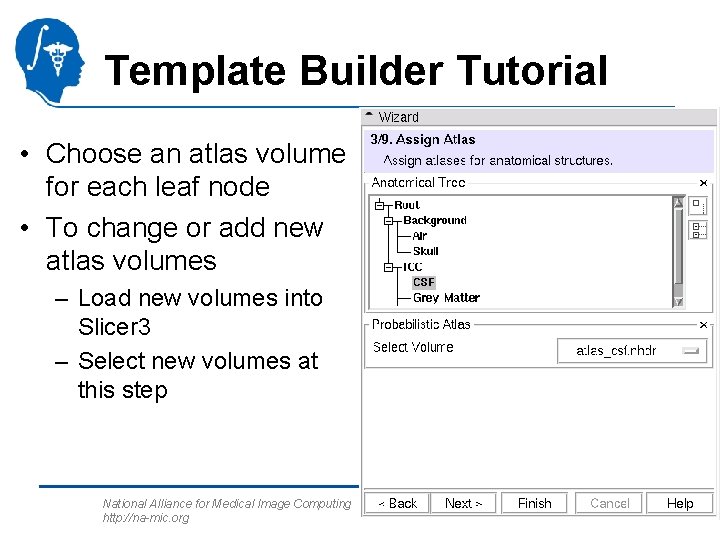
Template Builder Tutorial • Choose an atlas volume for each leaf node • To change or add new atlas volumes – Load new volumes into Slicer 3 – Select new volumes at this step National Alliance for Medical Image Computing http: //na-mic. org

Template Builder Tutorial • Choose target images • To change or add target images – Load new volumes into Slicer 3 – Select new volumes at this step • You can reorder target images; order is important National Alliance for Medical Image Computing http: //na-mic. org

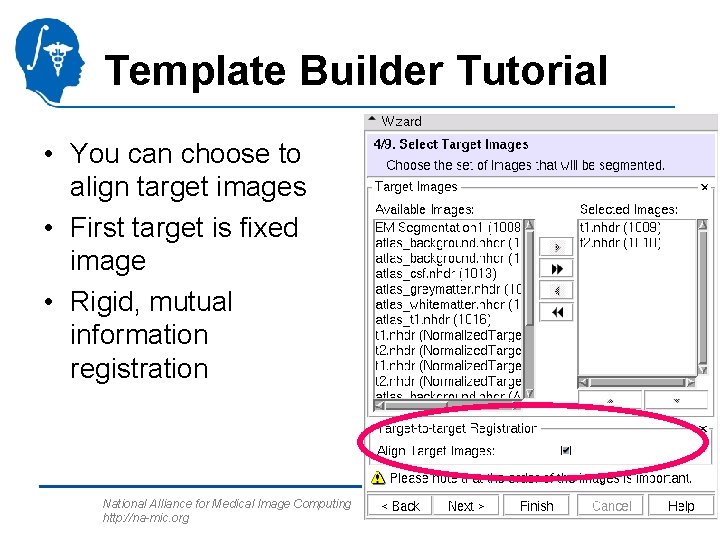
Template Builder Tutorial • You can choose to align target images • First target is fixed image • Rigid, mutual information registration National Alliance for Medical Image Computing http: //na-mic. org

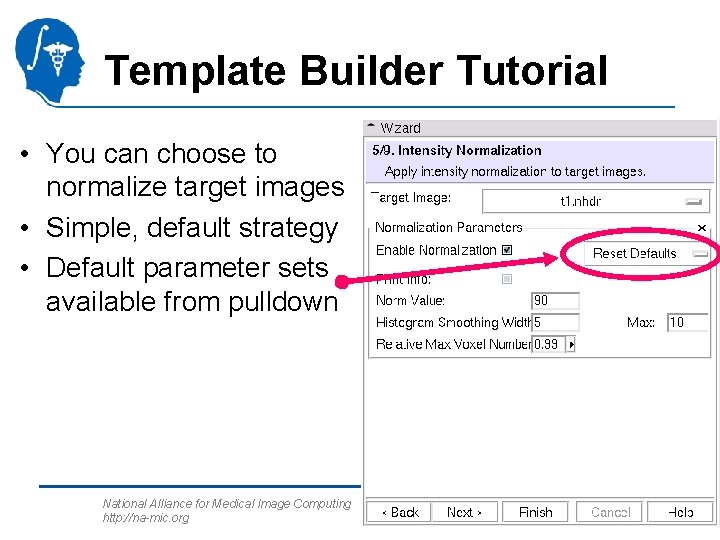
Template Builder Tutorial • You can choose to normalize target images • Simple, default strategy • Default parameter sets available from pulldown National Alliance for Medical Image Computing http: //na-mic. org

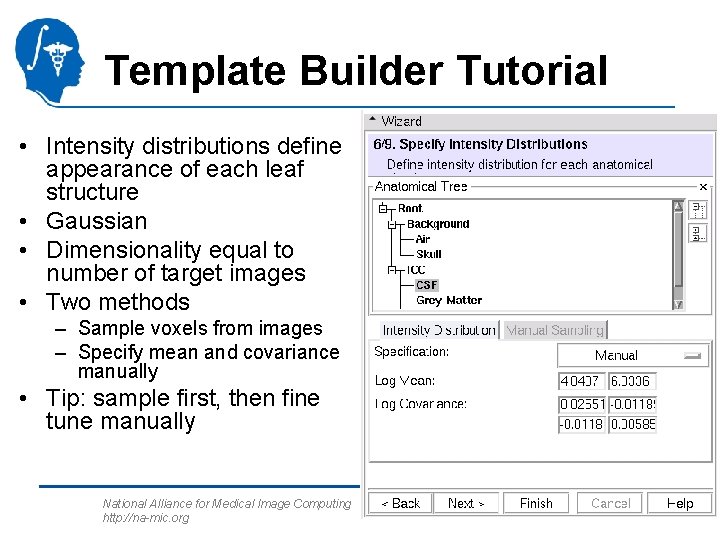
Template Builder Tutorial • Intensity distributions define appearance of each leaf structure • Gaussian • Dimensionality equal to number of target images • Two methods – Sample voxels from images – Specify mean and covariance manually • Tip: sample first, then fine tune manually National Alliance for Medical Image Computing http: //na-mic. org

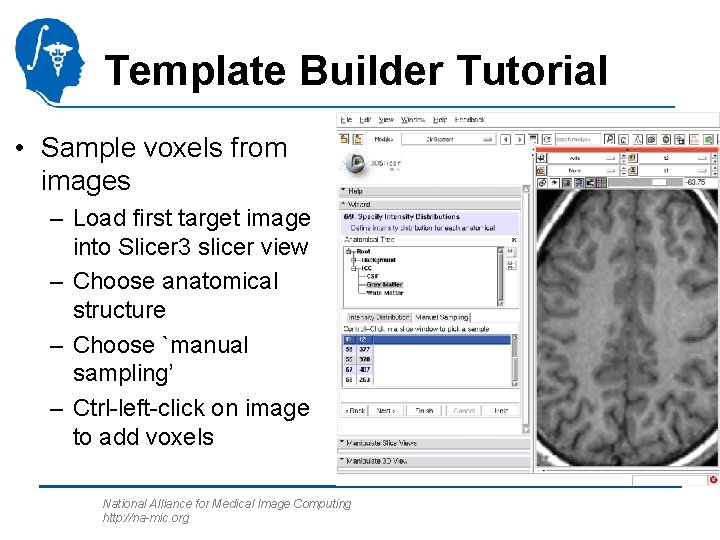
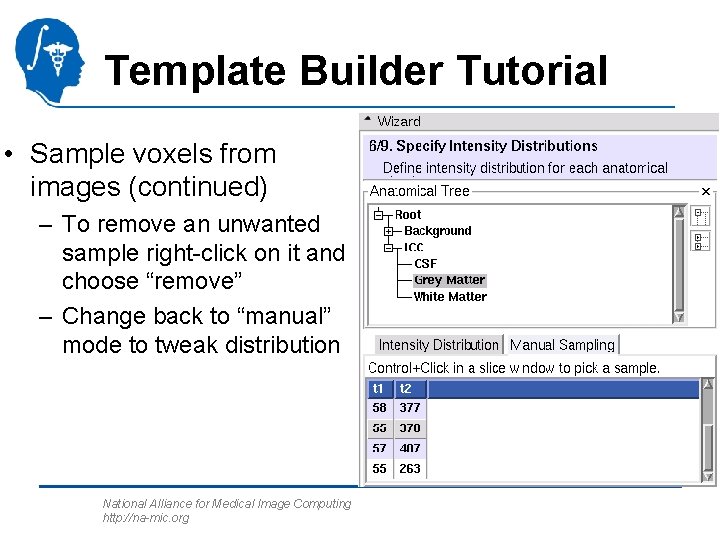
Template Builder Tutorial • Sample voxels from images – Load first target image into Slicer 3 slicer view – Choose anatomical structure – Choose `manual sampling’ – Ctrl-left-click on image to add voxels National Alliance for Medical Image Computing http: //na-mic. org

Template Builder Tutorial • Sample voxels from images (continued) – To remove an unwanted sample right-click on it and choose “remove” – Change back to “manual” mode to tweak distribution National Alliance for Medical Image Computing http: //na-mic. org

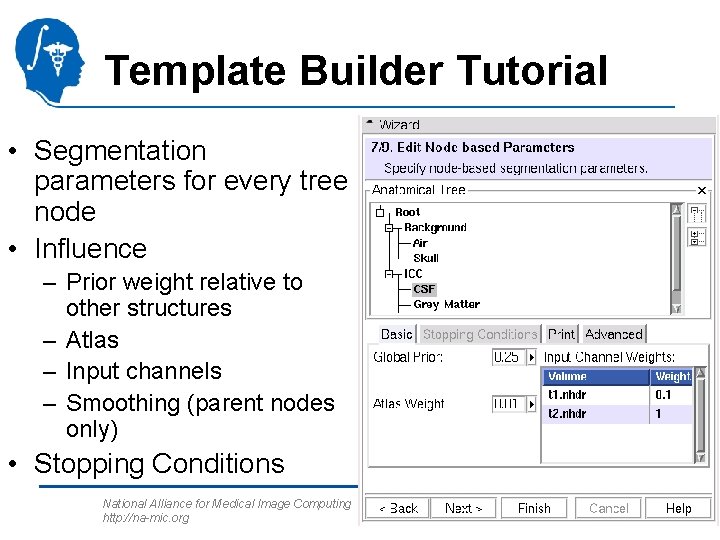
Template Builder Tutorial • Segmentation parameters for every tree node • Influence – Prior weight relative to other structures – Atlas – Input channels – Smoothing (parent nodes only) • Stopping Conditions National Alliance for Medical Image Computing http: //na-mic. org

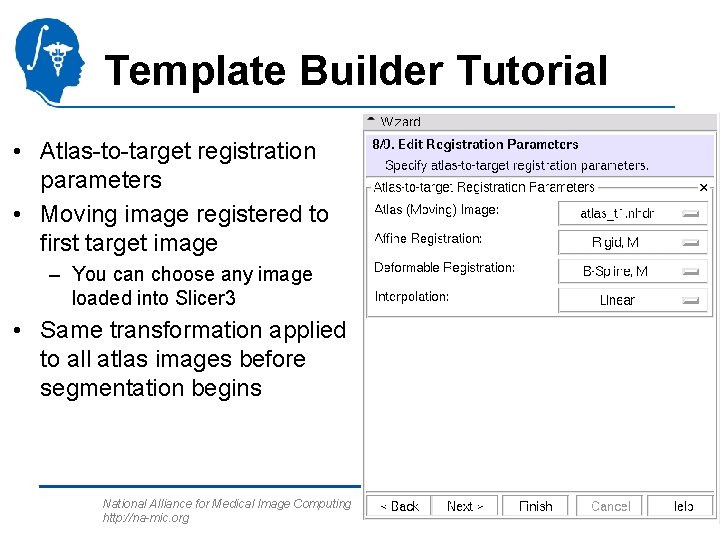
Template Builder Tutorial • Atlas-to-target registration parameters • Moving image registered to first target image – You can choose any image loaded into Slicer 3 • Same transformation applied to all atlas images before segmentation begins National Alliance for Medical Image Computing http: //na-mic. org

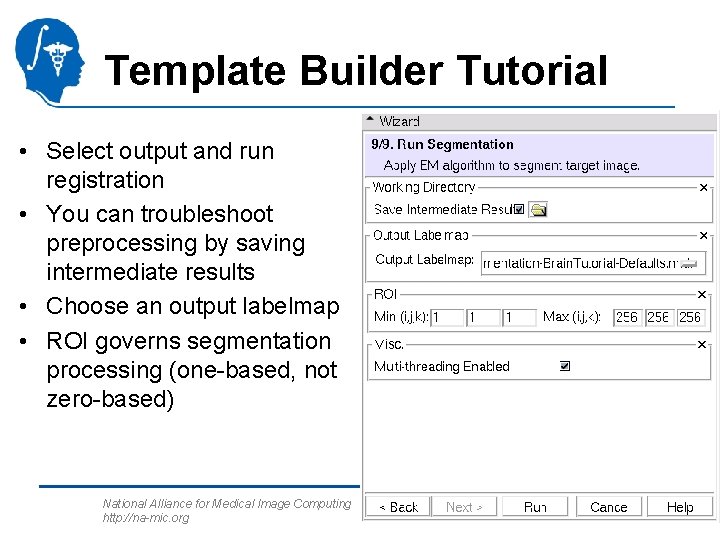
Template Builder Tutorial • Select output and run registration • You can troubleshoot preprocessing by saving intermediate results • Choose an output labelmap • ROI governs segmentation processing (one-based, not zero-based) National Alliance for Medical Image Computing http: //na-mic. org

Acknowledgements • • • Wendy Plesniak, SPL BWH Steve Pieper, Isomics Luis Ibáñez, Kitware Sylvain Bouix, PNL BWH William Wells, BWH Funding provided by NAMIC National Alliance for Medical Image Computing http: //na-mic. org

NA-MIC National Alliance for Medical Image Computing http: //na-mic. org Questions…

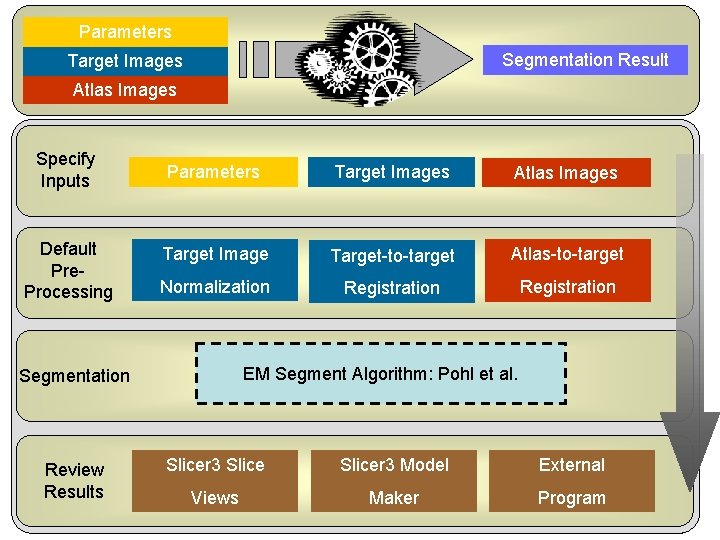
Slicer 3 EM Segment Workflow Specify Inputs Default Pre. Processing Segmentation Review Results Parameters Target Images Atlas Images Target Image Target-to-target Atlas-to-target Normalization Registration EM Segment Algorithm: Pohl et al. Slicer 3 Model National Alliance. Views for Medical Image Computing http: //na-mic. org Maker External Program

Parameters Segmentation Result Target Images Atlas Images Specify Inputs Default Pre. Processing Segmentation Review Results Parameters Target Images Atlas Images Target Image Target-to-target Atlas-to-target Normalization Registration EM Segment Algorithm: Pohl et al. Slicer 3 Model National Alliance. Views for Medical Image Computing http: //na-mic. org Maker External Program