MOLECULAR COMPUTING DNA COMPUTING Donald Riggs DNA Computing













- Slides: 65

MOLECULAR COMPUTING DNA COMPUTING Donald Riggs

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Two types of molecular computing: Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion ¨ Specially n n created molecules - Rotaxane Wire into grid - FPGA Configure to form logic gates ¨ DNA computing

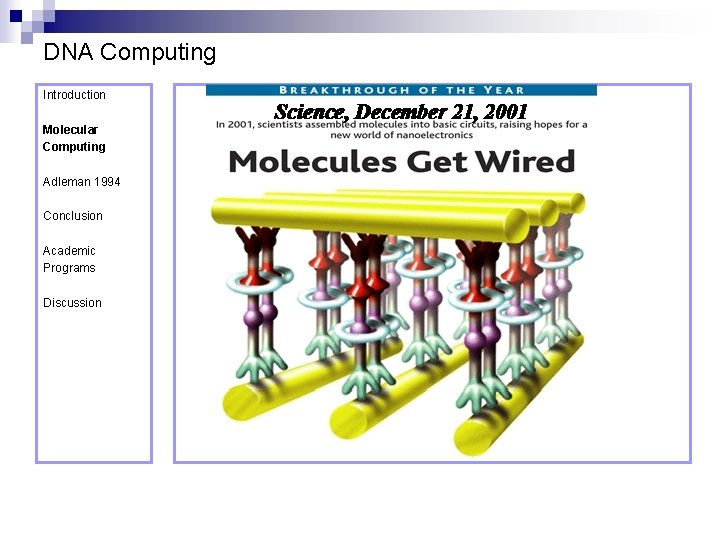
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

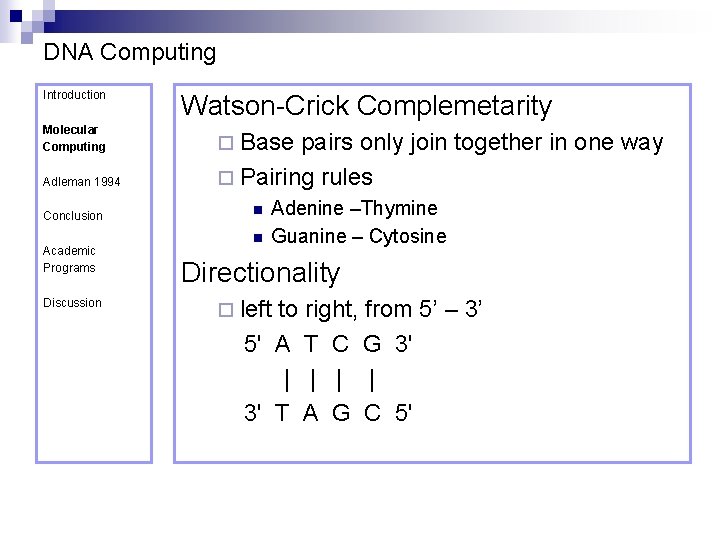
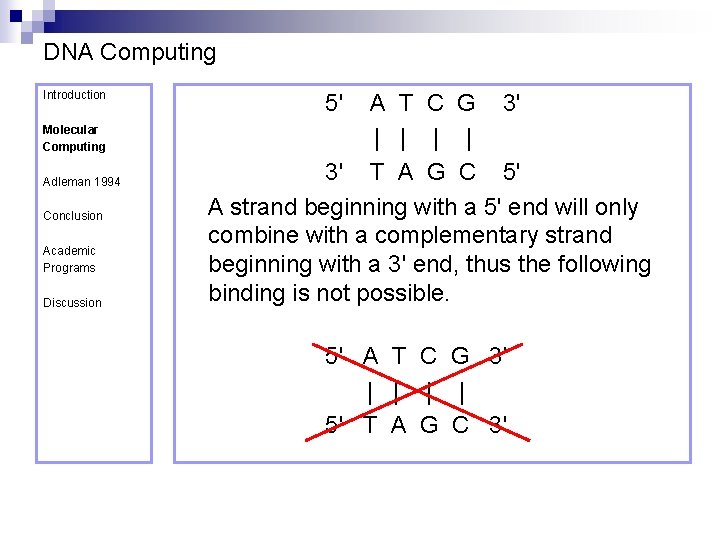
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Watson-Crick Complemetarity ¨ Base pairs only join together in one way ¨ Pairing rules n n Adenine –Thymine Guanine – Cytosine Directionality ¨ left to right, from 5’ – 3’ 5' A T C G 3' | | 3' T A G C 5'

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion 5' A T C G 3' | | 3' T A G C 5' A strand beginning with a 5' end will only combine with a complementary strand beginning with a 3' end, thus the following binding is not possible. 5' A T C G 3' | | 5' T A G C 3'

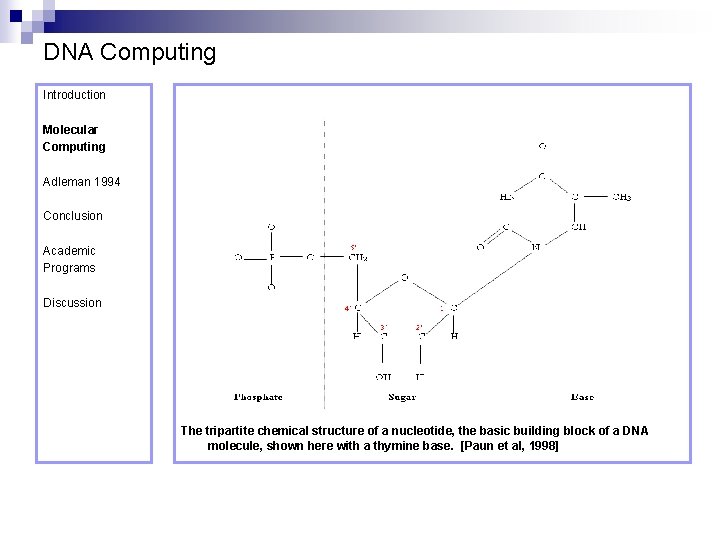
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion The tripartite chemical structure of a nucleotide, the basic building block of a DNA molecule, shown here with a thymine base. [Paun et al, 1998]

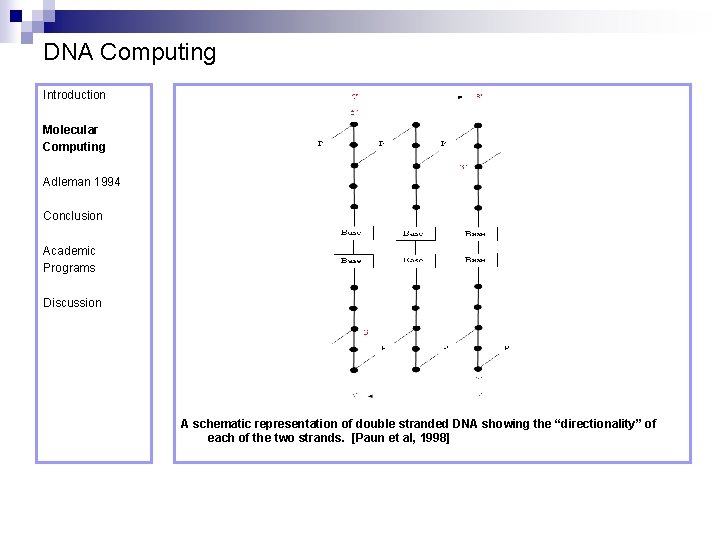
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion A schematic representation of double stranded DNA showing the “directionality” of each of the two strands. [Paun et al, 1998]

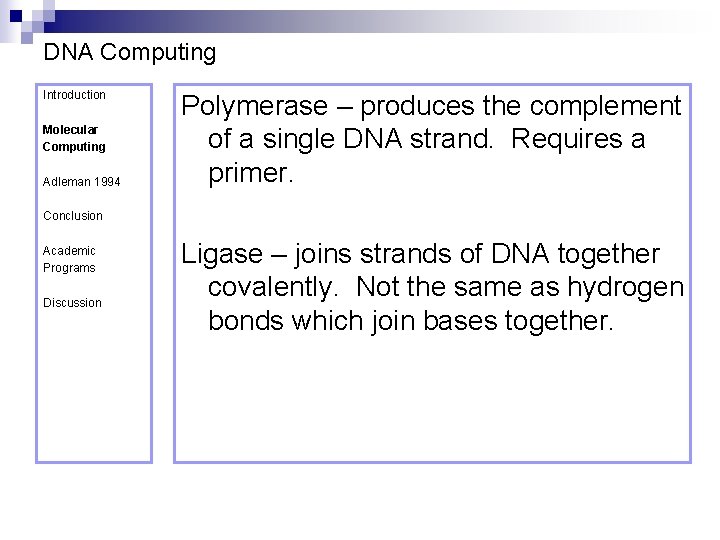
DNA Computing Introduction Molecular Computing Adleman 1994 Polymerase – produces the complement of a single DNA strand. Requires a primer. Conclusion Academic Programs Discussion Ligase – joins strands of DNA together covalently. Not the same as hydrogen bonds which join bases together.

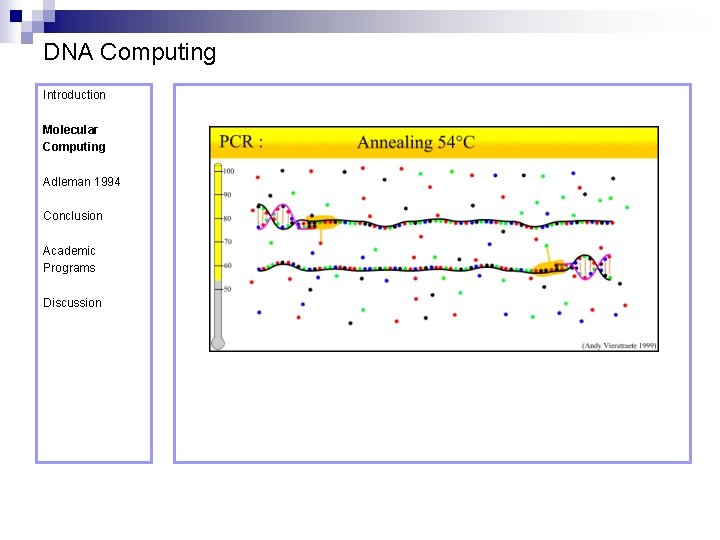
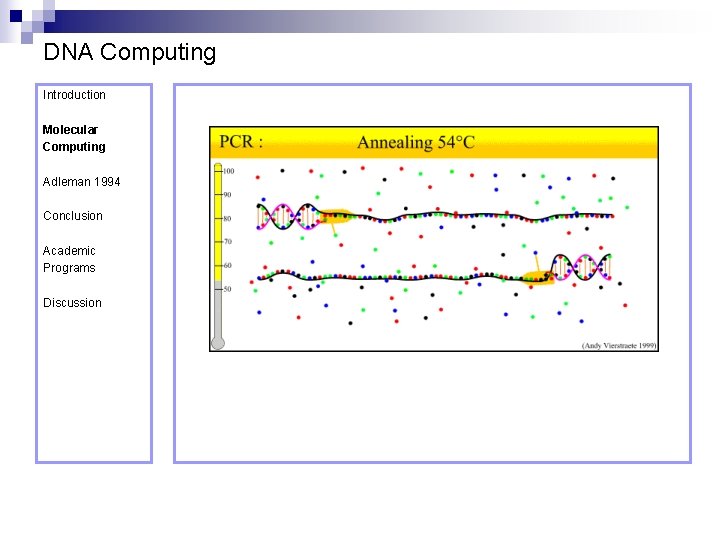
DNA Computing Introduction Molecular Computing Annealing – formation of doublestranded DNA from single strands. Adleman 1994 Conclusion Academic Programs Discussion Denaturation – separation of a doublestranded DNA molecule into two single-strands.

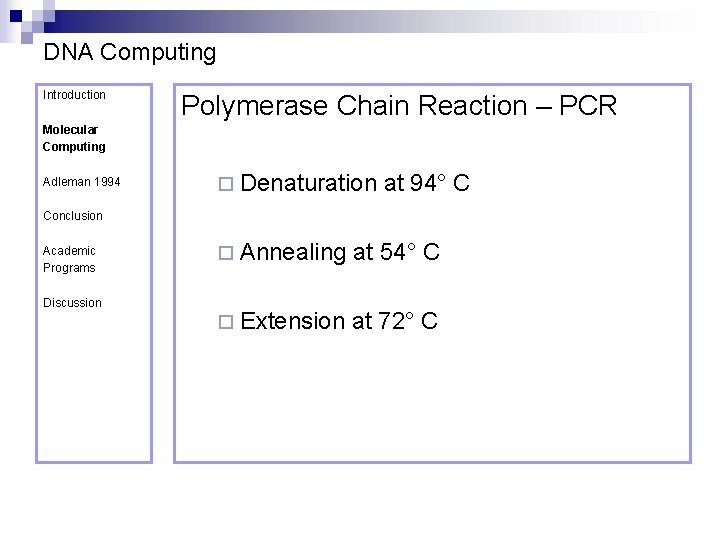
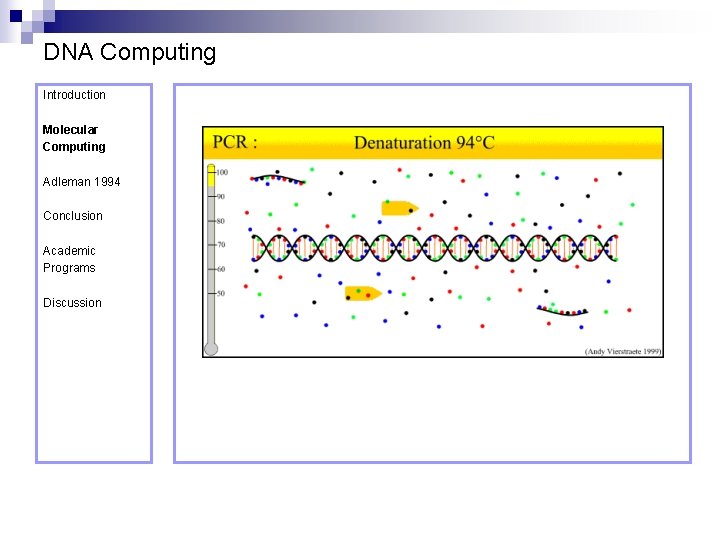
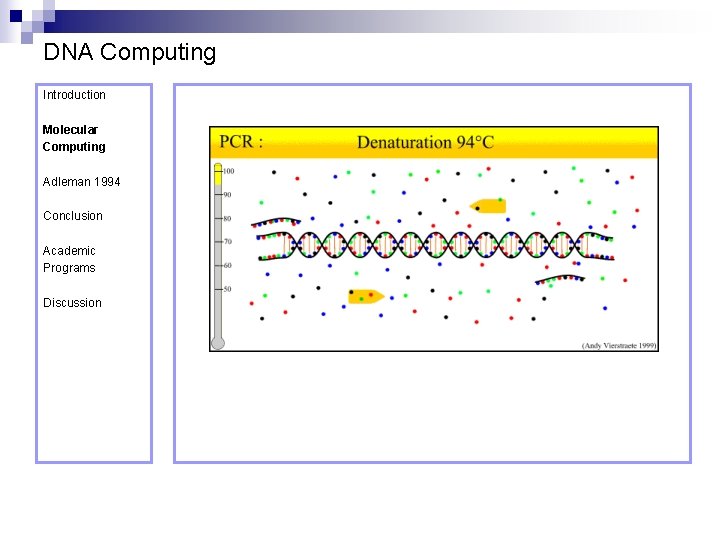
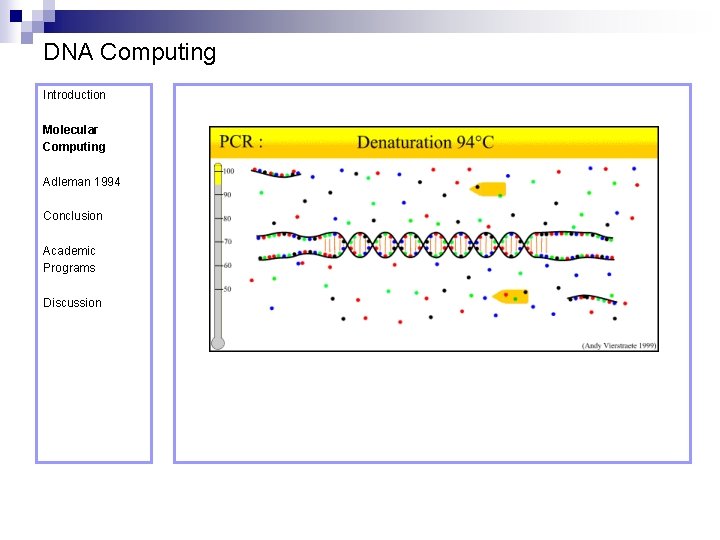
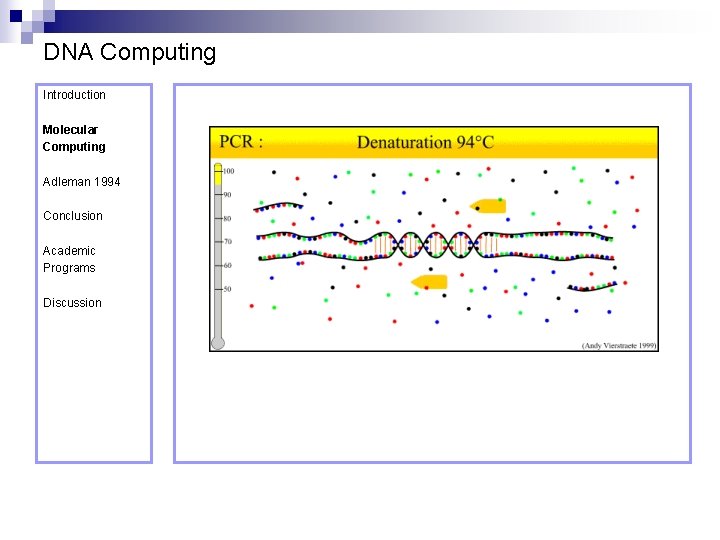
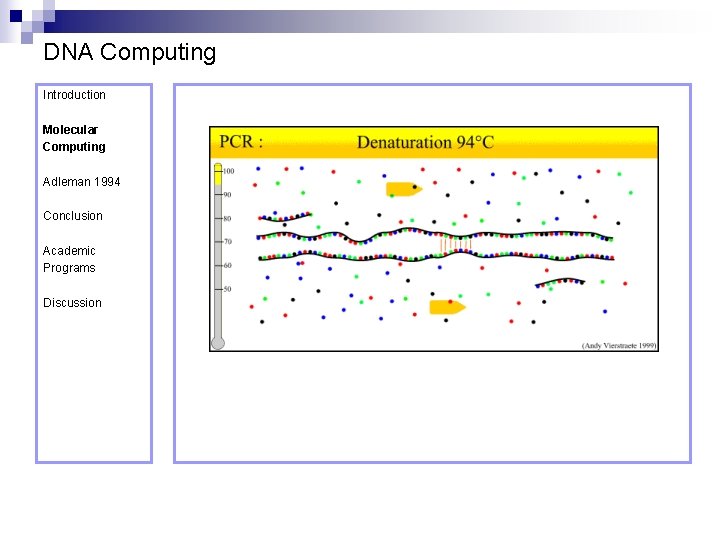
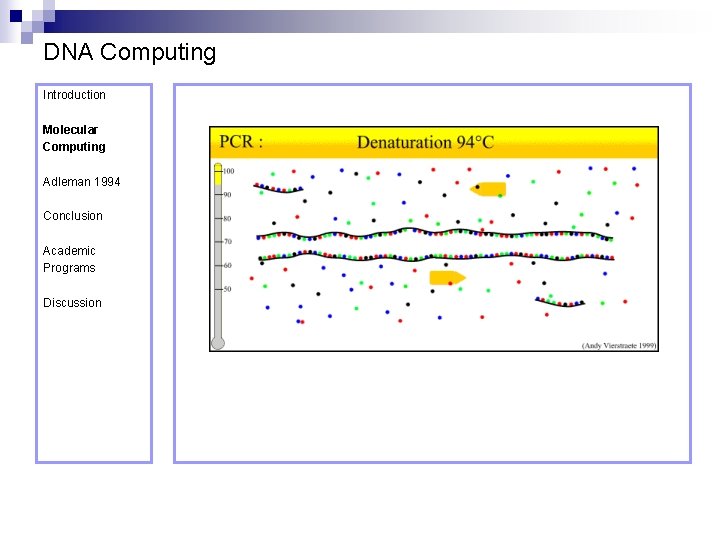
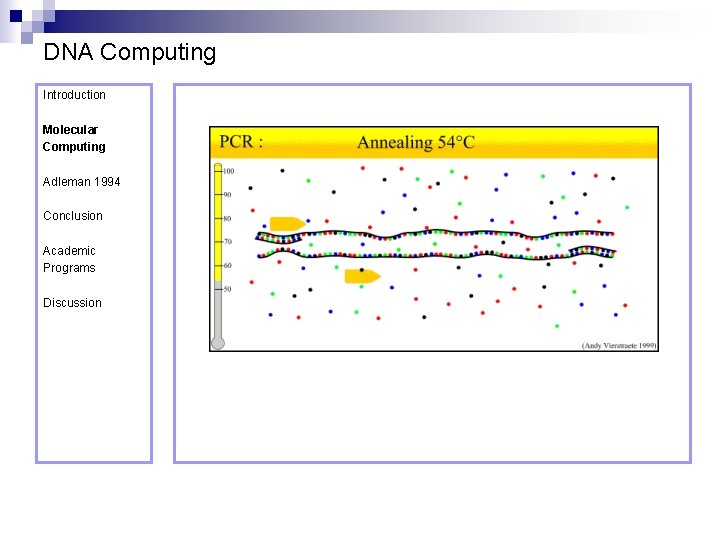
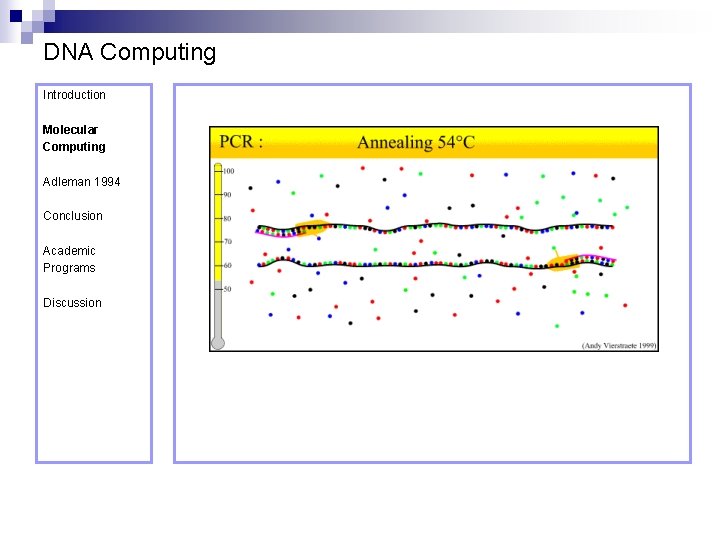
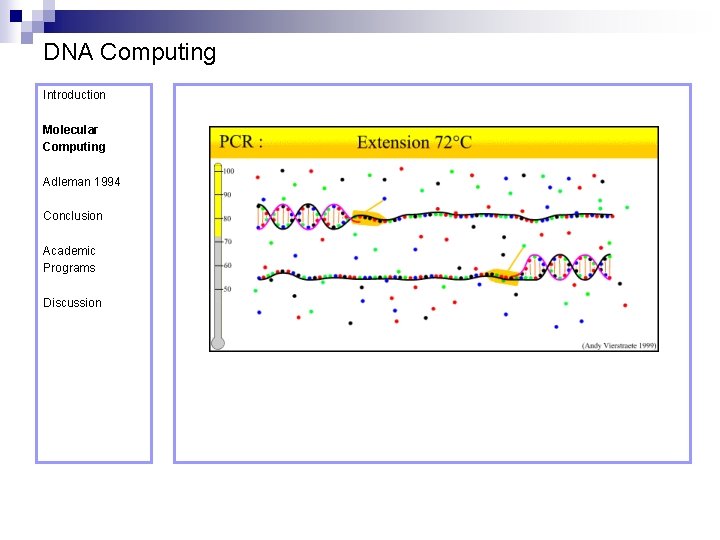
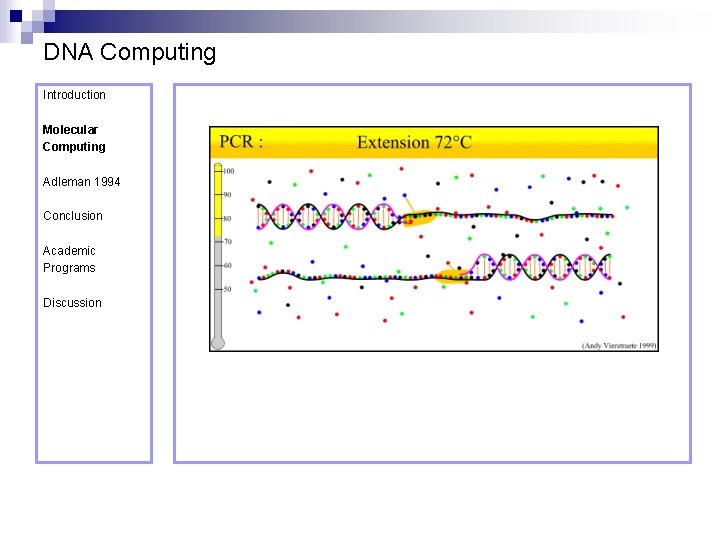
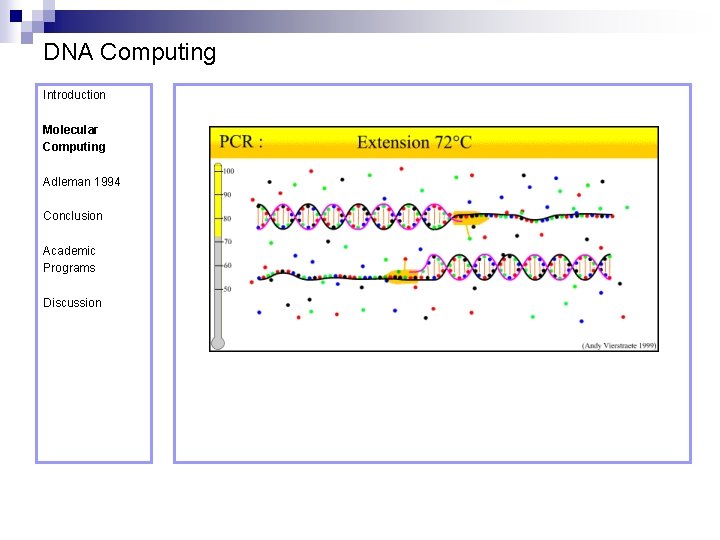
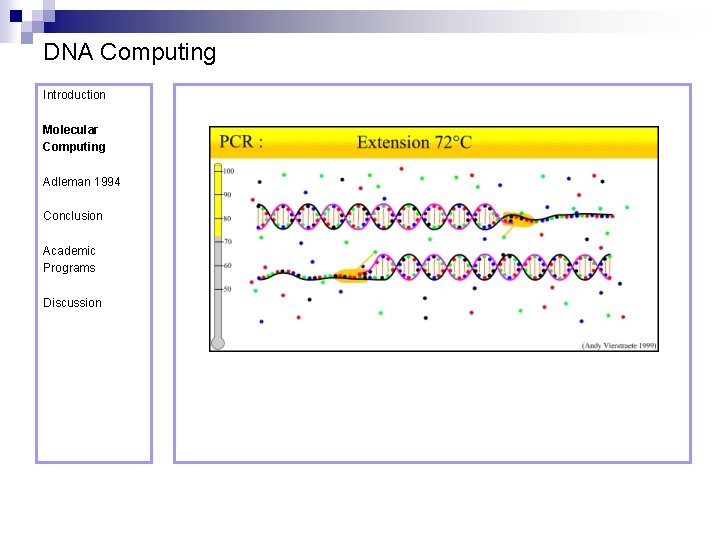
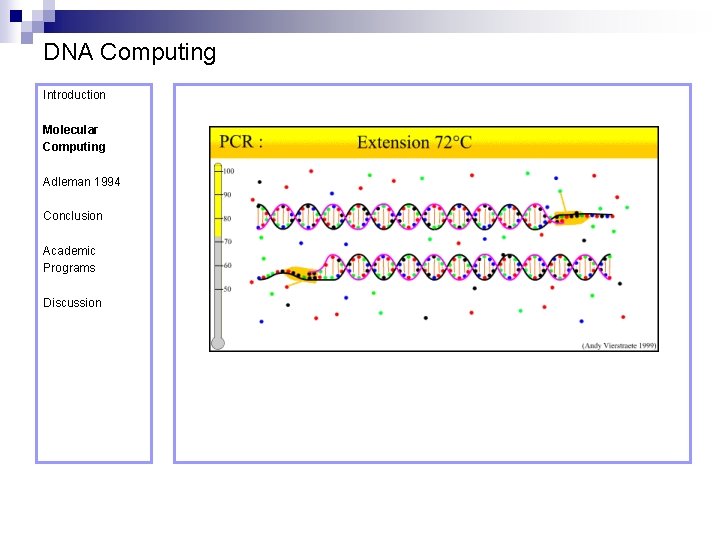
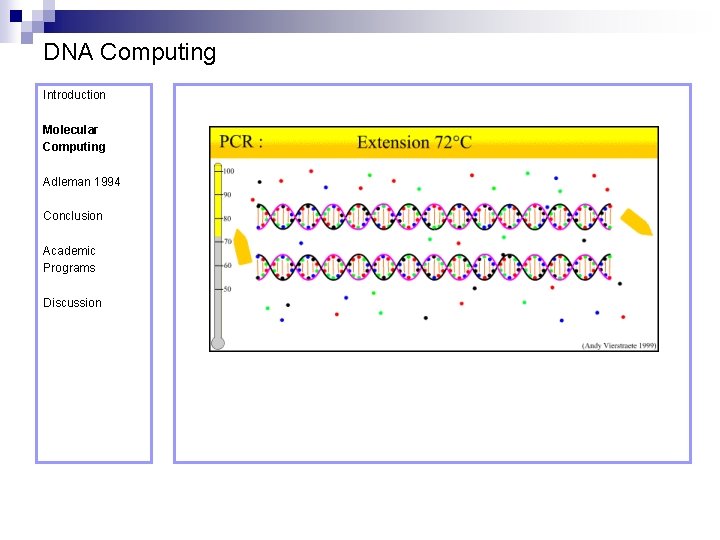
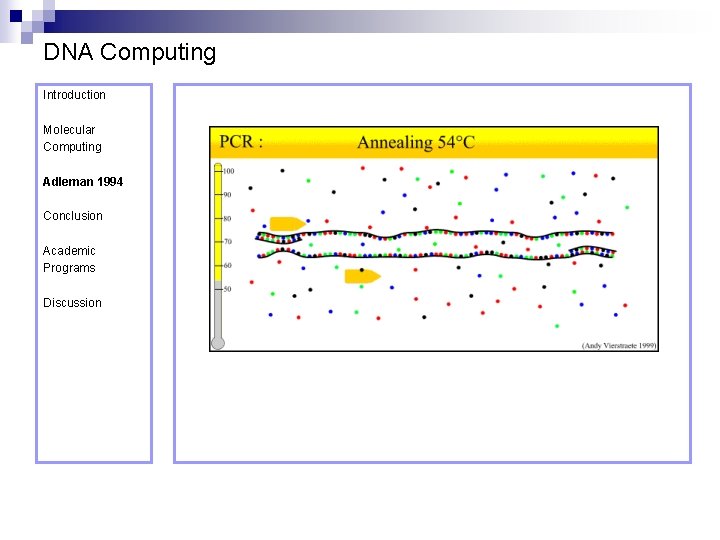
DNA Computing Introduction Polymerase Chain Reaction – PCR Molecular Computing Adleman 1994 ¨ Denaturation at 94° C Conclusion Academic Programs Discussion ¨ Annealing at 54° C ¨ Extension at 72° C

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

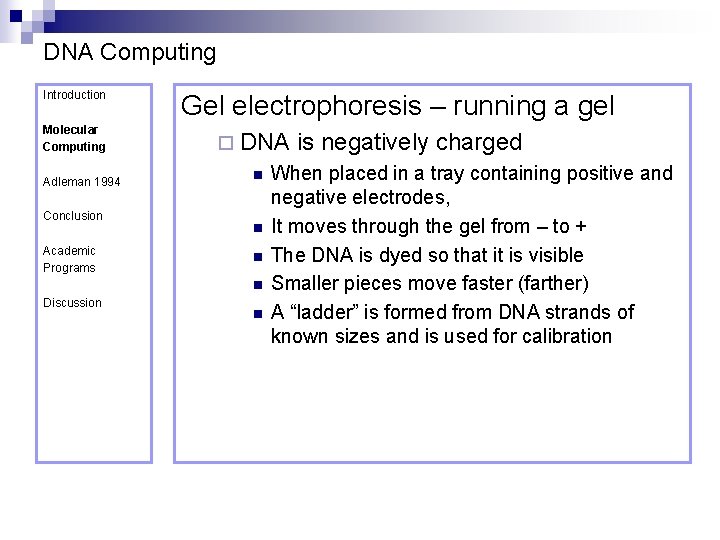
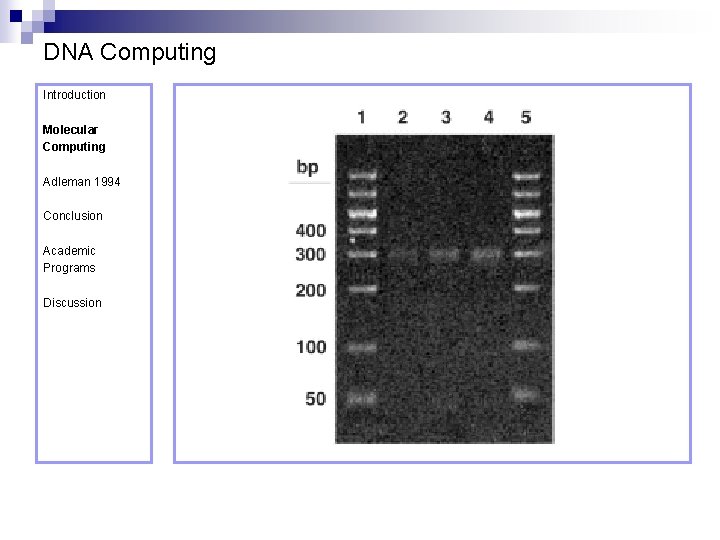
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Gel electrophoresis – running a gel ¨ DNA n n Discussion n is negatively charged When placed in a tray containing positive and negative electrodes, It moves through the gel from – to + The DNA is dyed so that it is visible Smaller pieces move faster (farther) A “ladder” is formed from DNA strands of known sizes and is used for calibration

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

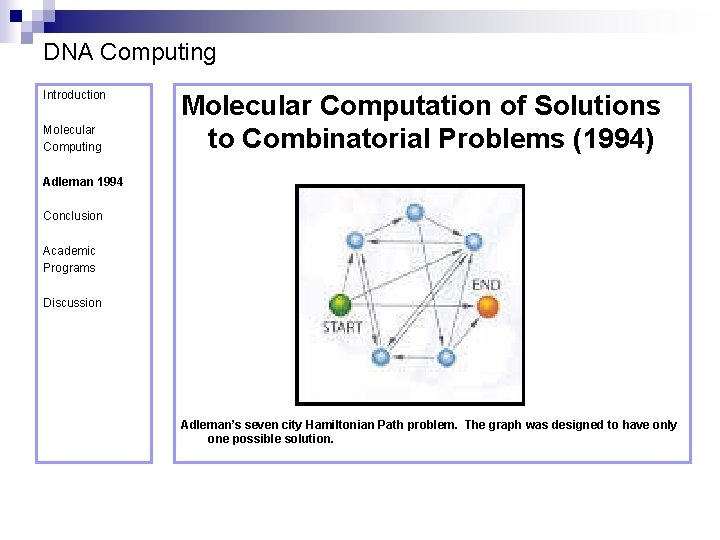
DNA Computing Introduction Molecular Computing Molecular Computation of Solutions to Combinatorial Problems (1994) Adleman 1994 Conclusion Academic Programs Discussion Adleman’s seven city Hamiltonian Path problem. The graph was designed to have only one possible solution.

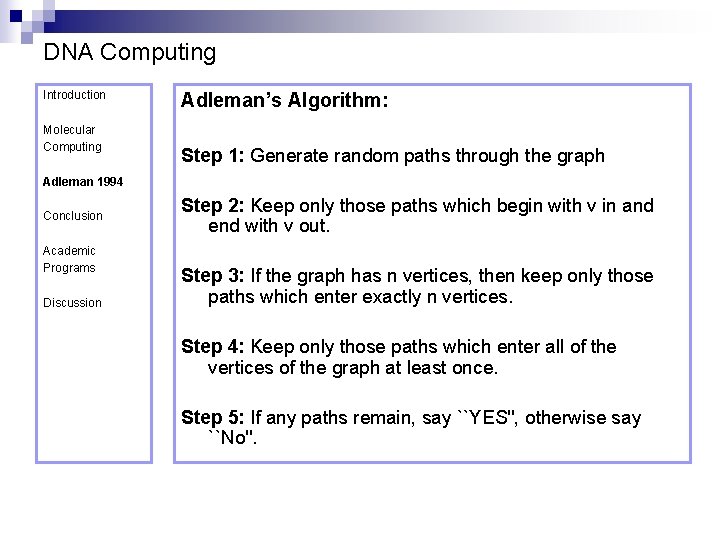
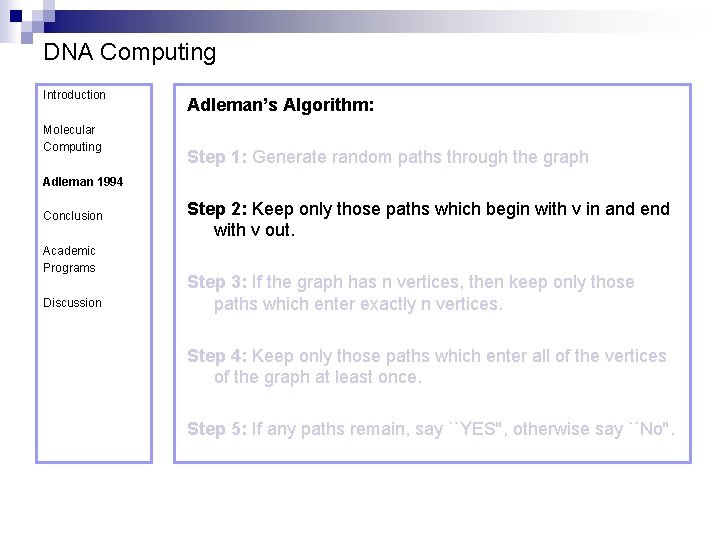
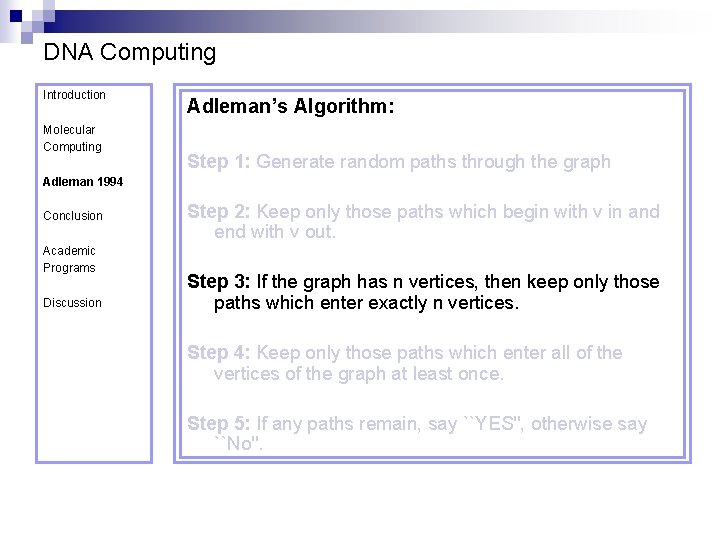
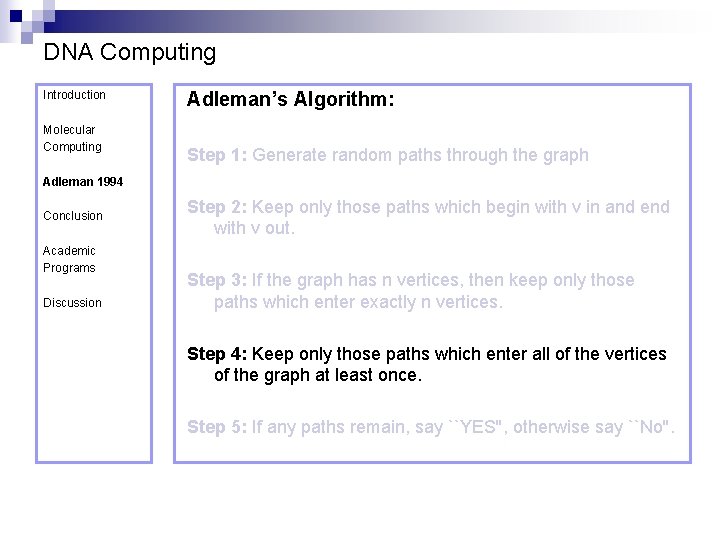
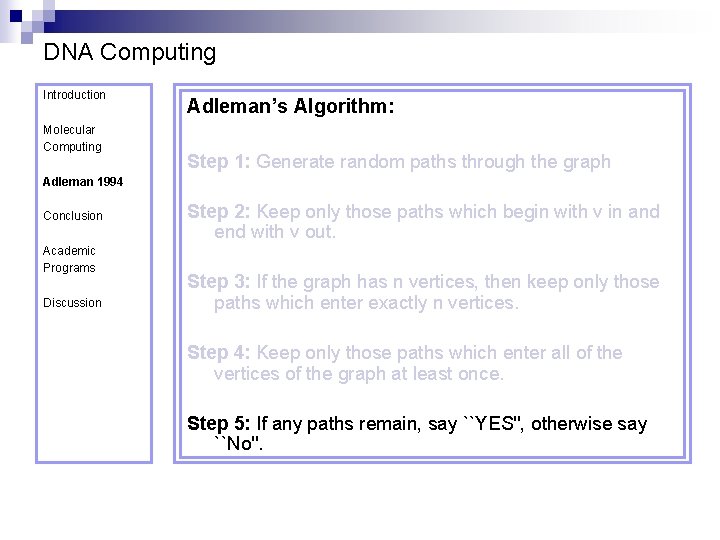
DNA Computing Introduction Molecular Computing Adleman’s Algorithm: Step 1: Generate random paths through the graph Adleman 1994 Conclusion Academic Programs Discussion Step 2: Keep only those paths which begin with v in and end with v out. Step 3: If the graph has n vertices, then keep only those paths which enter exactly n vertices. Step 4: Keep only those paths which enter all of the vertices of the graph at least once. Step 5: If any paths remain, say ``YES'', otherwise say ``No''.

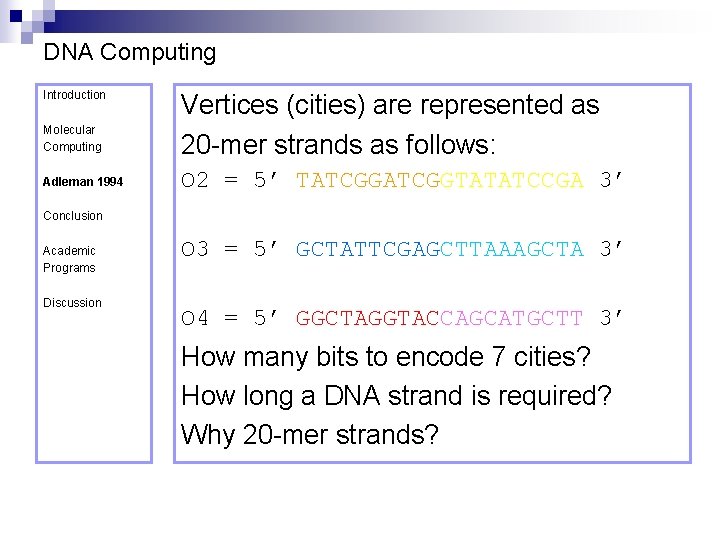
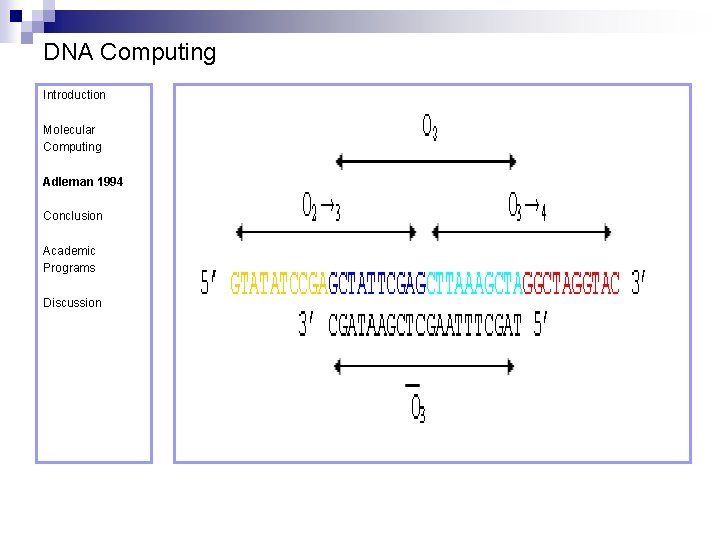
DNA Computing Introduction Molecular Computing Adleman 1994 Vertices (cities) are represented as 20 -mer strands as follows: O 2 = 5’ TATCGGTATATCCGA 3’ Conclusion Academic Programs Discussion O 3 = 5’ GCTATTCGAGCTTAAAGCTA 3’ O 4 = 5’ GGCTAGGTACCAGCATGCTT 3’

DNA Computing Introduction Molecular Computing Adleman 1994 Vertices (cities) are represented as 20 -mer strands as follows: O 2 = 5’ TATCGGTATATCCGA 3’ Conclusion Academic Programs Discussion O 3 = 5’ GCTATTCGAGCTTAAAGCTA 3’ O 4 = 5’ GGCTAGGTACCAGCATGCTT 3’ How many bits to encode 7 cities?

DNA Computing Introduction Molecular Computing Adleman 1994 Vertices (cities) are represented as 20 -mer strands as follows: O 2 = 5’ TATCGGTATATCCGA 3’ Conclusion Academic Programs Discussion O 3 = 5’ GCTATTCGAGCTTAAAGCTA 3’ O 4 = 5’ GGCTAGGTACCAGCATGCTT 3’ How many bits to encode 7 cities? How long a DNA strand is required?

DNA Computing Introduction Molecular Computing Adleman 1994 Vertices (cities) are represented as 20 -mer strands as follows: O 2 = 5’ TATCGGTATATCCGA 3’ Conclusion Academic Programs Discussion O 3 = 5’ GCTATTCGAGCTTAAAGCTA 3’ O 4 = 5’ GGCTAGGTACCAGCATGCTT 3’ How many bits to encode 7 cities? How long a DNA strand is required? Why 20 -mer strands?

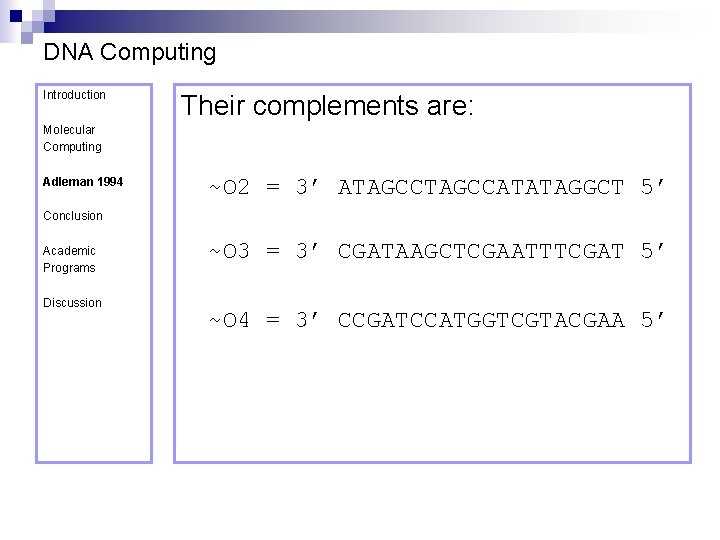
DNA Computing Introduction Their complements are: Molecular Computing Adleman 1994 ~O 2 = 3’ ATAGCCATATAGGCT 5’ Conclusion Academic Programs Discussion ~O 3 = 3’ CGATAAGCTCGAATTTCGAT 5’ ~O 4 = 3’ CCGATCCATGGTCGTACGAA 5’

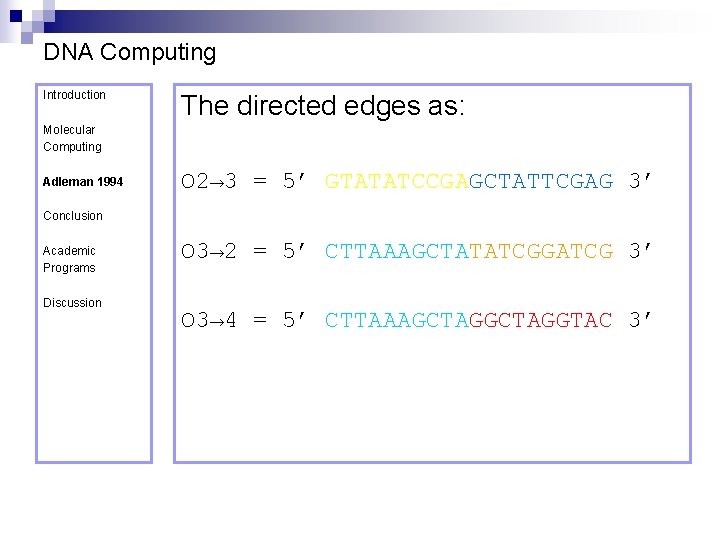
DNA Computing Introduction The directed edges as: Molecular Computing Adleman 1994 O 2→ 3 = 5’ GTATATCCGAGCTATTCGAG 3’ Conclusion Academic Programs Discussion O 3→ 2 = 5’ CTTAAAGCTATATCGGATCG 3’ O 3→ 4 = 5’ CTTAAAGCTAGGTAC 3’

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion “Unfortunately, although I held the solution in my hand, I also held about 100 trillion molecules that encoded paths that were not Hamiltonian. These had to be eliminated. ” [Adleman, 1998]

DNA Computing Introduction Molecular Computing Adleman’s Algorithm: Step 1: Generate random paths through the graph Adleman 1994 Conclusion Academic Programs Discussion Step 2: Keep only those paths which begin with v in and end with v out. Step 3: If the graph has n vertices, then keep only those paths which enter exactly n vertices. Step 4: Keep only those paths which enter all of the vertices of the graph at least once. Step 5: If any paths remain, say ``YES'', otherwise say ``No''.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Step 2 of the algorithm requires that only the DNA strands that begin at the first vertex and end at the last vertex be kept. This requirement was achieved using PCR. By adding the correct primers to the solution, the DNA strands beginning at city 0 and ending at city 6 can be made to multiply exponentially. After a sufficient amount of time, they will make up the vast majority of the solution.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion The PCR works as follows: A primer for the start city and another for the terminal city are added to the solution containing the result of the computation. DNA strands containing both the starting city and the terminal city are multiplied exponentially. Strands containing one of the two cities are multiplied linearly but none of the other strands are multiplied at all.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman’s Algorithm: Step 1: Generate random paths through the graph Adleman 1994 Conclusion Academic Programs Discussion Step 2: Keep only those paths which begin with v in and end with v out. Step 3: If the graph has n vertices, then keep only those paths which enter exactly n vertices. Step 4: Keep only those paths which enter all of the vertices of the graph at least once. Step 5: If any paths remain, say ``YES'', otherwise say ``No''.

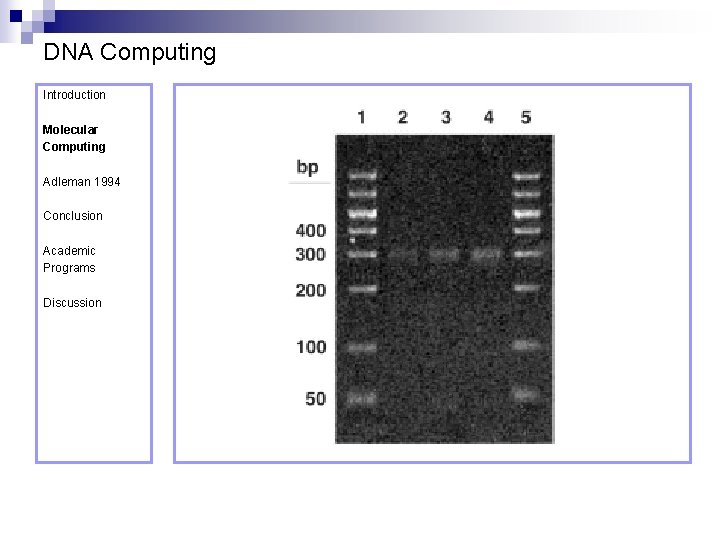
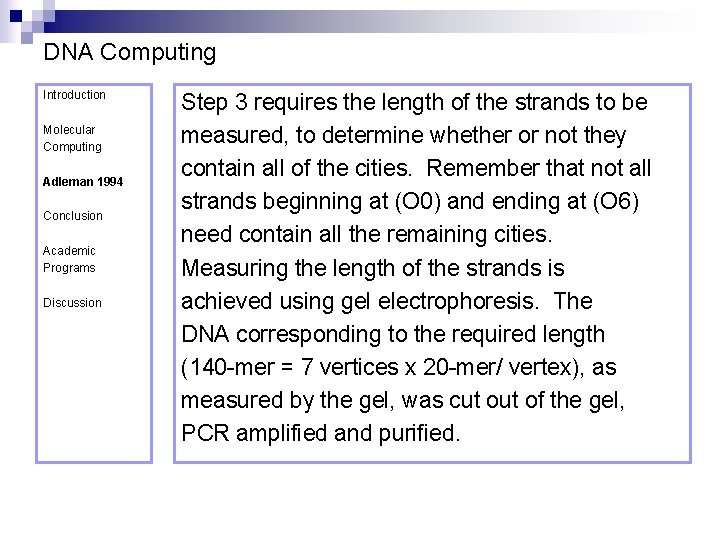
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Step 3 requires the length of the strands to be measured, to determine whether or not they contain all of the cities. Remember that not all strands beginning at (O 0) and ending at (O 6) need contain all the remaining cities. Measuring the length of the strands is achieved using gel electrophoresis. The DNA corresponding to the required length (140 -mer = 7 vertices x 20 -mer/ vertex), as measured by the gel, was cut of the gel, PCR amplified and purified.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman’s Algorithm: Step 1: Generate random paths through the graph Adleman 1994 Conclusion Academic Programs Discussion Step 2: Keep only those paths which begin with v in and end with v out. Step 3: If the graph has n vertices, then keep only those paths which enter exactly n vertices. Step 4: Keep only those paths which enter all of the vertices of the graph at least once. Step 5: If any paths remain, say ``YES'', otherwise say ``No''.

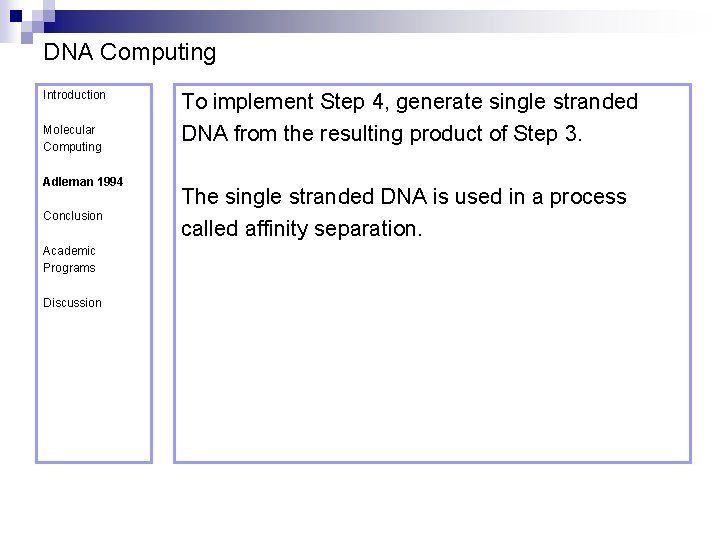
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion To implement Step 4, generate single stranded DNA from the resulting product of Step 3. The single stranded DNA is used in a process called affinity separation.

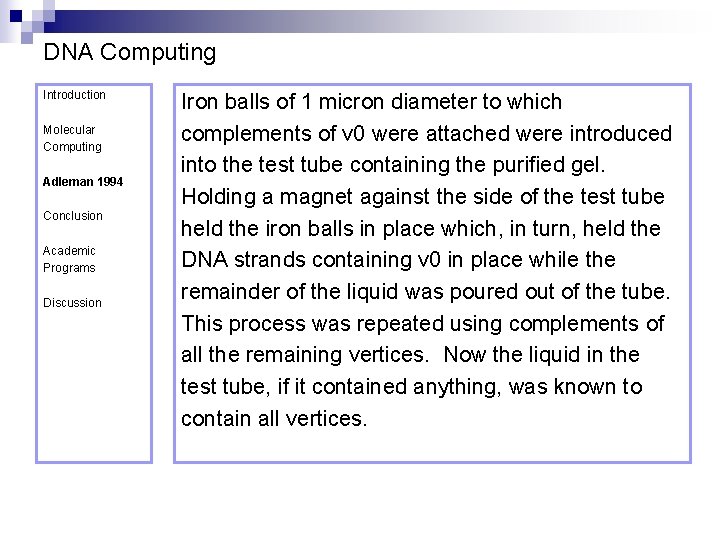
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Iron balls of 1 micron diameter to which complements of v 0 were attached were introduced into the test tube containing the purified gel. Holding a magnet against the side of the test tube held the iron balls in place which, in turn, held the DNA strands containing v 0 in place while the remainder of the liquid was poured out of the tube. This process was repeated using complements of all the remaining vertices. Now the liquid in the test tube, if it contained anything, was known to contain all vertices.

DNA Computing Introduction Molecular Computing Adleman’s Algorithm: Step 1: Generate random paths through the graph Adleman 1994 Conclusion Academic Programs Discussion Step 2: Keep only those paths which begin with v in and end with v out. Step 3: If the graph has n vertices, then keep only those paths which enter exactly n vertices. Step 4: Keep only those paths which enter all of the vertices of the graph at least once. Step 5: If any paths remain, say ``YES'', otherwise say ``No''.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Amplify the result from Step 4 using PCR and run it on a gel. An analysis of the results clearly shows the presence of a result, so the answer is “yes!”

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Adleman’s conclusions: Even a cursory reading his conclusions reveals his intense excitement about the results of this experiment. 3 main points n Speed n Storage density n Energy efficiency

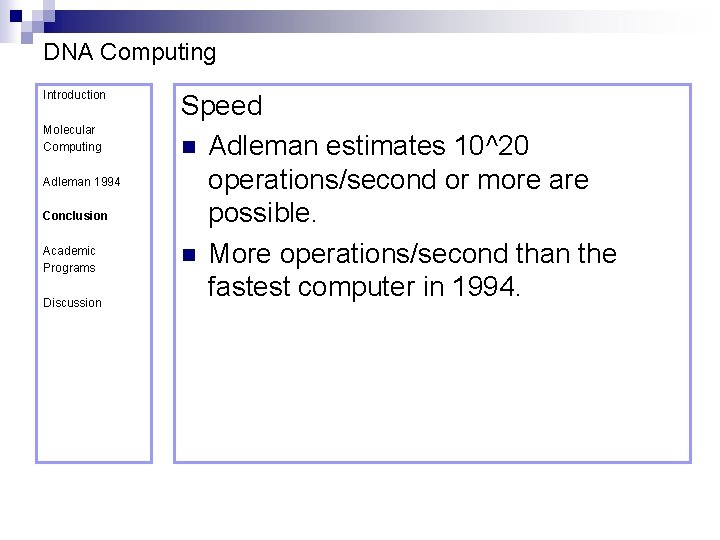
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Speed n Adleman estimates 10^20 operations/second or more are possible. n More operations/second than the fastest computer in 1994.

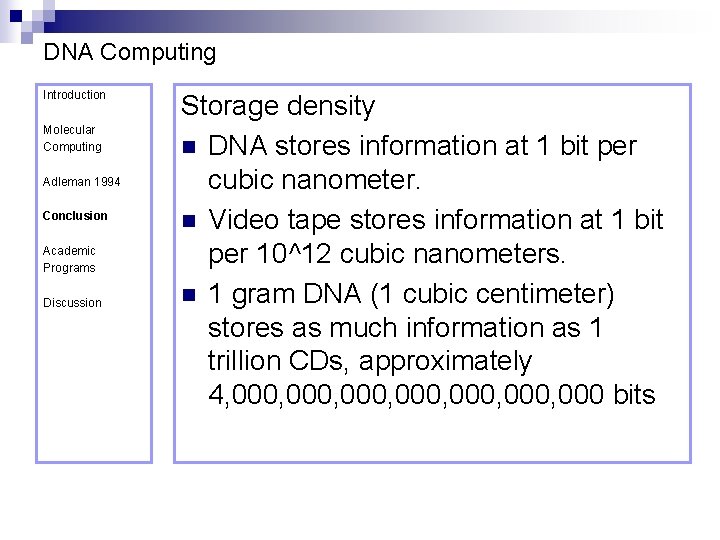
DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Storage density n DNA stores information at 1 bit per cubic nanometer. n Video tape stores information at 1 bit per 10^12 cubic nanometers. n 1 gram DNA (1 cubic centimeter) stores as much information as 1 trillion CDs, approximately 4, 000, 000, 000 bits

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Energy efficiency n Evolution has provided highly efficient DNA operations

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Alas, this excitement proves to be ephemeral. All is not well. n DNA computers don’t scale enough to perform really large calculations. n Despite advancements in technique, DNA computing remains labor intensive. n Automation of time-consuming lab work is partially successful.

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion “Despite our successes, and those of others, in the absence of technical breakthroughs, optimism regarding the creation of a molecular computer capable of competing with electronic computers on classical computational problems is not warranted. ” Adleman, 2002

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Current Research in DNA Computing Many colleges and universities offer some access to DNA computing. A few of the leading researchers are: n n n Len Adleman at the University of Southern California Eric Winfree at Caltech Laura Landweber at Princeton

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion The 9 th International Meeting on DNA Based Computers will be held on June 1 -4 in Madison, Wisconsin. For further information, try: http: //www. corninfo. chem. wisc. edu/writi ngs/DNAcomputing. html

DNA Computing Introduction Molecular Computing An excellent book on DNA computing is the aptly titled: Adleman 1994 Conclusion Academic Programs Discussion DNA Computing: New Computing Paradigms by Paun, Rozenberg and Salomaa Springer-Verlag, Berlin, 1998. ISBN: 3 -540 -64196 -3

DNA Computing Introduction Molecular Computing Adleman 1994 Conclusion Academic Programs Discussion Any questions?

DNA Computing Introduction Molecular Computing Adleman 1994 Adleman 2002 Conclusion Academic Programs Discussion

DNA Computing Introduction Molecular Computing Adleman 1994 Adleman 2002 Conclusion Academic Programs Discussion