Molecular Diagnostics 1 Molecular Diagnostics 2 Molecular Diagnostics






- Slides: 39

Molecular Diagnostics 1 Molecular Diagnostics

2 Molecular Diagnostics Textbook n MOLECULAR DIAGNOSTICS: Fundamentals, Methods, & Clinical Applications, 2007, F. A. Davis Company q Lela Buckingham, and Maribeth L. Flaws

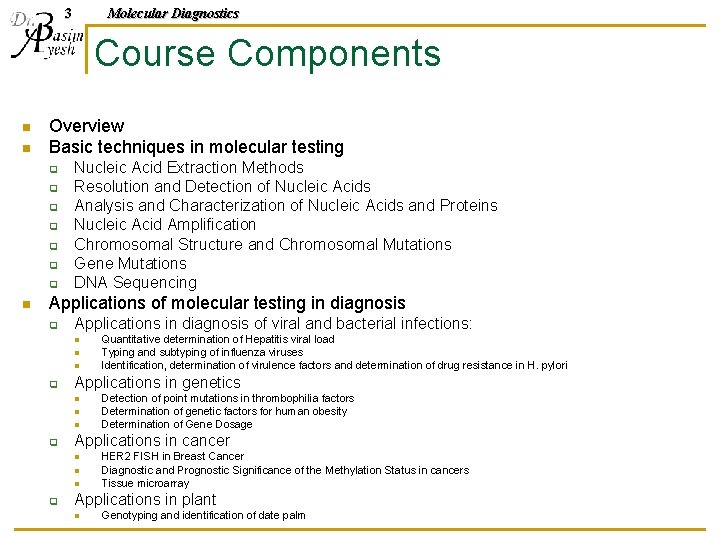
3 Molecular Diagnostics Course Components n n Overview Basic techniques in molecular testing q q q q n Nucleic Acid Extraction Methods Resolution and Detection of Nucleic Acids Analysis and Characterization of Nucleic Acids and Proteins Nucleic Acid Amplification Chromosomal Structure and Chromosomal Mutations Gene Mutations DNA Sequencing Applications of molecular testing in diagnosis q Applications in diagnosis of viral and bacterial infections: n n n q Applications in genetics n n n q Detection of point mutations in thrombophilia factors Determination of genetic factors for human obesity Determination of Gene Dosage Applications in cancer n n n q Quantitative determination of Hepatitis viral load Typing and subtyping of influenza viruses Identification, determination of virulence factors and determination of drug resistance in H. pylori HER 2 FISH in Breast Cancer Diagnostic and Prognostic Significance of the Methylation Status in cancers Tissue microarray Applications in plant n Genotyping and identification of date palm

4 Molecular Diagnostics

5 Molecular Diagnostics Don’t forget to read “Advanced Concepts” Concepts In the textbook MOLECULAR DIAGNOSTICS: Fundamentals, Methods, & Clinical Applications

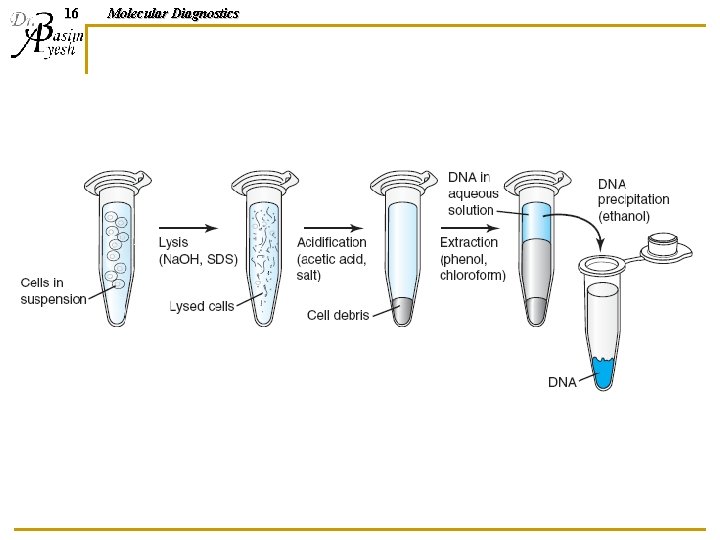
Nucleic Acid Extraction Methods 6 Molecular Diagnostics

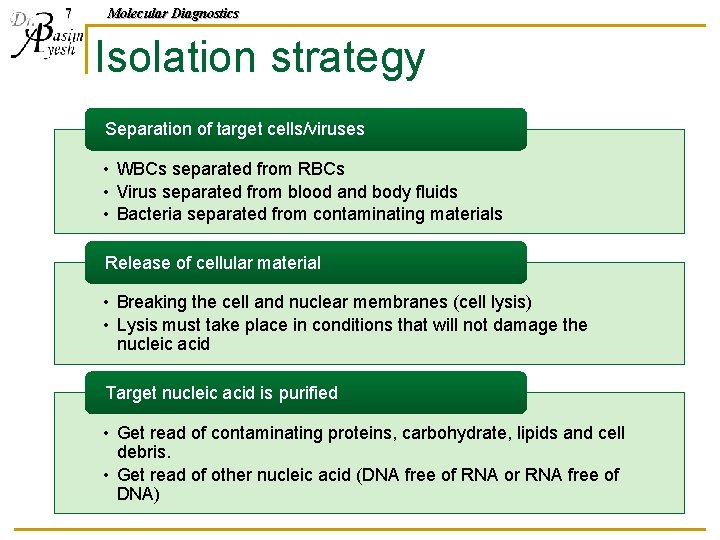
7 Molecular Diagnostics Isolation strategy Separation of target cells/viruses • WBCs separated from RBCs • Virus separated from blood and body fluids • Bacteria separated from contaminating materials Release of cellular material • Breaking the cell and nuclear membranes (cell lysis) • Lysis must take place in conditions that will not damage the nucleic acid Target nucleic acid is purified • Get read of contaminating proteins, carbohydrate, lipids and cell debris. • Get read of other nucleic acid (DNA free of RNA or RNA free of DNA)

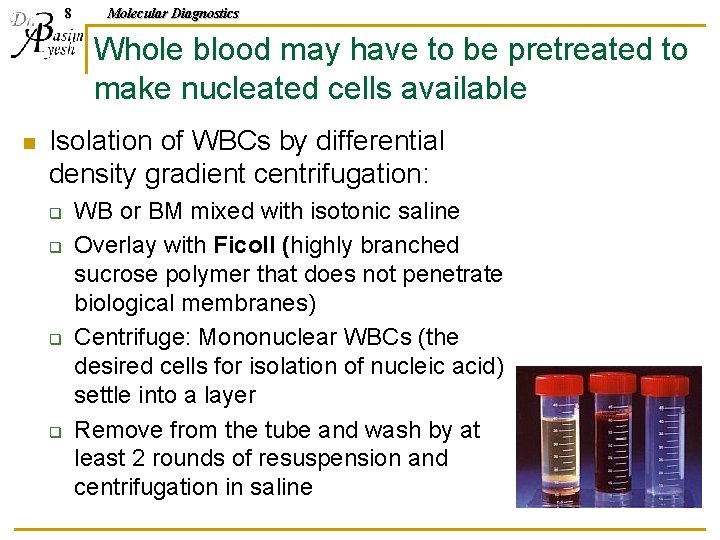
8 Molecular Diagnostics Whole blood may have to be pretreated to make nucleated cells available n Isolation of WBCs by differential density gradient centrifugation: q q WB or BM mixed with isotonic saline Overlay with Ficoll (highly branched sucrose polymer that does not penetrate biological membranes) Centrifuge: Mononuclear WBCs (the desired cells for isolation of nucleic acid) settle into a layer Remove from the tube and wash by at least 2 rounds of resuspension and centrifugation in saline

9 Molecular Diagnostics Whole blood may have to be pretreated to make nucleated cells available n Isolation of WBCs by differential lysis: q q q Takes advantage of the differences in the osmotic fragility of RBCs and WBCs. Hypotonic buffer or water will result in the lysis of the RBCs before the WBCs. The WBCs are then pelleted by centrifugation, leaving the empty RBC membranes (ghosts) and hemoglobin, respectively, in suspension and solution.

10 Molecular Diagnostics Fresh or frozen tissue samples must be dissociated n n n Whole tissue samples are disrupted by grinding the frozen tissue in liquid nitrogen, homogenizing the tissue, or simply mincing the tissue using a scalpel. Fixed embedded tissue has to be deparaffinized by soaking in xylene (a mixture of three isomers of dimethylbenzene). After xylene treatment, the tissue is usually rehydrated by soaking it in decreasing concentrations of ethanol.

11 Molecular Diagnostics Bacteria and fungi having tough cell walls must be broken to allow the release of nucleic acid n n By several enzyme products, e. g. , lyzozyme or zymolyase, that digest cell wall polymers Alternatively, cell walls can be broken mechanically by grinding or by vigorously mixing with glass beads. Treatment with detergent (1% sodium dodecyl sulfate) and strong base (0. 2 M Na. OH) in the presence of Tris base, ethylenediaminetetraacetic acid (EDTA), and glucose can also break bacterial cell walls. Boiling in 8% sucrose, 8% Triton X-100 detergent, Tris buffer, and EDTA after lysozyme treatment releases DNA that can be immediately precipitated with alcohol

12 n n n Molecular Diagnostics Gentler enzymatic methods are less likely to damage chromosomal DNA and thus are preferred for methods involving larger chromosomal targets as opposed to plasmid DNA extracted with Na. OH or boiling procedures is denatured (single-stranded) and may not be suitable for methods such as restriction enzyme analysis that require double-stranded DNA. The advantage of these types of extraction is their speed and simplicity. Amplification methods will work with this type of DNA isolation.

13 Molecular Diagnostics DNA ORGANIC ISOLATION METHODS Further purification of DNA from contaminating proteins, lipids, carbohydrates, and cell debris using a combination of high salt, low p. H, and an organic mixture of phenol and chloroform.

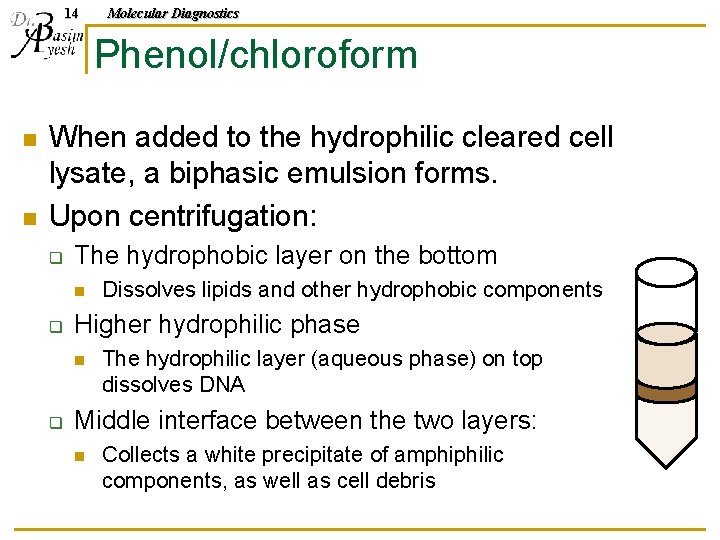
14 Molecular Diagnostics Phenol/chloroform n n When added to the hydrophilic cleared cell lysate, a biphasic emulsion forms. Upon centrifugation: q The hydrophobic layer on the bottom n q Higher hydrophilic phase n q Dissolves lipids and other hydrophobic components The hydrophilic layer (aqueous phase) on top dissolves DNA Middle interface between the two layers: n Collects a white precipitate of amphiphilic components, as well as cell debris

15 Molecular Diagnostics DNA-precipitation n n The upper phase containing the DNA is collected DNA is precipitated using ethanol (at 2: 1 ratio) or isopropanol (at 1: 1 ratio) in a high concentration of salt (ammonium, potassium or sodium acetate, or lithium or sodium chloride). Upon centrifugation, DNA forms a solid precipitate. Excess salt is removed by rinsing the pellet in 70% ethanol, centrifuging and discarding the ethanol supernatant DNA pellet is dissolved in rehydration buffer, usually 10 m. M Tris, 1 m. M EDTA (TE), or water.

16 Molecular Diagnostics

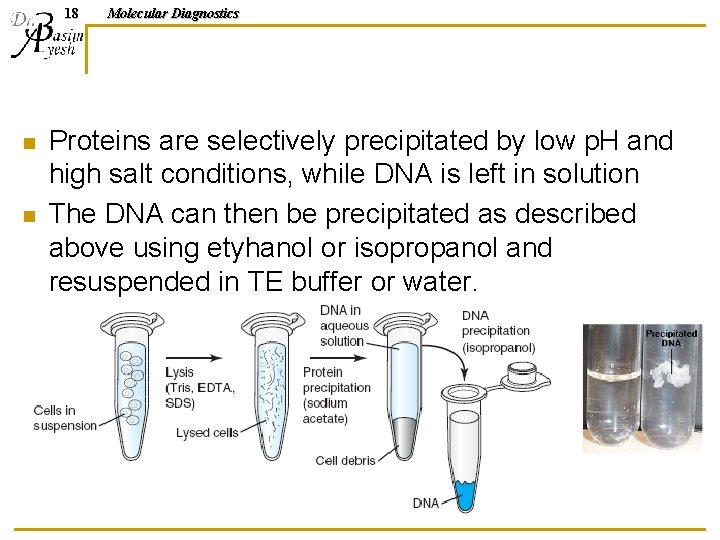
17 Molecular Diagnostics DNA INORGANIC ISOLATION METHODS “SALTING OUT” Have been developed to avoid use of toxic reagents such as phenol. Initially, these methods did not provide the efficient recovery of clean DNA achieved with phenol extraction; however, newer methods have proven to produce high quality DNA preparations in good yields.

18 n n Molecular Diagnostics Proteins are selectively precipitated by low p. H and high salt conditions, while DNA is left in solution The DNA can then be precipitated as described above using etyhanol or isopropanol and resuspended in TE buffer or water.

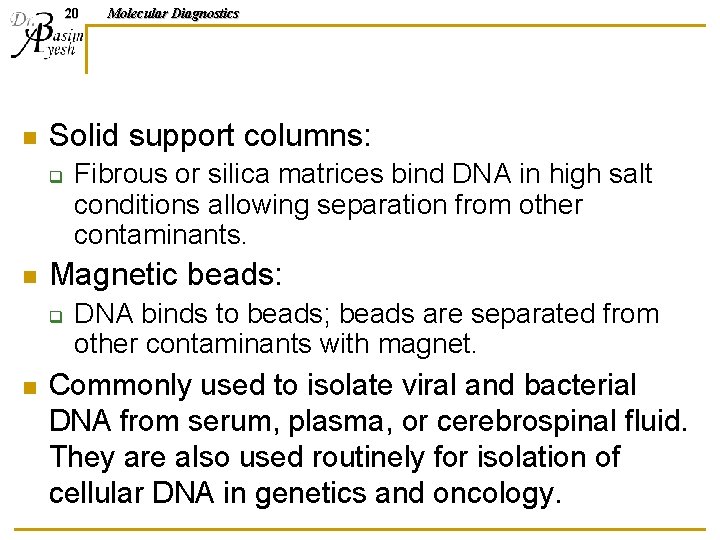
19 Molecular Diagnostics DNA SOLID-PHASE ISOLATION DNA extraction using solid matrices to bind and wash the DNA.

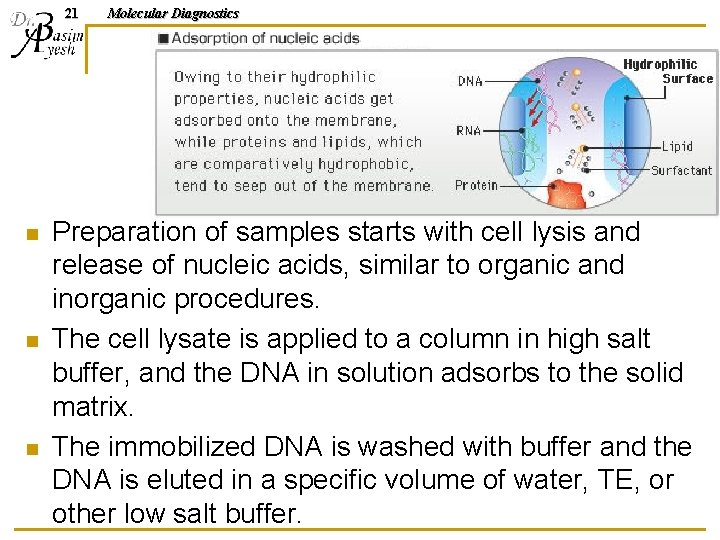
20 n Solid support columns: q n Fibrous or silica matrices bind DNA in high salt conditions allowing separation from other contaminants. Magnetic beads: q n Molecular Diagnostics DNA binds to beads; beads are separated from other contaminants with magnet. Commonly used to isolate viral and bacterial DNA from serum, plasma, or cerebrospinal fluid. They are also used routinely for isolation of cellular DNA in genetics and oncology.

21 n n n Molecular Diagnostics Preparation of samples starts with cell lysis and release of nucleic acids, similar to organic and inorganic procedures. The cell lysate is applied to a column in high salt buffer, and the DNA in solution adsorbs to the solid matrix. The immobilized DNA is washed with buffer and the DNA is eluted in a specific volume of water, TE, or other low salt buffer.

22 n Molecular Diagnostics DNA absorbed to magnetic beads is washed by suspension of the beads in buffer and collection of the beads using a magnet applied to the outside of the tube while the buffer is aspirated or poured off.

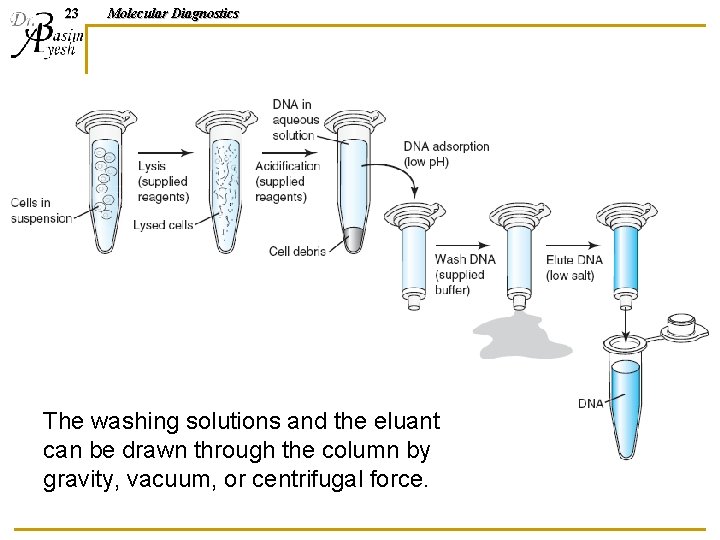
23 Molecular Diagnostics The washing solutions and the eluant can be drawn through the column by gravity, vacuum, or centrifugal force.

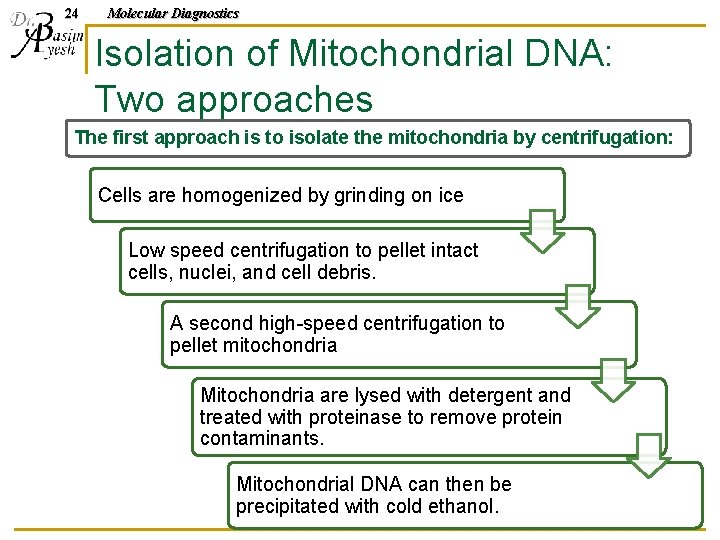
24 Molecular Diagnostics Isolation of Mitochondrial DNA: Two approaches The first approach is to isolate the mitochondria by centrifugation: Cells are homogenized by grinding on ice Low speed centrifugation to pellet intact cells, nuclei, and cell debris. A second high-speed centrifugation to pellet mitochondria Mitochondria are lysed with detergent and treated with proteinase to remove protein contaminants. Mitochondrial DNA can then be precipitated with cold ethanol.

25 Molecular Diagnostics The second approach is to isolate total DNA q The preparation will contain mitochondrial DNA that can be analyzed within the total DNA background by hybridization or PCR.

26 Molecular Diagnostics Isolation of RNA n Requires STRICT precautions to avoid sample degradation. q q RNA especially labile. RNases are naturally occurring enzymes that degrade RNA n n q Common laboratory contaminant (from bacterial and human sources) Also released from cellular compartments during isolation of RNA from biological samples RNases can be difficult to inactivate n Small proteins that can renature and become active.

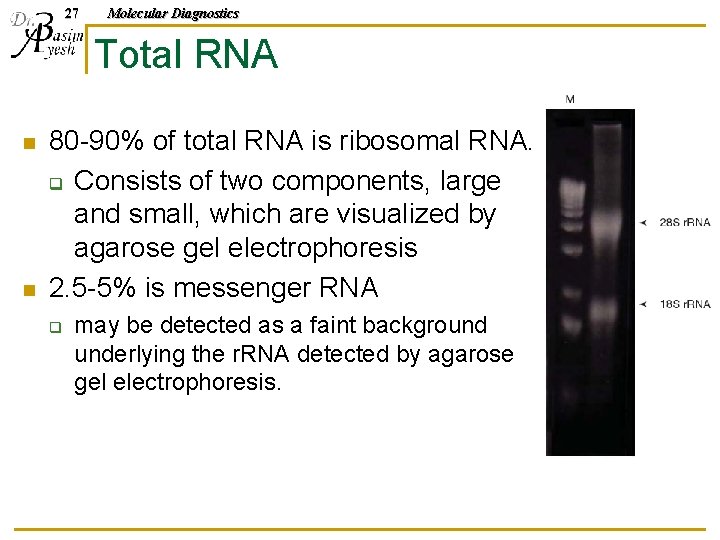
27 Molecular Diagnostics Total RNA n n 80 -90% of total RNA is ribosomal RNA. q Consists of two components, large and small, which are visualized by agarose gel electrophoresis 2. 5 -5% is messenger RNA q may be detected as a faint background underlying the r. RNA detected by agarose gel electrophoresis.

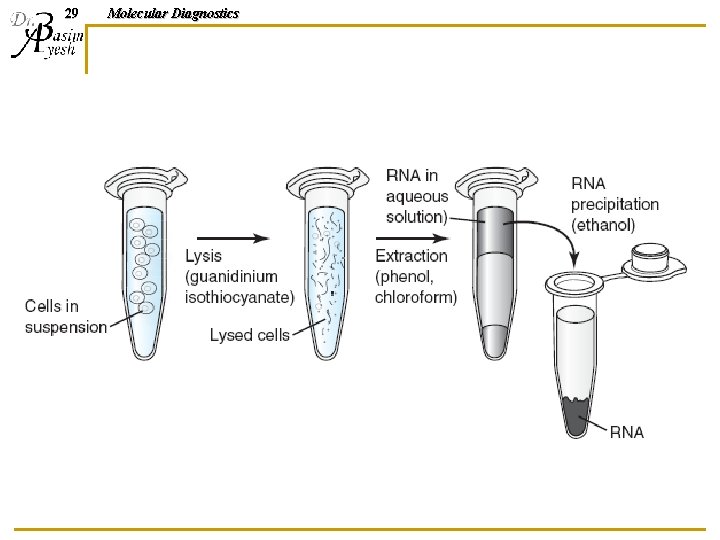
28 Molecular Diagnostics Organic RNA Extraction n n Lyse/homogenize cells Add phenol: chloroform: isoamyl alcohol (25: 24: 1) to lysed sample, and centrifuge q q q n n Organic phase separates from aqueous phase Organic solvents on bottom (must be acidic: ph 4– 5) Aqueous phase on top (contains total RNA) Cellular debris and genomic DNA appears as a “film” of debris at the interface of the two solutions Remove RNA solution to a clean tube; precipitate RNA and wash with ethanol, then resuspend RNA in water

29 Molecular Diagnostics

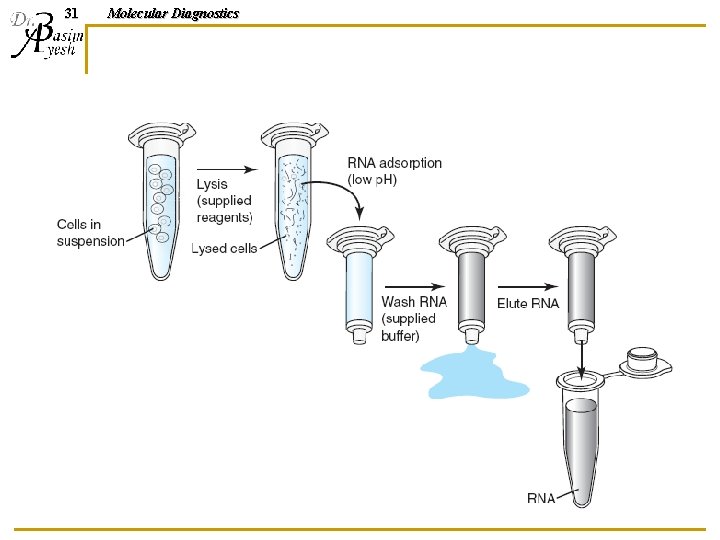
30 Molecular Diagnostics Affinity Purification of RNA 1. 2. 3. 4. Lyse cells, and spin to remove large particulates/cell debris Apply lysate (containing nucleic acids and cellular contaminants) to column with glass membrane Wash with alcohol to remove contaminants; nucleic acids stick to glass membrane while contaminants wash through. Treat with DNase enzyme to remove contaminating DNA. Apply water to the column; purified RNA washes off the glass and is collected

31 Molecular Diagnostics

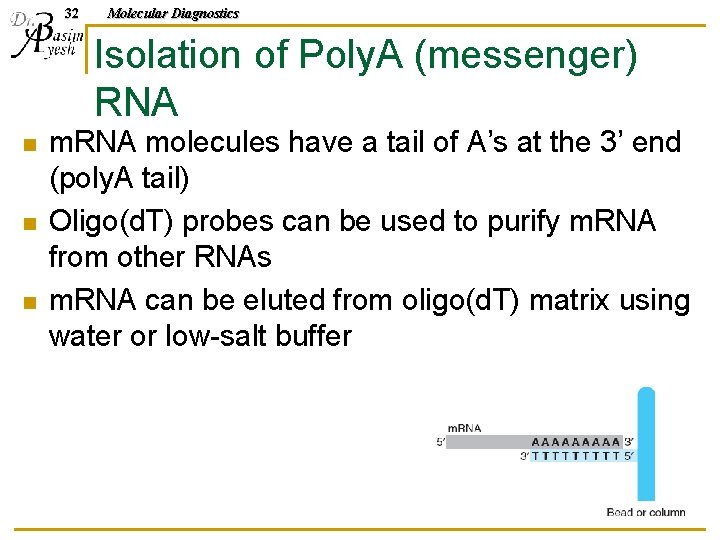
32 Molecular Diagnostics Isolation of Poly. A (messenger) RNA n n n m. RNA molecules have a tail of A’s at the 3’ end (poly. A tail) Oligo(d. T) probes can be used to purify m. RNA from other RNAs m. RNA can be eluted from oligo(d. T) matrix using water or low-salt buffer

33 Molecular Diagnostics MEASUREMENT OF NUCLEIC ACID QUALITY AND QUANTITY

34 Molecular Diagnostics Electrophoresis n n Analyze DNA and RNA for quality by electrophoresis. Fluorescent dyes q q n Ethidium bromide Sybre Green Appearance depends on type of DNA isolated.

35 Molecular Diagnostics Spectrophotometry n Sample absorbances are determined on the spectrophotometer at 260 nm and 280 nm q q n n nucleic acid (DNA, RNA, nucleotides) absorb light at 260 nm protein absorbs light at 280 nm 230 nm: guanidine A 260/A 280 ratio is a measure of DNA purity The absorbance wavelength is directly proportional to the concentration of nucleic acid.

36 Molecular Diagnostics Determining Concentration n Formula: Concentration = OD 260 nm X OD unit X Dilution factor n One OD or absorbance unit at 260 nm is equal to q q 50 ug/m. L for DNA 40 ug/m. L for RNA.

37 Molecular Diagnostics Determining Purity n n The OD 260 nm should be 1. 6 -2. 00 times more than OD 280 nm Divide the OD 260 nm by the OD 280 nm to get the ratio. q q n n If the OD 260 nm / OD 280 nm ratio is less than 1. 6 for DNA, 2. 02. 3 for RNA this indicates contamination, usually with protein. DNA -If the OD ratio is higher than 2. 0 it may be contaminated with RNA. Ratio of the readings : OD 260 nm / OD 280 nm is a measure of purity. Pure preparations of DNA and RNA have OD 260 nm / OD 280 nm of 1. 8 and 2. 0 respectively.

38 Molecular Diagnostics Fluorometry n n n Fluorometry utilizes fluorescent dyes which specifically bind DNA or RNA. It requires a negative control (to set the zero point on the fluorometer) and a standard of known concentration. The fluorometer shines light on the sample (excitation) and then measures level of fluorescent light being emitted to the side (at a 90º angle) of the excitation light beam. The fluorescent dyes are relatively specific to nucleic acids as opposed to protein and other cellular components. The fluorescence of the dyes increases when they bind nucleic acids.

39 Molecular Diagnostics Fluorometry n n n Fluorometry is about 1, 000 x more sensitive than spectrophotometric absorbance (i. e. measurement of A 260) and less susceptible to protein and RNA contamination. However it also does not give a crude measurement of purity (like an A 260/A 280 ratio) nor does it assure that the DNA or RNA is not degraded (e. g. like size determination by gel electrophoesis). Do not use glass (spectrophotmetry) cuvettes in a fluorometer because the frosted glass on the side of the cuvette interferes with detection of fluorescent light.