Heuristic Approaches for Sequence Alignments courseeleg 667 01














- Slides: 37

Heuristic Approaches for Sequence Alignments /course/eleg 667 -01 -f/Topic 2 b. ppt

Outline w Sequence Alignment w Database Search w FASTA w BLAST /course/eleg 667 -01 -f/Topic-2 b. ppt 2

Sequence Alignment Dynamic Programming (give optimal solution(s)) w Needleman-Wunsch (Global Alignment) w Smith-Waterman (Local Alignment) Heuristics (give approximate solution(s)) Trade speed for precision (good for DB search) w FASTA (finds local alignments) w BLAST (Basic Local Alignment Search Tool) /course/eleg 667 -01 -f/Topic-2 b. ppt 3

Database Search w One of the major uses of alignments is to find similar sequences in a database, i. e. compare one input sequence with all sequences in the database and obtain the most similar ones; w Current databases contain massive number of sequences; w Finding homologies in these databases optimally with dynamic programming can take long. /course/eleg 667 -01 -f/Topic-2 b. ppt 4

Database Search using Heuristic Sequence Comparison Algorithms w Most database search algorithms relay on heuristic procedures w These are not guaranteed to find the best match w Sometimes, they will completely miss a high-scoring match /course/eleg 667 -01 -f/Topic-2 b. ppt 5

Database Search and PAM Matrices - Motivation w Simple scoring scheme (e. g. +1 for match, 0 for mismatch, -1 for mismatch) is not enough, especially for protein sequences w Amino Acids: must consider their relative replacement features in an evolutionary scenario /course/eleg 667 -01 -f/Topic-2 b. ppt 6

(Cont’d) w Factors affecting such mutual substitution are numerous (size, chemical properties, etc. ) w PAM (Point Accepted Mutations) matrices are widely used – they are derived by direct observation of actual substitution rates. /course/eleg 667 -01 -f/Topic-2 b. ppt 7

PAM Matrices (Contn’d) w 1 -PAM Matrix: reflect an amount of evolution producing on average one mutation per hundred amino acids w How to build a 1 -PAM matrices? n A probability transition matrix M: each entry Mab denotes the probability of a changing into b n A scoring matrix S n S is derived from M /course/eleg 667 -01 -f/Topic-2 b. ppt 8

How to Build a Probability Transition Matrix M? w We need: n n A list of accepted mutations The probability of occurrence Pa for each amino acid a w M 1 (M for 1 -PAM) can be computed by simple probability arguments w Mk (M for K-PAM) = M 1 k /course/eleg 667 -01 -f/Topic-2 b. ppt 9

How to Derive S from M? Question: Assuming pairing an amino acid a with b what is the probability (called a likelihood ratio) this pair is a mutation, not a random occurrence? Answer: Mab This ratio = P b Where Pb is the probability of a random occurrence of b. /course/eleg 667 -01 -f/Topic-2 b. ppt 10

How to Pick Up a PAM Matrix to Use w Use default one – but should know what it is w Select several to cover a wide range if little is known for the sequences w In general low PAM numbers are good for finding local, strong similarities, while large PAM numbers good for detecting long, weak ones. /course/eleg 667 -01 -f/Topic-2 b. ppt 11

A Note on FAST Algorithms w Fast is a family of algorithm, e. g. FASTP, FASTA, TFASTA, LFASTA, . . . w In this lecture we use FAST or FASTA interchangeably w References: [Pearson 90, 91, Pearson. Lipman 88, etc. ] /course/eleg 667 -01 -f/Topic-2 b. ppt 12

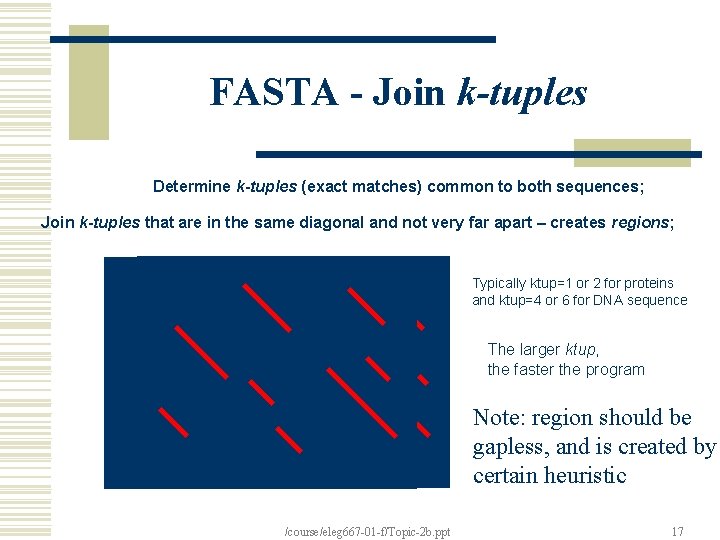
FASTA (Pearson and Lipman, 1988) w Determine k-tuples (exact matches) common to both sequences (with two parameters: ktup and offset). w Join k-tuples that are in the same diagonal and not very far apart – creates regions; w Find region with best score – “initial score” to rank the sequences; w Compute an “optimized score”, using DP, restricted to a band around the region. /course/eleg 667 -01 -f/Topic-2 b. ppt 13

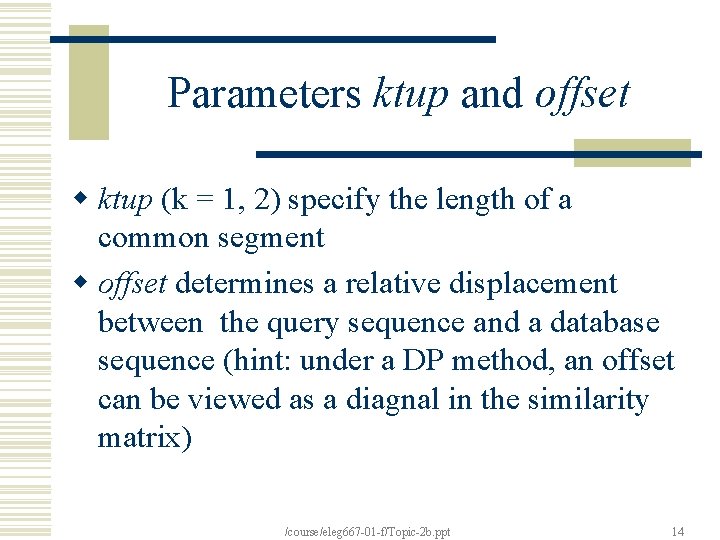
Parameters ktup and offset w ktup (k = 1, 2) specify the length of a common segment w offset determines a relative displacement between the query sequence and a database sequence (hint: under a DP method, an offset can be viewed as a diagnal in the similarity matrix) /course/eleg 667 -01 -f/Topic-2 b. ppt 14

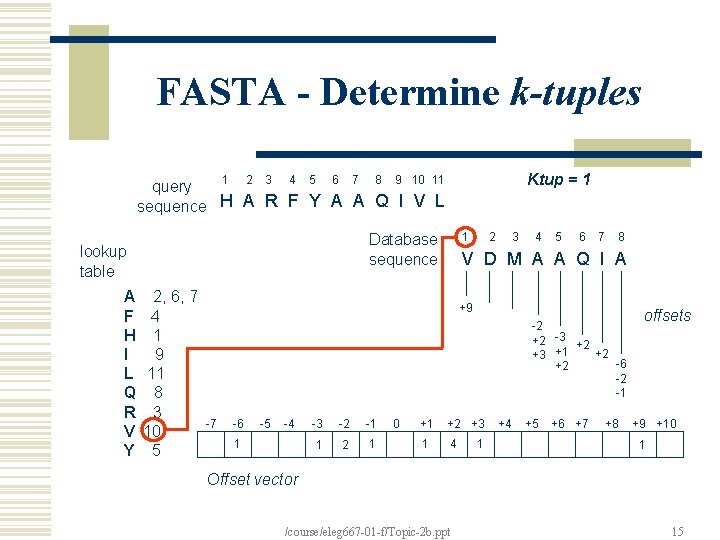
FASTA - Determine k-tuples 1 2 3 4 5 6 7 8 Ktup = 1 9 10 11 query sequence H A R F Y A A Q I V L lookup table A F H I L Q R V Y 1 Database sequence 2, 6, 7 4 1 9 11 8 3 10 5 2 3 4 5 6 7 8 V D M A A Q I A +9 -2 +2 -3 +2 +2 +3 +1 -6 +2 -2 -1 -7 -6 -5 -4 1 -3 -2 -1 1 2 1 0 +1 +2 +3 1 4 1 +4 +5 +6 +7 +8 offsets +9 +10 1 Offset vector /course/eleg 667 -01 -f/Topic-2 b. ppt 15

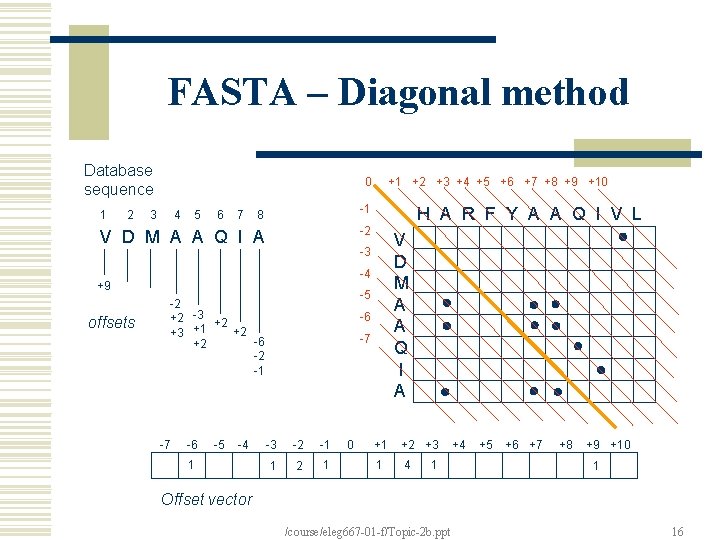
FASTA – Diagonal method Database sequence 0 8 -1 V D M A A Q I A -2 1 2 3 4 5 6 7 H A R F Y A A Q I V L V D M A A Q I A -3 -4 +9 offsets +1 +2 +3 +4 +5 +6 +7 +8 +9 +10 -5 -2 +2 -3 +2 +2 +3 +1 -6 +2 -2 -1 -7 -6 -5 -4 1 -6 -7 -3 -2 -1 1 2 1 0 +1 +2 +3 1 4 1 +4 +5 +6 +7 +8 +9 +10 1 Offset vector /course/eleg 667 -01 -f/Topic-2 b. ppt 16

FASTA - Join k-tuples Determine k-tuples (exact matches) common to both sequences; Join k-tuples that are in the same diagonal and not very far apart – creates regions; Typically ktup=1 or 2 for proteins and ktup=4 or 6 for DNA sequence The larger ktup, the faster the program Note: region should be gapless, and is created by certain heuristic /course/eleg 667 -01 -f/Topic-2 b. ppt 17

FASTA - Compute an optimized score for highest score region Find region with best score – “initial score”; Compute an “optimized score”, using DP, restricted to a band around the region. /course/eleg 667 -01 -f/Topic-2 b. ppt 18

Some Issues of FAST Algorithms w Selectivity vs. Sensitivity n n Ktup selectivity sensitivity w Statistical significance of the scores /course/eleg 667 -01 -f/Topic-2 b. ppt 19

BLAST (Altschul et al, 1990) w Compile list of high-scoring words based on the query sequence; w Scanning the database to search for hits – each hit gives a seed; w Extend seeds for each sequence; w Report high scoring segments /course/eleg 667 -01 -f/Topic-2 b. ppt 20

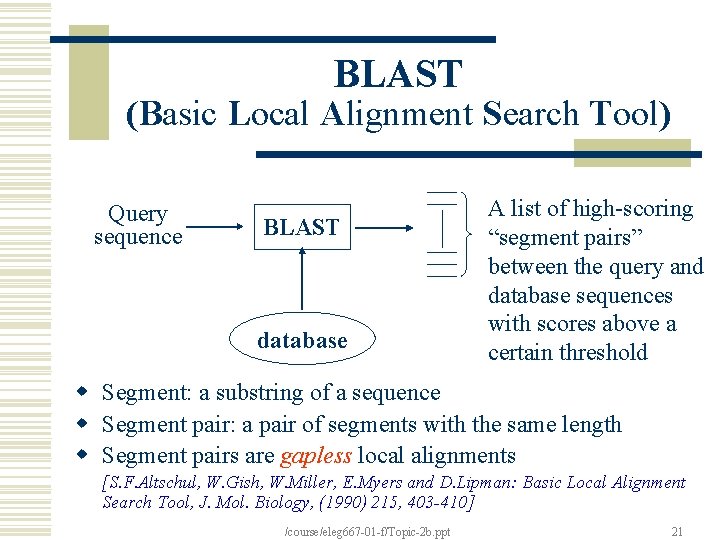
BLAST (Basic Local Alignment Search Tool) Query sequence BLAST database A list of high-scoring “segment pairs” between the query and database sequences with scores above a certain threshold w Segment: a substring of a sequence w Segment pair: a pair of segments with the same length w Segment pairs are gapless local alignments [S. F. Altschul, W. Gish, W. Miller, E. Myers and D. Lipman: Basic Local Alignment Search Tool, J. Mol. Biology, (1990) 215, 403 -410] /course/eleg 667 -01 -f/Topic-2 b. ppt 21

Maximum segment pair (MSP) – is a segment pair of maximum score. /course/eleg 667 -01 -f/Topic-2 b. ppt 22

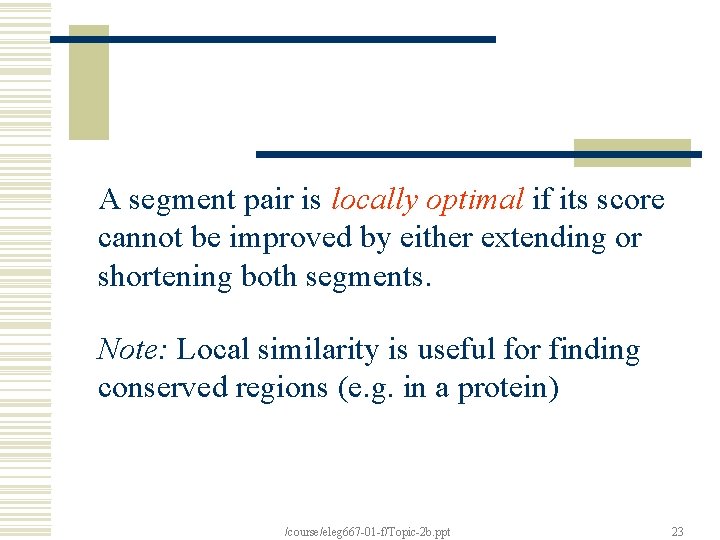
A segment pair is locally optimal if its score cannot be improved by either extending or shortening both segments. Note: Local similarity is useful for finding conserved regions (e. g. in a protein) /course/eleg 667 -01 -f/Topic-2 b. ppt 23

w BLAST is interested in finding only those sequences with MSP scores over some cutoff score S. w The main strategy of BLAST is to seek only segment pairs that contain a word pair with a score of at least T. /course/eleg 667 -01 -f/Topic-2 b. ppt 24

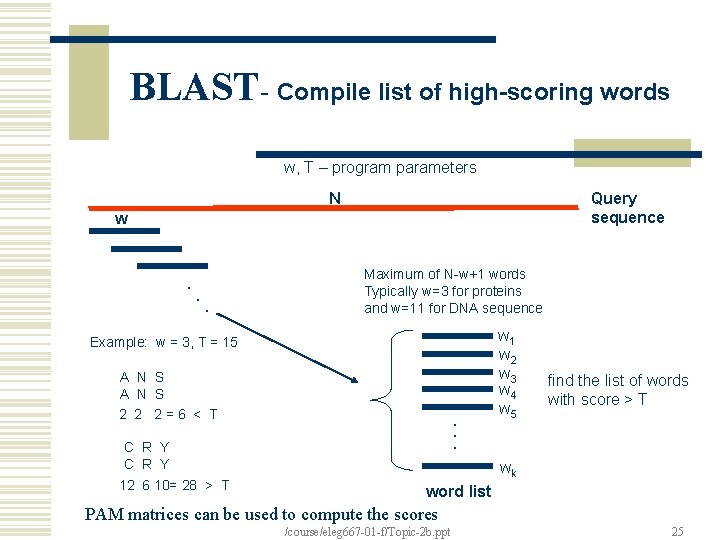
BLAST- Compile list of high-scoring words w, T – program parameters N Query sequence w . . . Maximum of N-w+1 words Typically w=3 for proteins and w=11 for DNA sequence Example: w = 3, T = 15 A N S 2 2 2=6 < T . . . C R Y 12 6 10= 28 > T w 1 w 2 w 3 w 4 w 5 find the list of words with score > T wk word list PAM matrices can be used to compute the scores /course/eleg 667 -01 -f/Topic-2 b. ppt 25

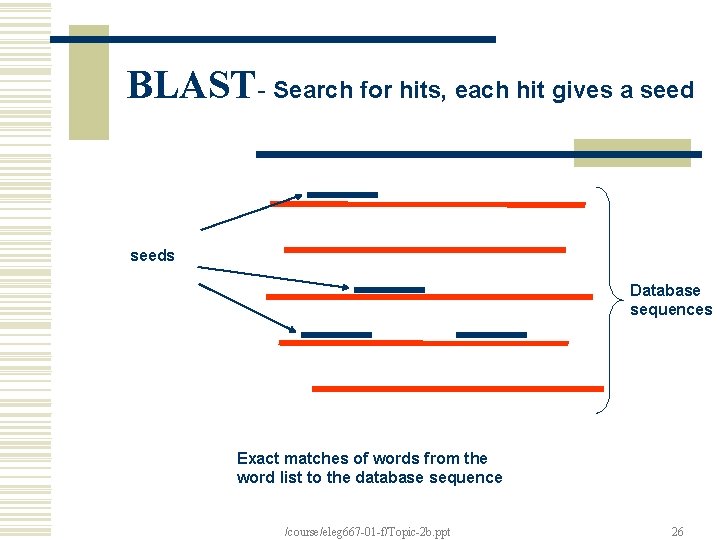
BLAST- Search for hits, each hit gives a seeds Database sequences Exact matches of words from the word list to the database sequence /course/eleg 667 -01 -f/Topic-2 b. ppt 26

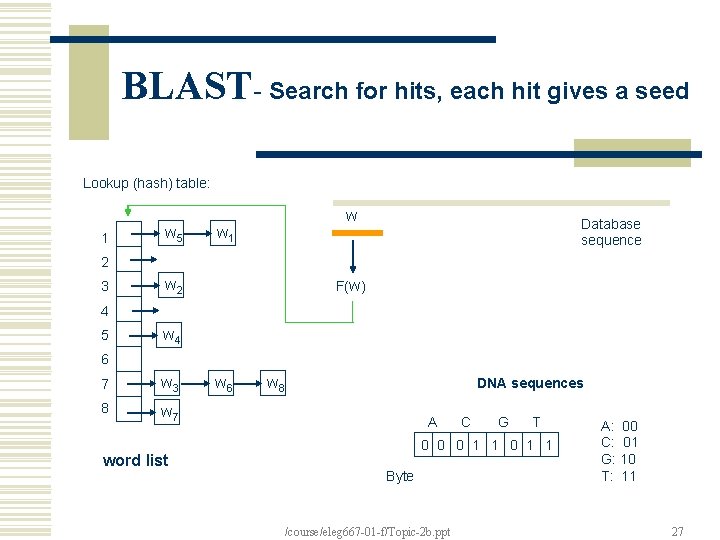
BLAST- Search for hits, each hit gives a seed Lookup (hash) table: 1 w 5 w w 1 Database sequence 2 3 w 2 F(w) 4 5 w 4 6 7 w 3 8 w 7 word list w 6 w 8 DNA sequences A C G T 0 0 0 1 1 Byte /course/eleg 667 -01 -f/Topic-2 b. ppt A: 00 C: 01 G: 10 T: 11 27

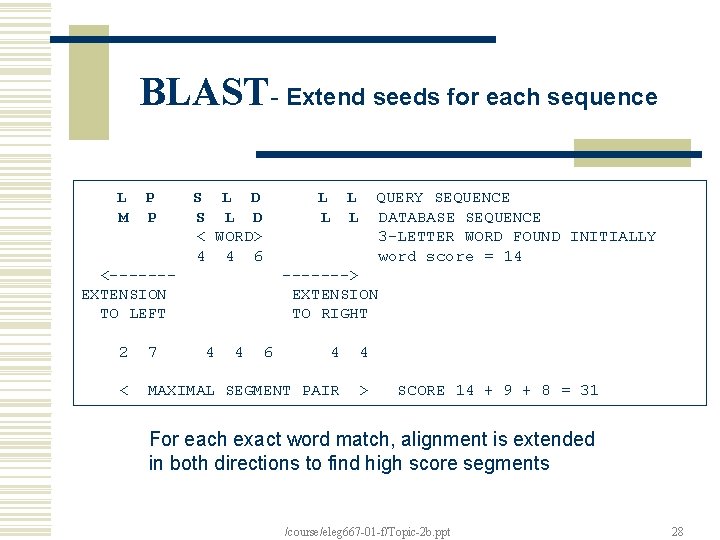
BLAST- Extend seeds for each sequence L M P P <------EXTENSION TO LEFT S L D < WORD> 4 4 6 L L QUERY SEQUENCE DATABASE SEQUENCE 3 -LETTER WORD FOUND INITIALLY word score = 14 -------> EXTENSION TO RIGHT 2 7 4 Maximum 4 6 4 4 Pairs (MSPs) Segment < MAXIMAL SEGMENT PAIR > SCORE 14 + 9 + 8 = 31 For each exact word match, alignment is extended in both directions to find high score segments /course/eleg 667 -01 -f/Topic-2 b. ppt 28

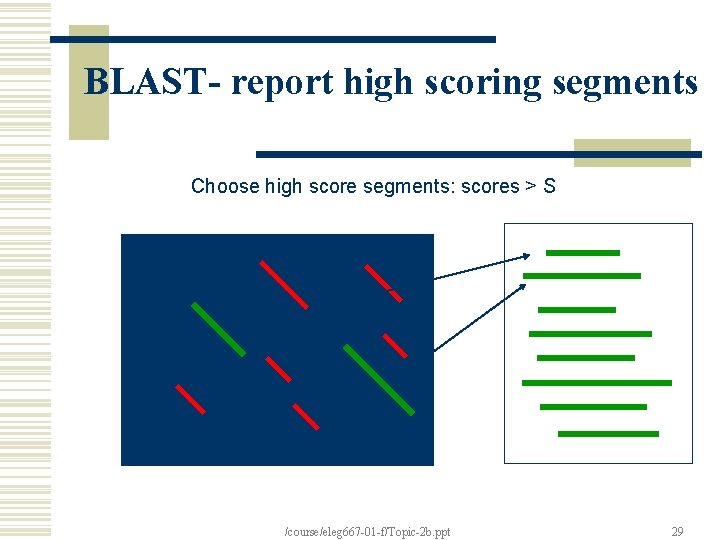
BLAST- report high scoring segments Choose high score segments: scores > S /course/eleg 667 -01 -f/Topic-2 b. ppt 29

Why BLAST is Fast? Because: the alignments are gapless! /course/eleg 667 -01 -f/Topic-2 b. ppt 30

Statistical Significance of BLAST Results Question: If a match found by BLAST – what is the probability that such match is due to chance alone? A well-funded statistical theory is used by BLAST in determine the matching scores. /course/eleg 667 -01 -f/Topic-2 b. ppt 31

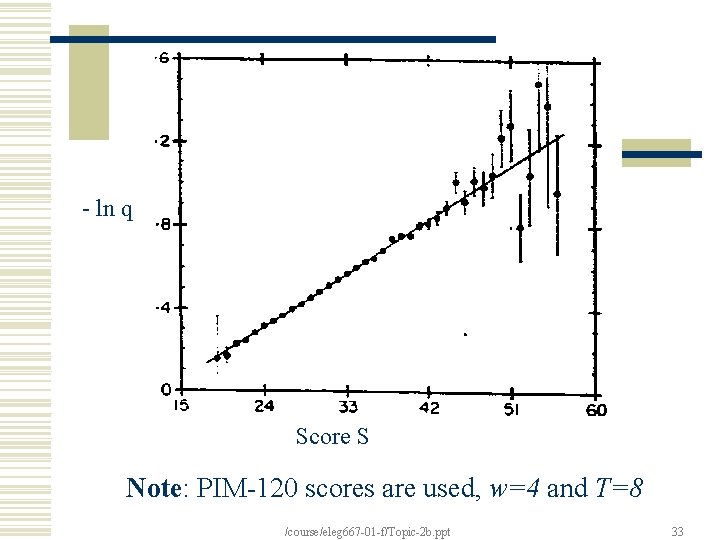
Questions Q 1: What proportion of segment pairs with a given score contain a word pair with a score at least T? Answer: [Karlin 91] Q 2: What probability q of a MSP pair found (under a threshold score S) will fail to contain a seed word W (of score >= T)? Answer: See Plot [Alschul et. al. 90] /course/eleg 667 -01 -f/Topic-2 b. ppt 32

- ln q Score S Note: PIM-120 scores are used, w=4 and T=8 /course/eleg 667 -01 -f/Topic-2 b. ppt 33

Improvement of The Basic BLASTGapped BLAST and PSI-BLAST [S. F. Altschul, et. al. , Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Algorithms, Nucleic Acids Research, 1997, Vol 25, No. 17, 3389 -3402] w Objectives n n Speedup the execution substantially Enhance the sensitivity to weak similarities /course/eleg 667 -01 -f/Topic-2 b. ppt 34

Major Extensions/Changes to BLAST w Add ability to generate gapped alignment using dynamic programming to extend a seed in both directions w Using a “two-hit” method to “filter” out the candidate pairs for extension w The search may be iterated: round i will generate a new position-specific score matrix from significant alignments found to be used for round i+1 (this process involves the construction of a multiple sequence alignment – see Topic 2 C) /course/eleg 667 -01 -f/Topic-2 b. ppt 35

The Two-Hit Method w Observation: an HSP of interest is much longer than a single word pair, thus may contain multiple hits on the same diagonal within a relatively short distance apart. w Methods: Choose a “window” , and do extension only when two non-overlapping hits are found within distance A of one another on the same diagonal w Effectiveness: reduce candidate pairs for extension substantially (by 86%) /course/eleg 667 -01 -f/Topic-2 b. ppt 36

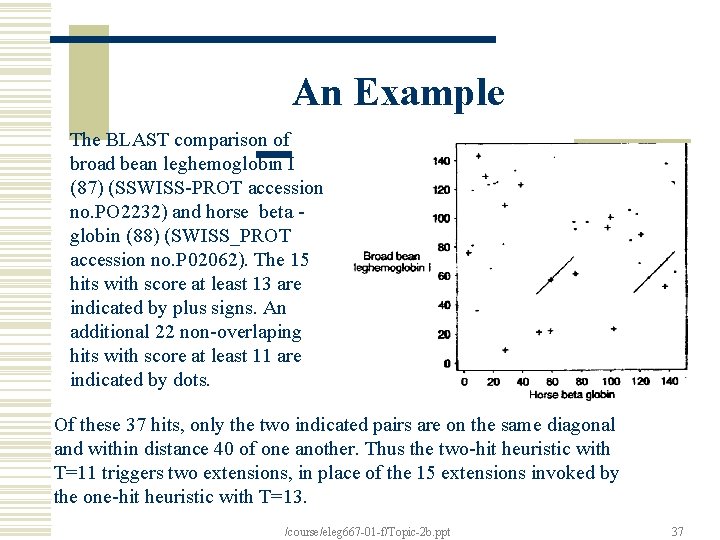
An Example The BLAST comparison of broad bean leghemoglobin I (87) (SSWISS-PROT accession no. PO 2232) and horse beta globin (88) (SWISS_PROT accession no. P 02062). The 15 hits with score at least 13 are indicated by plus signs. An additional 22 non-overlaping hits with score at least 11 are indicated by dots. Of these 37 hits, only the two indicated pairs are on the same diagonal and within distance 40 of one another. Thus the two-hit heuristic with T=11 triggers two extensions, in place of the 15 extensions invoked by the one-hit heuristic with T=13. /course/eleg 667 -01 -f/Topic-2 b. ppt 37