Primer Design Restriction Analysis 2 nd April 2014





















- Slides: 111

Primer Design & Restriction Analysis 2 nd April 2014 Carrie Iwema, Ph. D, MLS, AHIP Information Specialist in Molecular Biology Health Sciences Library System University of Pittsburgh iwema@pitt. edu http: //www. hsls. pitt. edu/molbio

Goals: n PCR primer construction & analysis n Restriction digestion & mapping http: //www. hsls. pitt. edu/molbio

Tools: n Primer Analysis & Design q q q n Net. Primer 3 Plus Primer-BLAST Restriction Mapping q q NEBcutter Webcutter http: //www. hsls. pitt. edu/molbio

Primer Analysis & Design A little something to get you in the mood… http: //www. hsls. pitt. edu/molbio

Polymerase Chain Reaction (PCR) 1983 -Kary Mullis very simple n n n exponential amplification similar to natural DNA replication The primary reagents, used in PCR are: n n Template DNA–DNA sequence to amplify DNA nucleotides–building blocks for new DNA Taq polymerase–heat stable enzyme catalyzes new DNA Primers–single-stranded DNA, ~20 -50 nucleotides, complimentary to a short region on either side of template DNA http: //www. hsls. pitt. edu/molbio

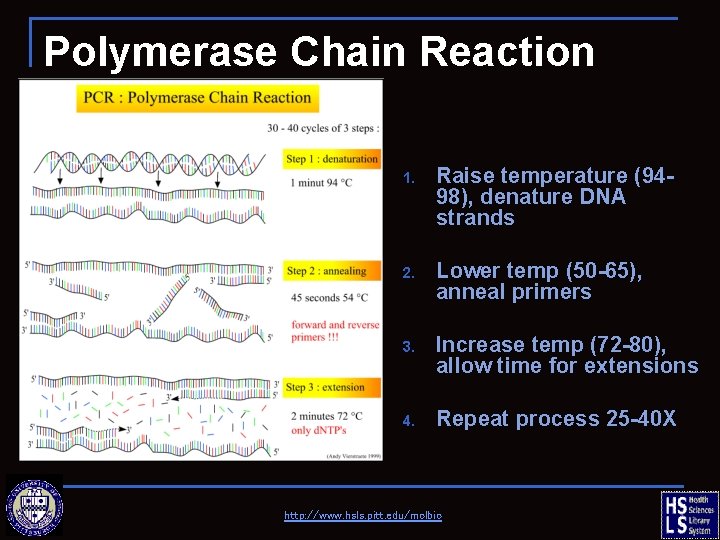
Polymerase Chain Reaction (PCR) 1. Raise temperature (9498), denature DNA strands 2. Lower temp (50 -65), anneal primers 3. Increase temp (72 -80), allow time for extensions 4. Repeat process 25 -40 X http: //www. hsls. pitt. edu/molbio

Things to consider for primer design… n Primer-Dimer formation n Secondary Structures in Primers n Illegitimate Priming in Template DNA due to repeated sequences n Incompatibility with PCR conditions SOURCE: NCBI http: //www. hsls. pitt. edu/molbio

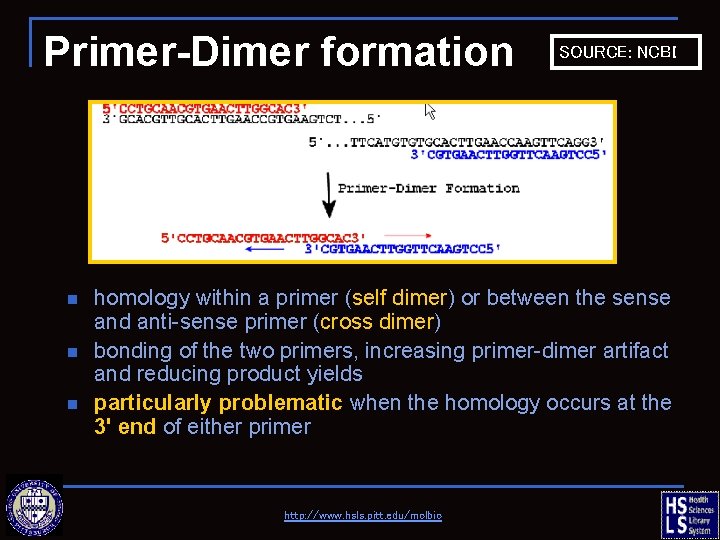
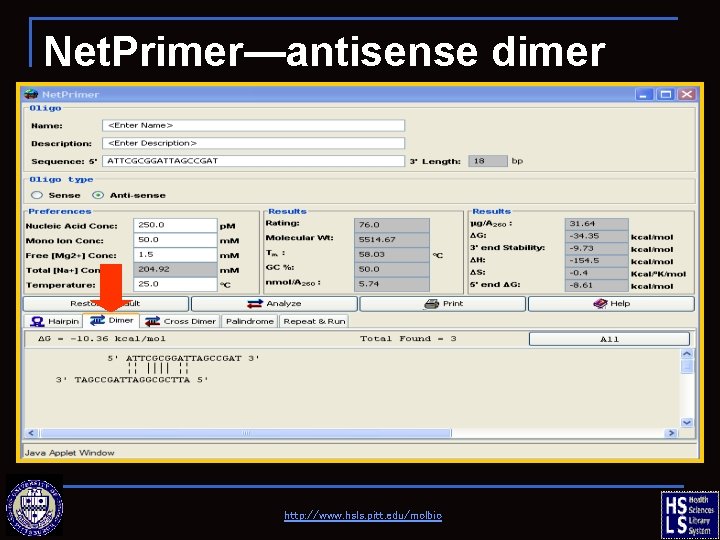
Primer-Dimer formation n SOURCE: NCBI homology within a primer (self dimer) or between the sense and anti-sense primer (cross dimer) bonding of the two primers, increasing primer-dimer artifact and reducing product yields particularly problematic when the homology occurs at the 3' end of either primer http: //www. hsls. pitt. edu/molbio

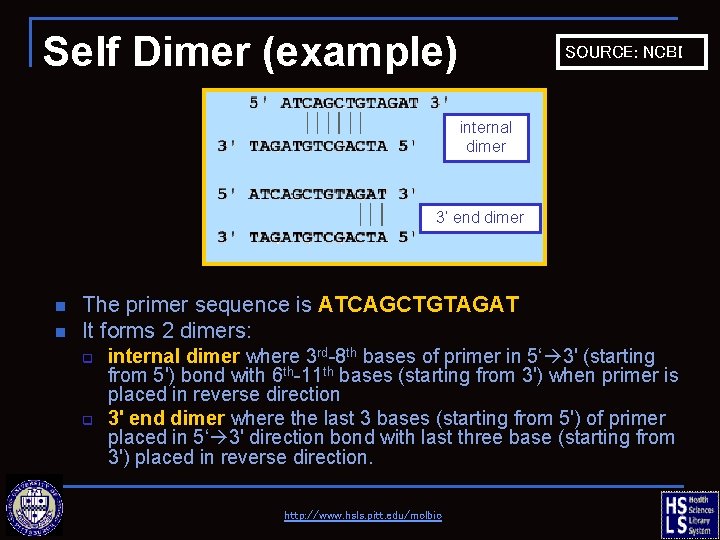
Self Dimer (example) SOURCE: NCBI internal dimer 3’ end dimer n n The primer sequence is ATCAGCTGTAGAT It forms 2 dimers: q q internal dimer where 3 rd-8 th bases of primer in 5‘ 3' (starting from 5') bond with 6 th-11 th bases (starting from 3') when primer is placed in reverse direction 3' end dimer where the last 3 bases (starting from 5') of primer placed in 5‘ 3' direction bond with last three base (starting from 3') placed in reverse direction. http: //www. hsls. pitt. edu/molbio

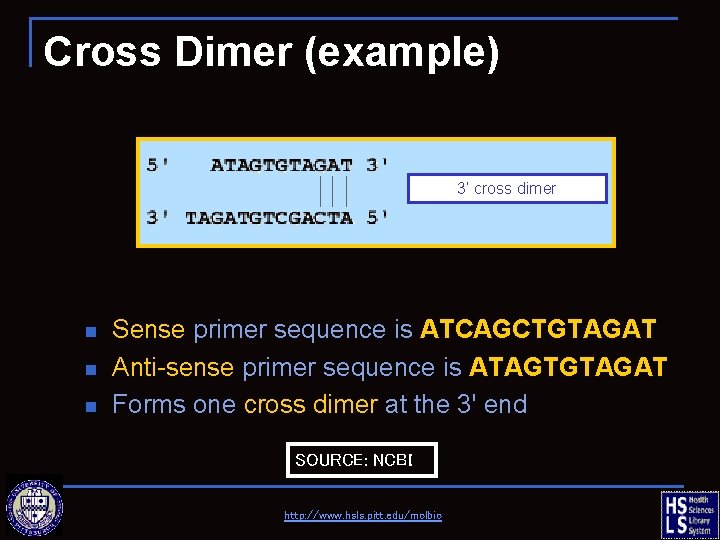
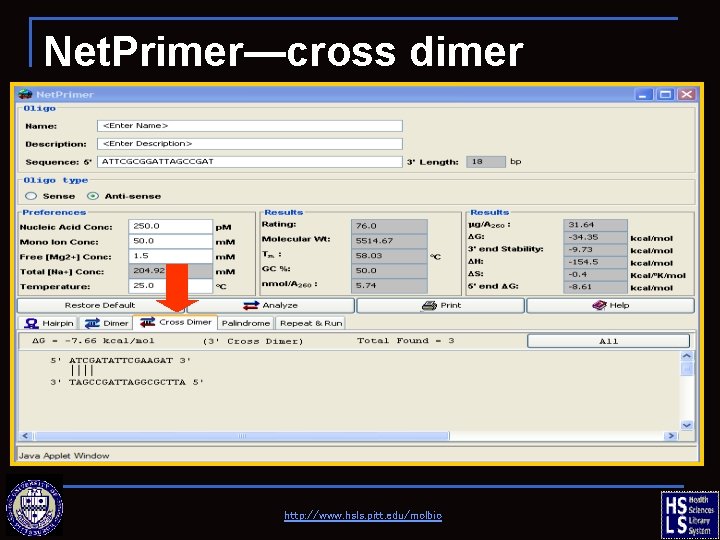
Cross Dimer (example) 3’ cross dimer n n n Sense primer sequence is ATCAGCTGTAGAT Anti-sense primer sequence is ATAGTGTAGAT Forms one cross dimer at the 3' end SOURCE: NCBI http: //www. hsls. pitt. edu/molbio

Secondary Structure in Primers n Hairpin loop q formed when primer folds back upon itself q held in place by intramolecular bonding q can occur with as few as 3 consecutive homologous bases q stability measured by the free energy The free energy of the loop is based upon the energy of the intramolecular bond and the energy needed to twist the DNA to form the loop. q If free energy >0, the loop is too unstable to interfere with the reaction q If free energy <0, the loop could reduce the efficiency of amplification http: //www. hsls. pitt. edu/molbio

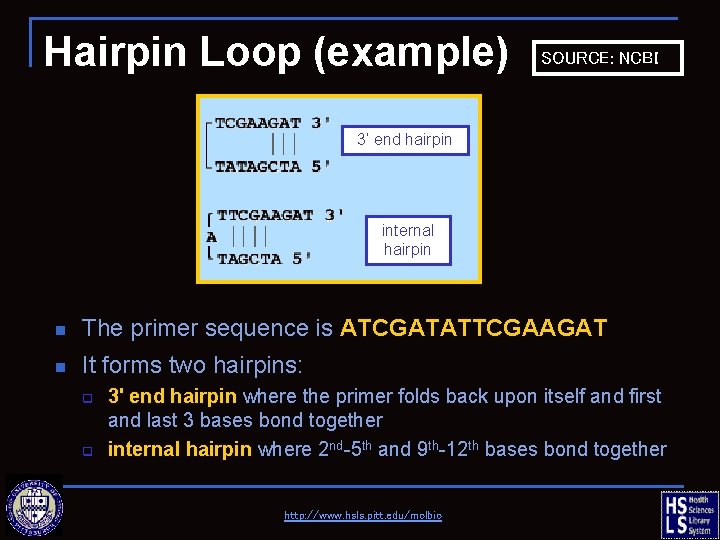
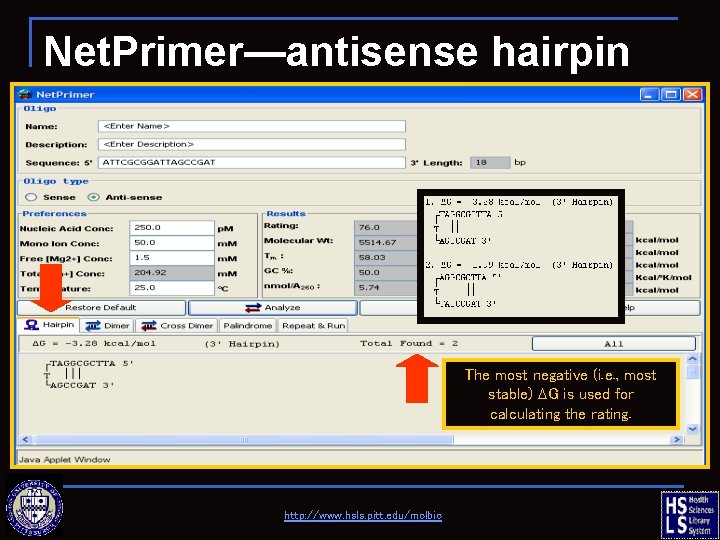
Hairpin Loop (example) SOURCE: NCBI 3’ end hairpin internal hairpin n The primer sequence is ATCGATATTCGAAGAT n It forms two hairpins: q q 3' end hairpin where the primer folds back upon itself and first and last 3 bases bond together internal hairpin where 2 nd-5 th and 9 th-12 th bases bond together http: //www. hsls. pitt. edu/molbio

Basic Primer Analysis & Design Software n Net. Primer q n Primer 3 Plus q n http: //www. premierbiosoft. com/netprimer/ http: //www. bioinformatics. nl/cgi-bin/primer 3 plus. cgi Primer-BLAST q http: //www. ncbi. nlm. nih. gov/tools/primer-blast/ http: //www. hsls. pitt. edu/molbio

Net. Primer n http: //www. premierbiosoft. com/netprimer/ n From PREMIER Biosoft Free Major features: n n q q q Primer properties: Tm , molecular weight, GC%, optical activity (both in nmol/A 260 & µg/A 260), DG, 3' end stability, DH, DS, and 5' end DG Secondary structures: Hairpins, dimers, cross dimers, palindromes, repeats and runs Primer rating: Quantitative prediction of the efficiency of a primer Comprehensive report: Prints complete primer analysis for an individual primer or primer pair Primer pairs: Analyze individual primers or primer pairs Comprehensive help: Details all the formulas and references used in primer analysis algorithm http: //www. hsls. pitt. edu/molbio

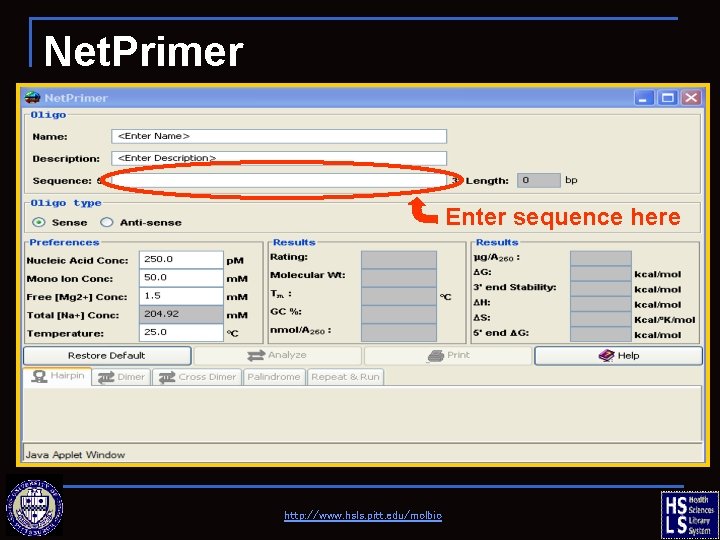
Net. Primer Enter sequence here http: //www. hsls. pitt. edu/molbio

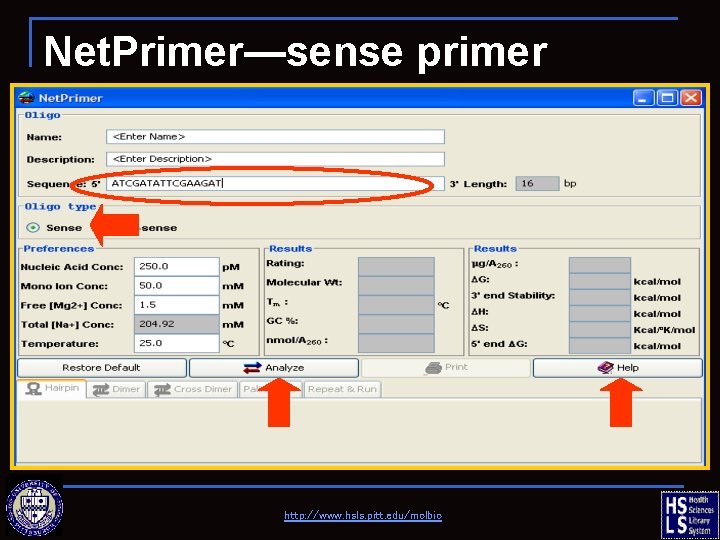
Net. Primer—sense primer http: //www. hsls. pitt. edu/molbio

Net. Primer—help http: //www. hsls. pitt. edu/molbio

Net. Primer—theories & formulas http: //www. hsls. pitt. edu/molbio

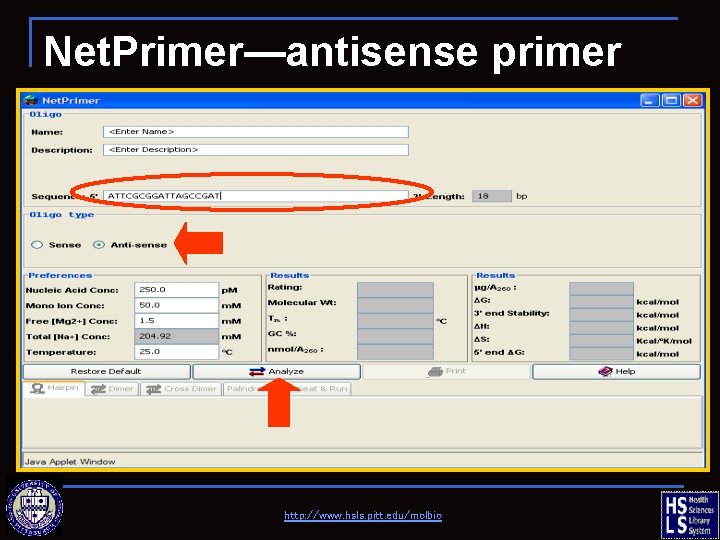
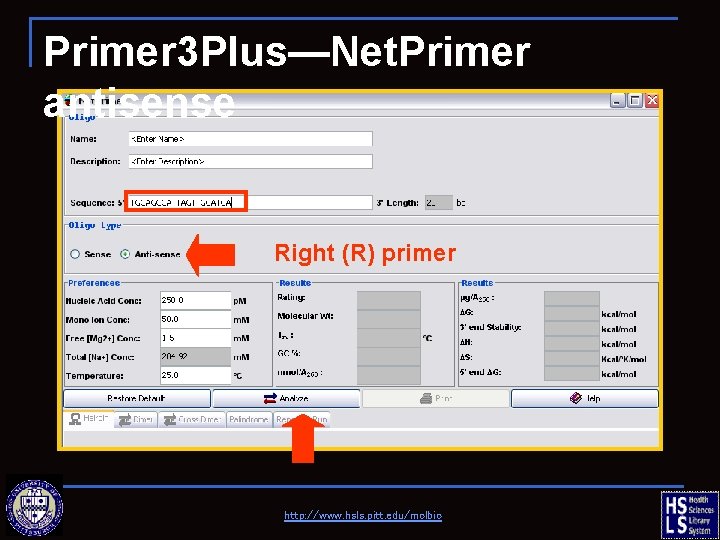
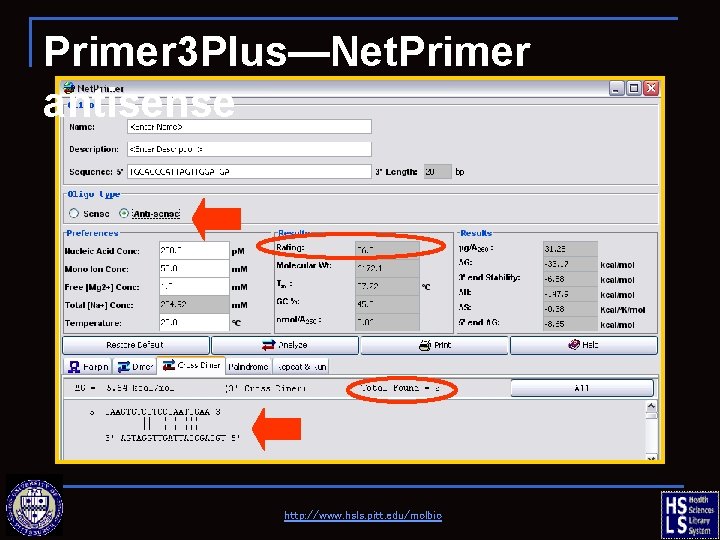
Net. Primer—antisense primer http: //www. hsls. pitt. edu/molbio

Net. Primer—antisense hairpin The most negative (i. e. , most stable) DG is used for calculating the rating. http: //www. hsls. pitt. edu/molbio

Net. Primer—antisense dimer http: //www. hsls. pitt. edu/molbio

Net. Primer—cross dimer http: //www. hsls. pitt. edu/molbio

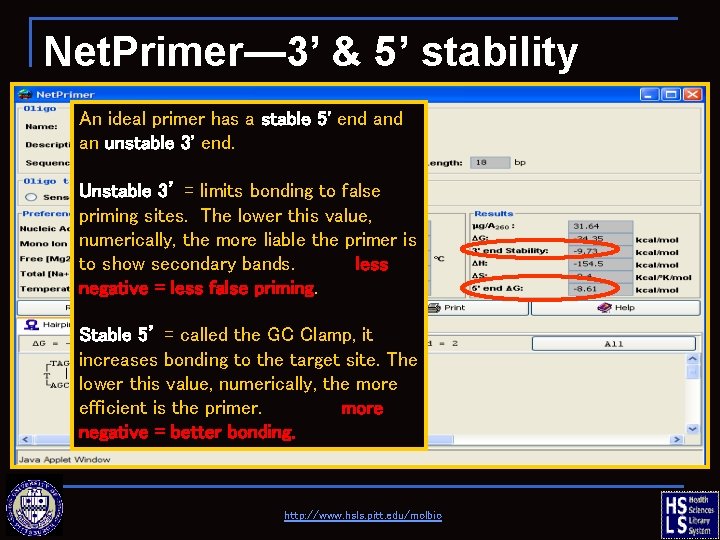
Net. Primer— 3’ & 5’ stability An ideal primer has a stable 5' end an unstable 3' end. Unstable 3’ = limits bonding to false priming sites. The lower this value, numerically, the more liable the primer is to show secondary bands. less negative = less false priming. Stable 5’ = called the GC Clamp, it increases bonding to the target site. The lower this value, numerically, the more efficient is the primer. more negative = better bonding. http: //www. hsls. pitt. edu/molbio

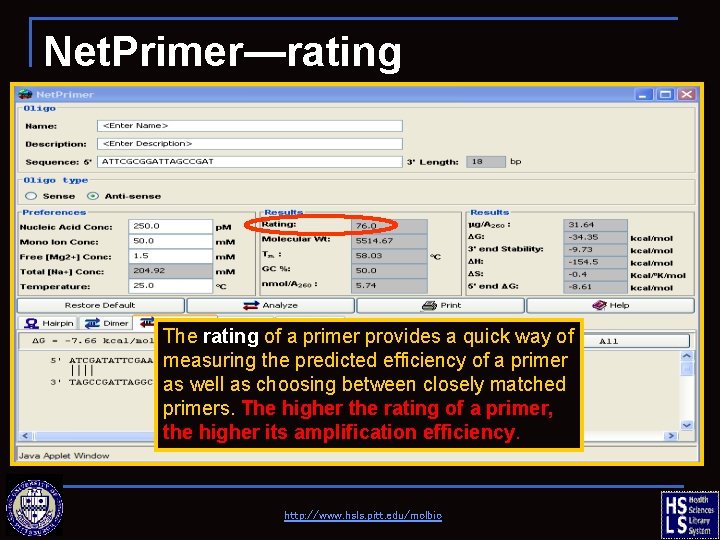
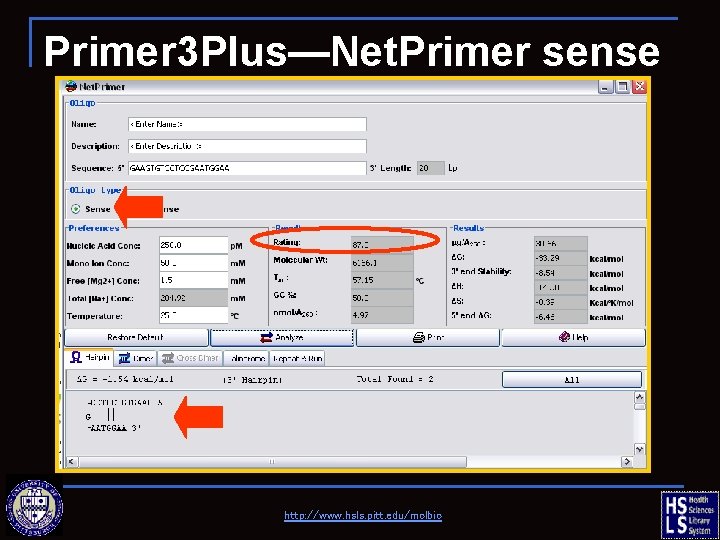
Net. Primer—rating The rating of a primer provides a quick way of measuring the predicted efficiency of a primer as well as choosing between closely matched primers. The higher the rating of a primer, the higher its amplification efficiency. http: //www. hsls. pitt. edu/molbio

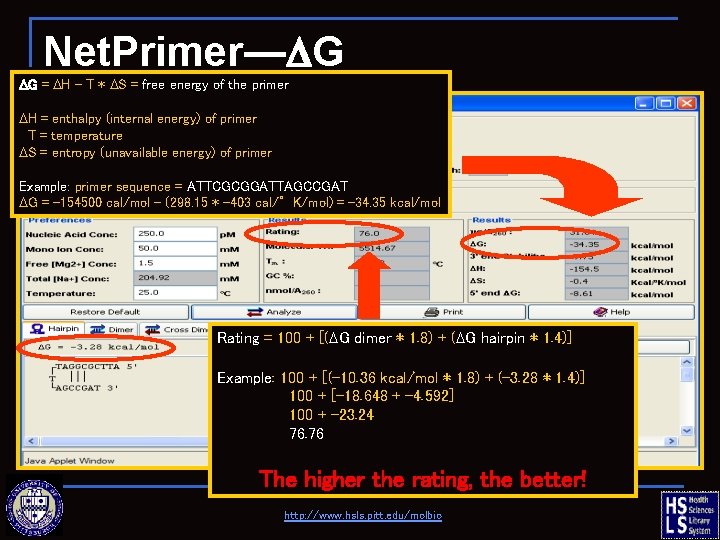
Net. Primer—DG DG = DH – T * DS = free energy of the primer DH = enthalpy (internal energy) of primer T = temperature DS = entropy (unavailable energy) of primer Example: primer sequence = ATTCGCGGATTAGCCGAT DG = -154500 cal/mol – (298. 15 * -403 cal/°K/mol) = -34. 35 kcal/mol Rating = 100 + [(DG dimer * 1. 8) + (DG hairpin * 1. 4)] Example: 100 + [(-10. 36 kcal/mol * 1. 8) + (-3. 28 * 1. 4)] 100 + [-18. 648 + -4. 592] 100 + -23. 24 76. 76 The higher the rating, the better! http: //www. hsls. pitt. edu/molbio

Net. Primer—practice primers Rank these primers with attention to rating, 5’ end DG, and 3’ end stability 1. 2. 3. 4. 5. 6. atgtgcgaggagaaagtgct acaaaccctggacttgcatc cgacttgtcccaggtgtttt ctgaaaccattggcacacac ggctgtgaacatggacattg ggctgaagccaaagctacac http: //www. hsls. pitt. edu/molbio

Net. Primer n Ideal for checking primers n To create primers, try Primer 3 Plus http: //www. hsls. pitt. edu/molbio

Primer 3 Plus n http: //www. bioinformatics. nl/cgi-bin/primer 3 plus. cgi n Select primer pairs to detect a given template sequence n Targets and included/excluded regions can be specified n Steve Rozen and Helen J. Skaletsky (2000) Primer 3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics Methods and Protocols: Methods in Molecular Biology. Human Press, Totowa, NJ, pp 365 -386 http: //www. hsls. pitt. edu/molbio

Primer 3 Plus http: //www. hsls. pitt. edu/molbio

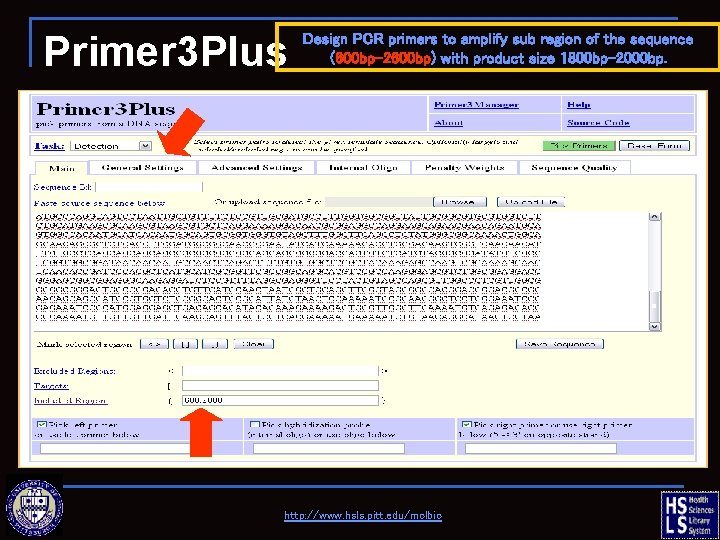
Primer 3 Plus Design PCR primers to amplify sub region of the sequence (600 bp-2600 bp) with product size 1800 bp-2000 bp. http: //www. hsls. pitt. edu/molbio

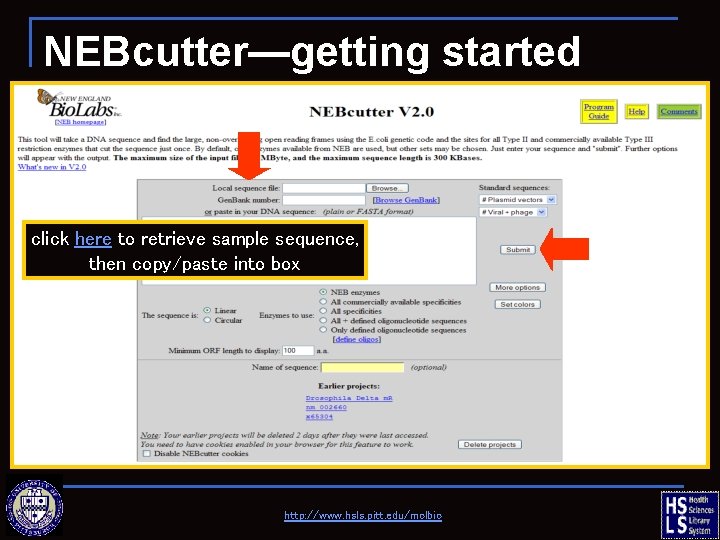
Primer 3 Plus—getting started click here to retrieve sample sequence, then copy/paste into box http: //www. hsls. pitt. edu/molbio

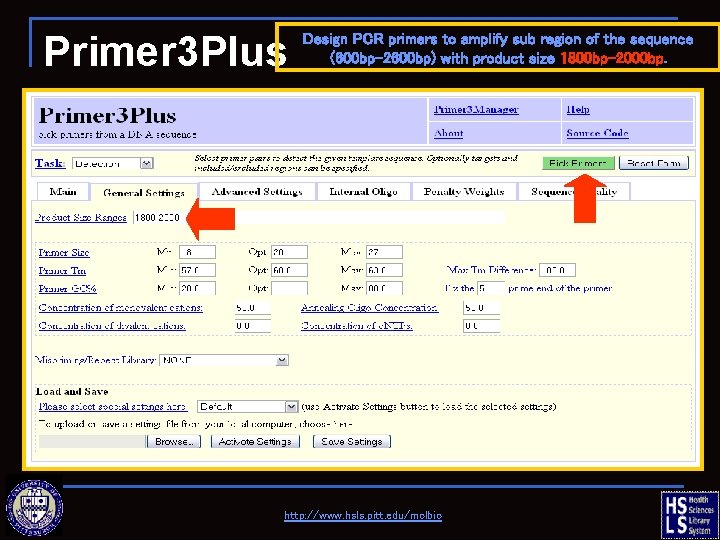
Primer 3 Plus Design PCR primers to amplify sub region of the sequence (600 bp-2600 bp) with product size 1800 bp-2000 bp. http: //www. hsls. pitt. edu/molbio

Primer 3 Plus Design PCR primers to amplify sub region of the sequence (600 bp-2600 bp) with product size 1800 bp-2000 bp. http: //www. hsls. pitt. edu/molbio

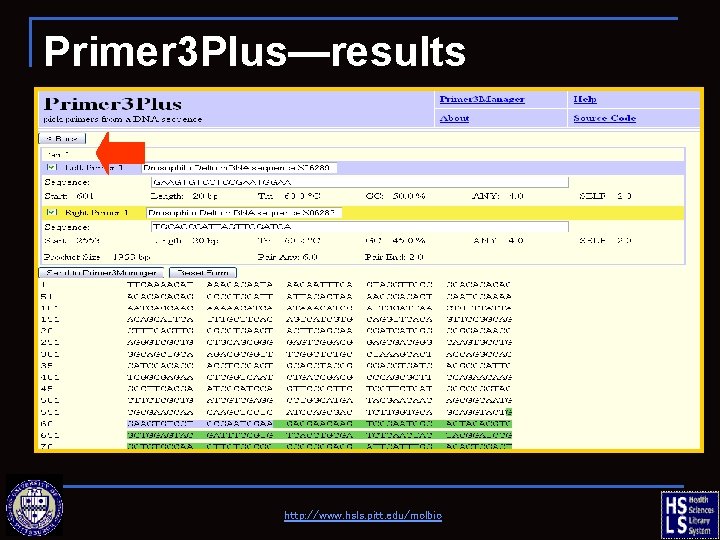
Primer 3 Plus—results http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—results http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—results http: //www. hsls. pitt. edu/molbio

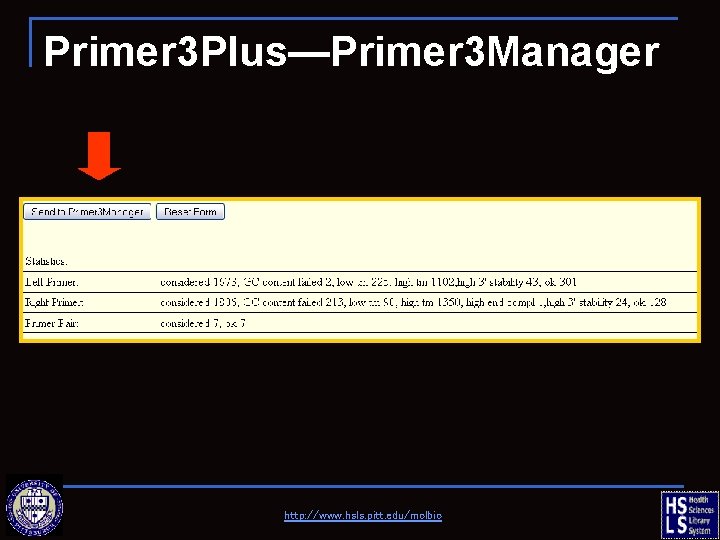
Primer 3 Plus—Primer 3 Manager http: //www. hsls. pitt. edu/molbio

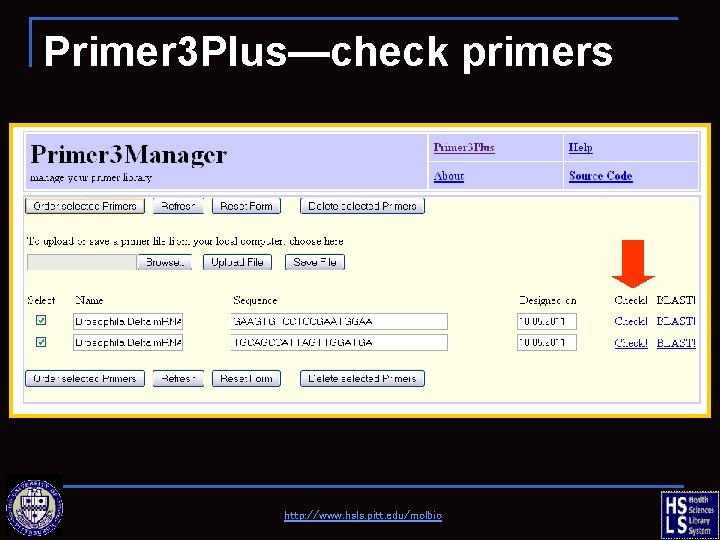
Primer 3 Plus—check primers http: //www. hsls. pitt. edu/molbio

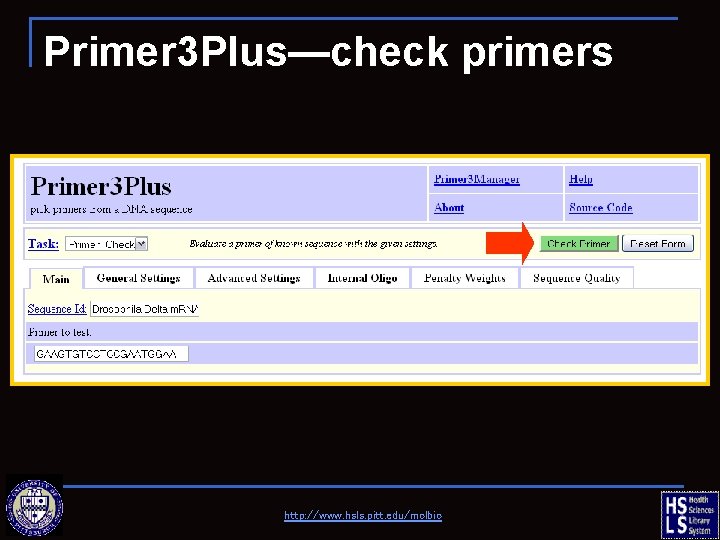
Primer 3 Plus—check primers http: //www. hsls. pitt. edu/molbio

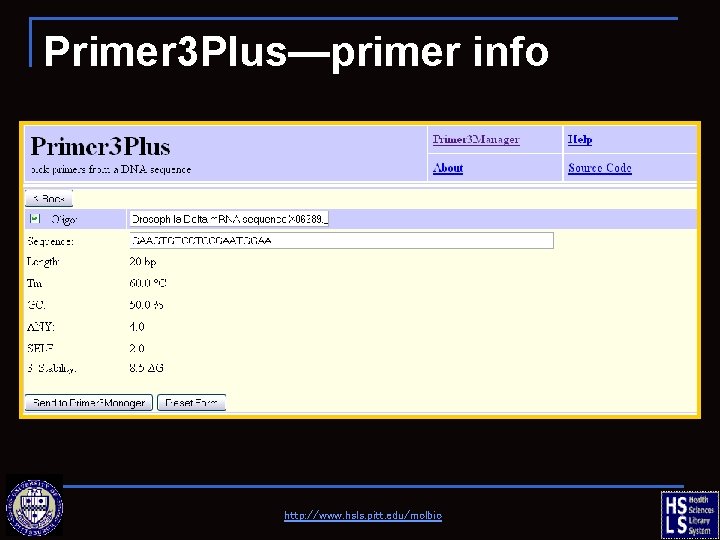
Primer 3 Plus—primer info http: //www. hsls. pitt. edu/molbio

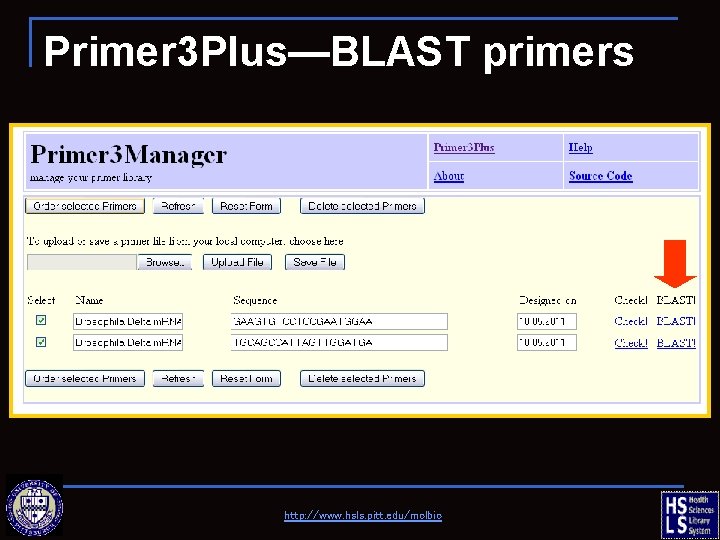
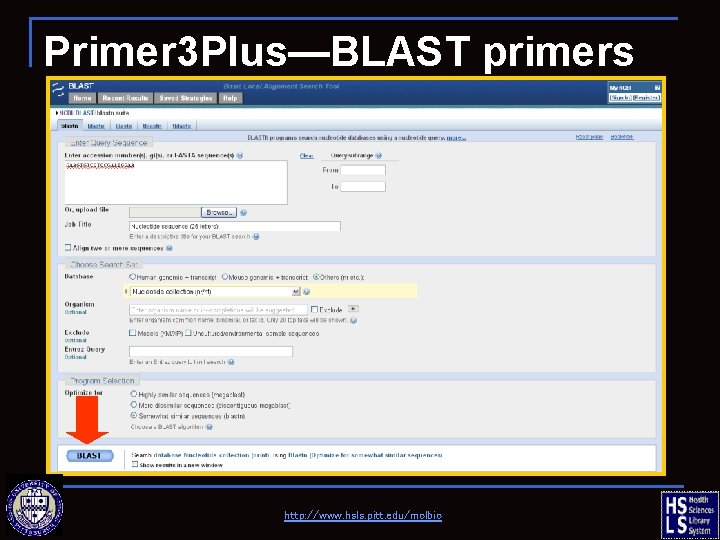
Primer 3 Plus—BLAST primers http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—BLAST primers http: //www. hsls. pitt. edu/molbio

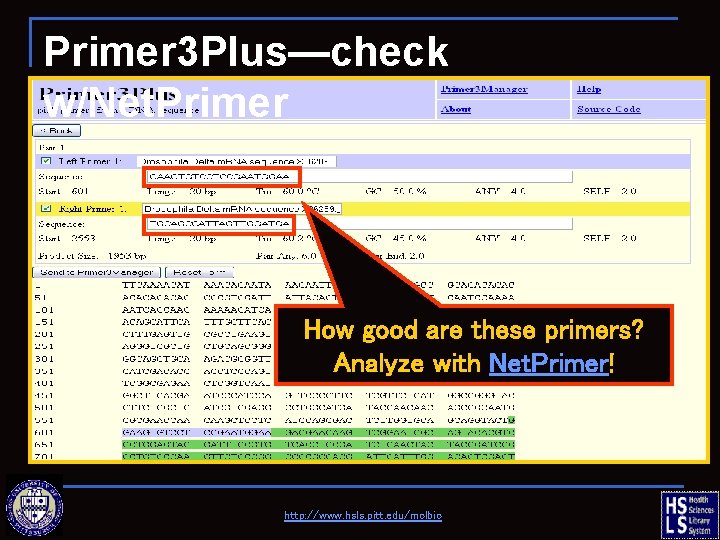
Primer 3 Plus—check w/Net. Primer How good are these primers? Analyze with Net. Primer! http: //www. hsls. pitt. edu/molbio

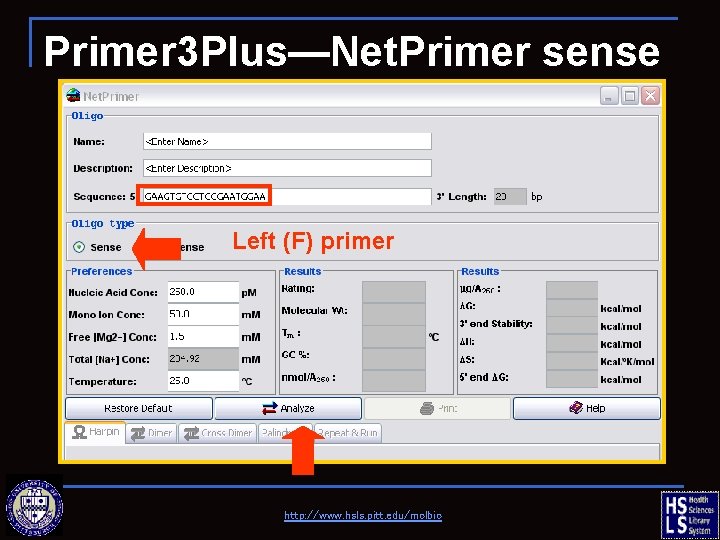
Primer 3 Plus—Net. Primer sense Left (F) primer http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—Net. Primer sense http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—Net. Primer antisense Right (R) primer http: //www. hsls. pitt. edu/molbio

Primer 3 Plus—Net. Primer antisense http: //www. hsls. pitt. edu/molbio

Primer-BLAST n http: //www. ncbi. nlm. nih. gov/tools/primer-blast/ n Combines primer design (Primer 3) and a specificity check (BLAST) n Can also be used w/pre-designed primers n ref: http: //www. ncbi. nlm. nih. gov/pmc/articles/PMC 3412702/ http: //www. hsls. pitt. edu/molbio

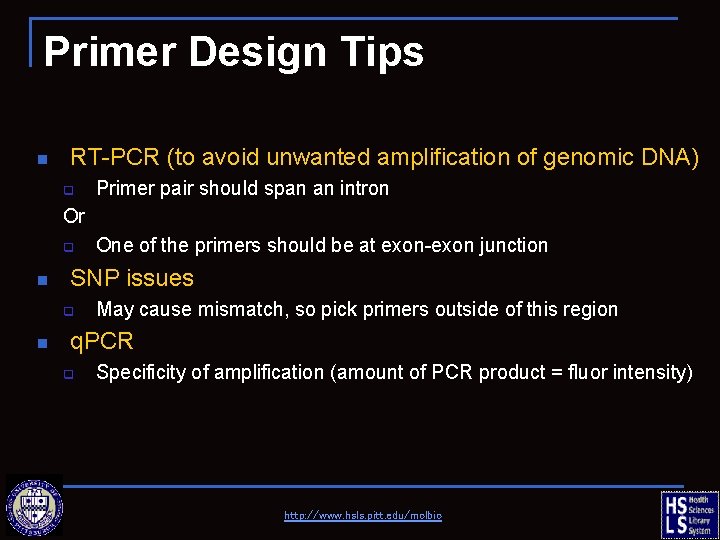
Primer Design Tips n RT-PCR (to avoid unwanted amplification of genomic DNA) q Primer pair should span an intron Or q n SNP issues q n One of the primers should be at exon-exon junction May cause mismatch, so pick primers outside of this region q. PCR q Specificity of amplification (amount of PCR product = fluor intensity) http: //www. hsls. pitt. edu/molbio

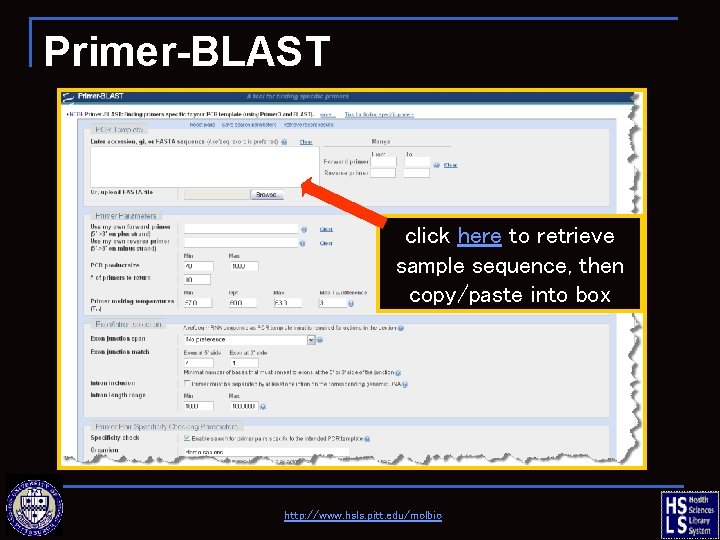
Primer-BLAST click here to retrieve sample sequence, then copy/paste into box http: //www. hsls. pitt. edu/molbio

Primer-BLAST results http: //www. hsls. pitt. edu/molbio

HSLS Mol. Bio Primer Design Tools http: //www. hsls. pitt. edu/molbio

Finding Primer Resources… search. HSLS. Mol. Bio http: //www. hsls. pitt. edu/molbio

More Primer Databases http: //www. hsls. pitt. edu/molbio

Restriction Mapping www. biologyreference. com http: //www. hsls. pitt. edu/molbio

Restriction Mapping—for your sequence n n n Determine the # of restriction sites Determine the nucleotide position of each cut List the enzymes that do not cut List the enzymes that cut only once Graphical representation of the restriction sites Textual representation of the restriction sites http: //www. hsls. pitt. edu/molbio

Restriction Mapping Tools n NEBcutter q n http: //tools. neb. com/NEBcutter 2/index. php Webcutter q http: //bio. biomedicine. gu. se/cutter 2/ http: //www. hsls. pitt. edu/molbio

NEBcutter V 2. 0 n n n From New England Bio. Labs Free Major features: q q Takes a DNA sequence and finds the large, non-overlapping open reading frames using the E. coli genetic code and the sites for all Type II and commercially available Type III restriction enzymes that cut the sequence just once. By default, only enzymes from NEB are used, but other sets may be chosen. Further options appear in the output. Maximum size of input file = 1 MB; maximum sequence length = 300 KB. http: //www. hsls. pitt. edu/molbio

NEBcutter http: //www. hsls. pitt. edu/molbio

NEBcutter—program guide http: //www. hsls. pitt. edu/molbio

NEBcutter http: //www. hsls. pitt. edu/molbio

NEBcutter—help http: //www. hsls. pitt. edu/molbio

NEBcutter—getting started click here to retrieve sample sequence, then copy/paste into box http: //www. hsls. pitt. edu/molbio

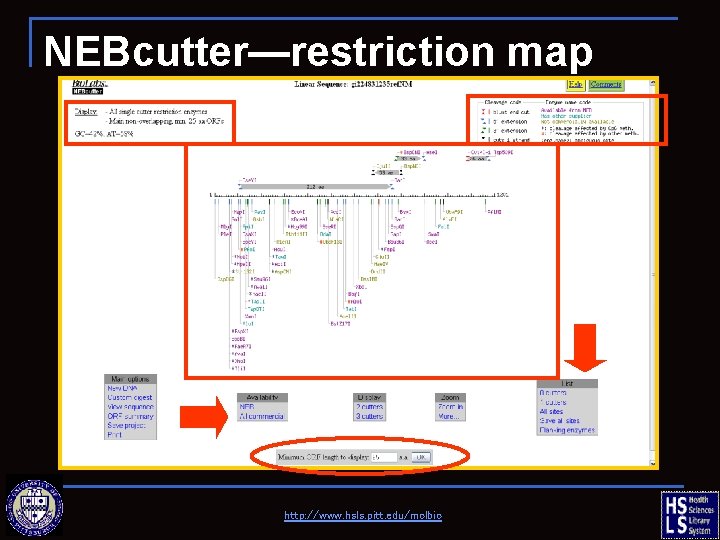
NEBcutter—restriction map http: //www. hsls. pitt. edu/molbio

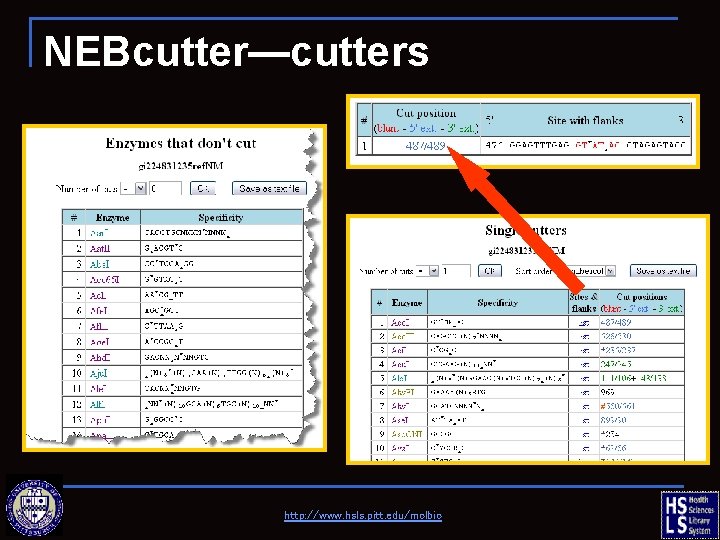
NEBcutter—cutters http: //www. hsls. pitt. edu/molbio

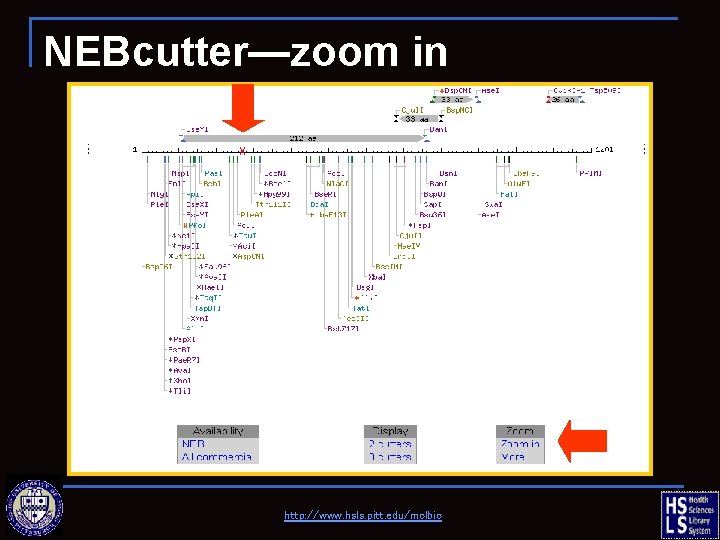
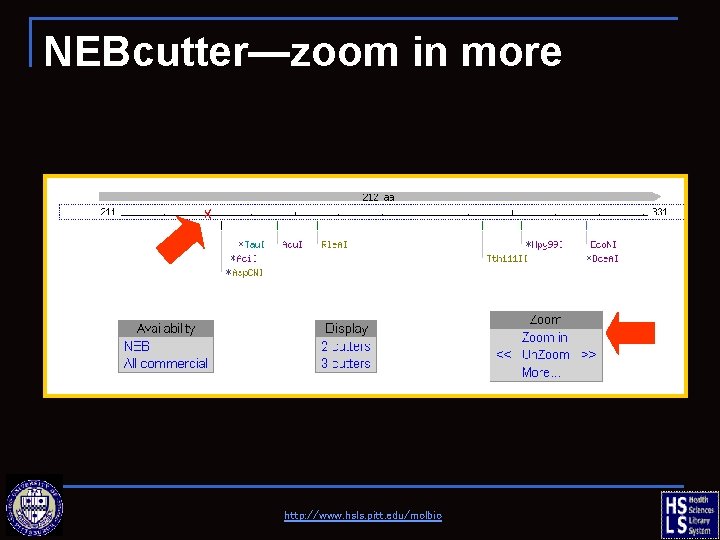
NEBcutter—zoom in http: //www. hsls. pitt. edu/molbio

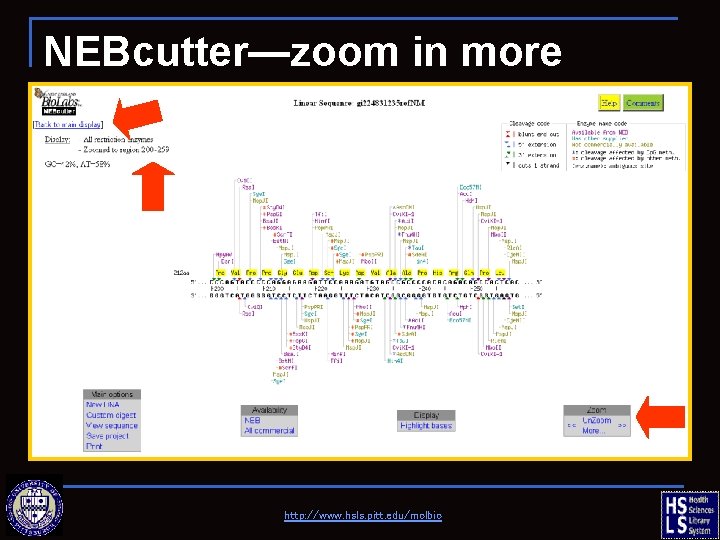
NEBcutter—zoom in more http: //www. hsls. pitt. edu/molbio

NEBcutter—zoom in more http: //www. hsls. pitt. edu/molbio

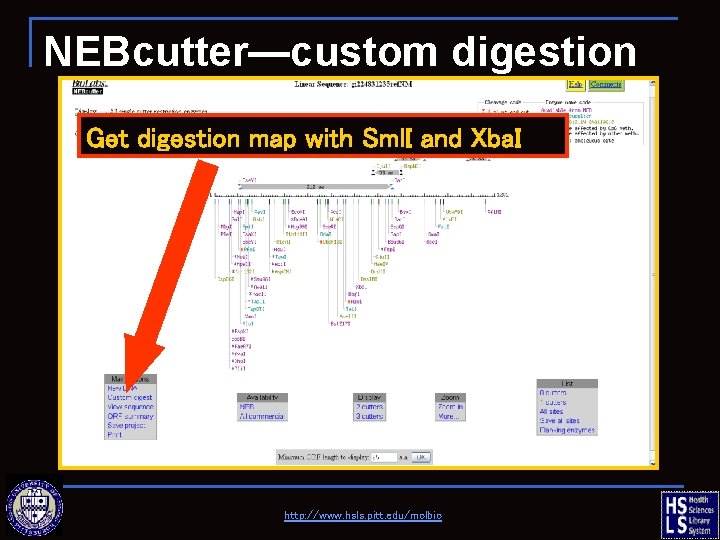
NEBcutter—custom digestion Get digestion map with Sml. I and Xba. I http: //www. hsls. pitt. edu/molbio

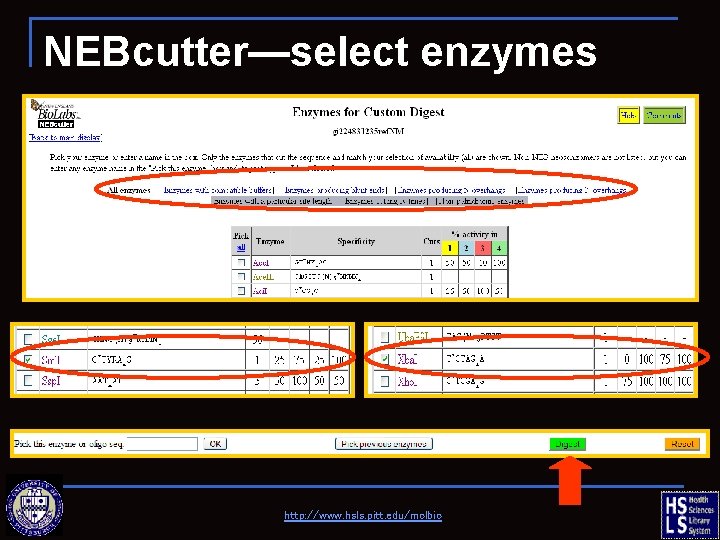
NEBcutter—select enzymes http: //www. hsls. pitt. edu/molbio

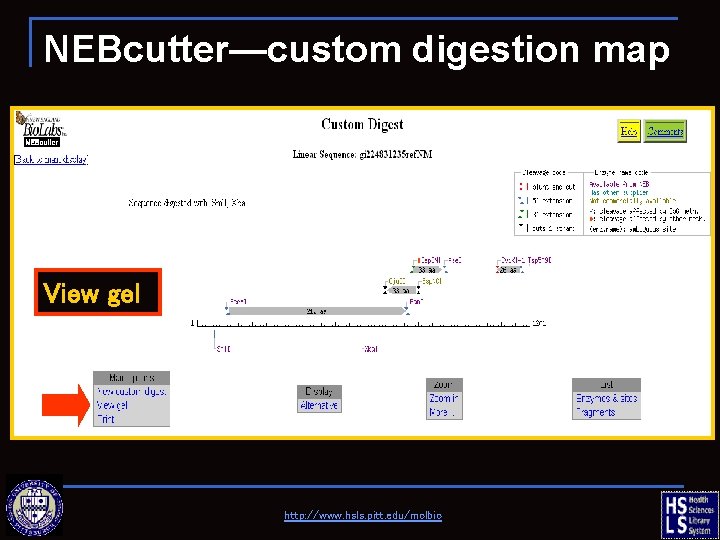
NEBcutter—custom digestion map View gel http: //www. hsls. pitt. edu/molbio

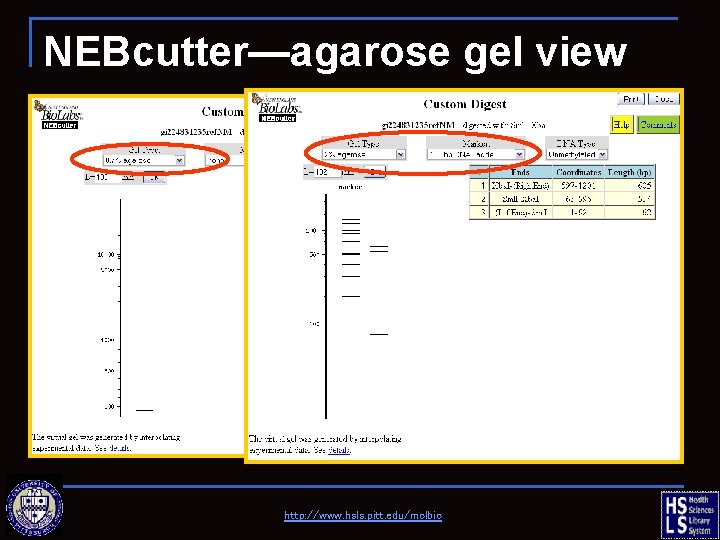
NEBcutter—agarose gel view http: //www. hsls. pitt. edu/molbio

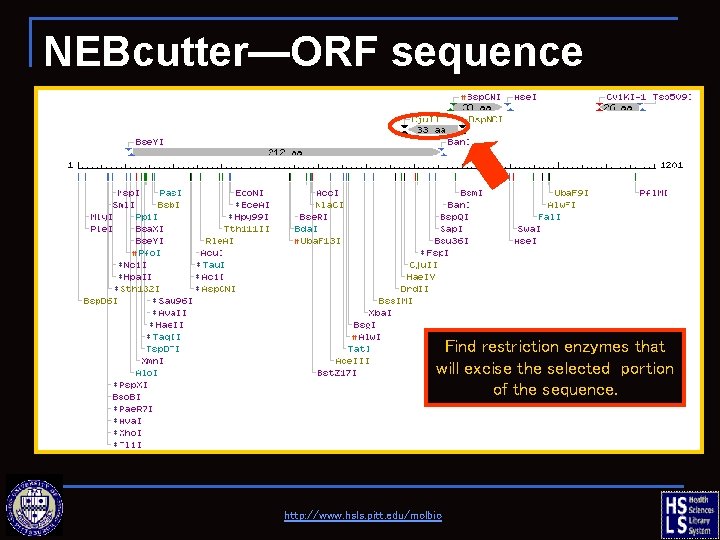
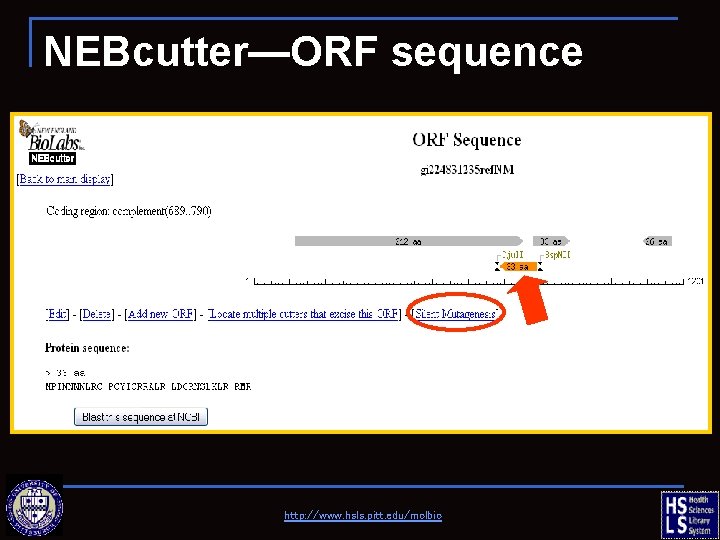
NEBcutter—ORF sequence Find restriction enzymes that will excise the selected portion of the sequence. http: //www. hsls. pitt. edu/molbio

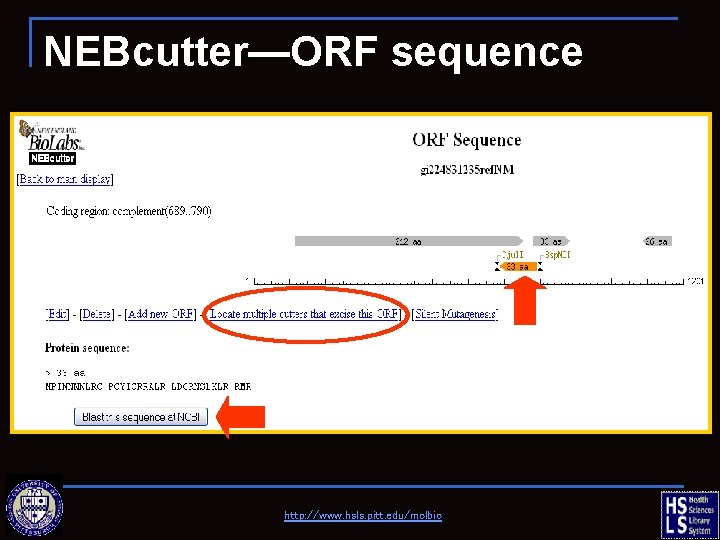
NEBcutter—ORF sequence http: //www. hsls. pitt. edu/molbio

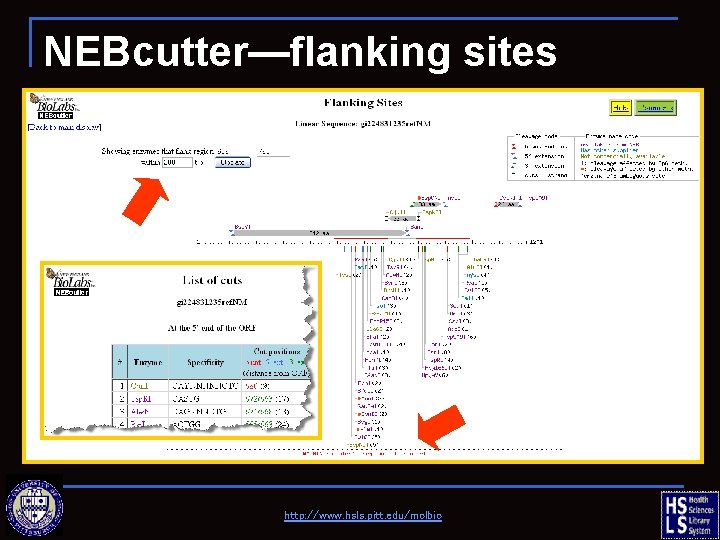
NEBcutter—flanking sites http: //www. hsls. pitt. edu/molbio

NEBcutter—ORF sequence http: //www. hsls. pitt. edu/molbio

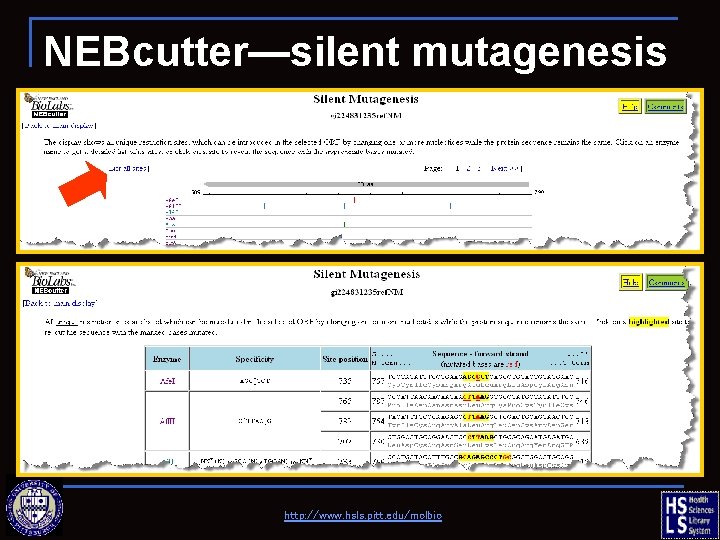
NEBcutter—silent mutagenesis http: //www. hsls. pitt. edu/molbio

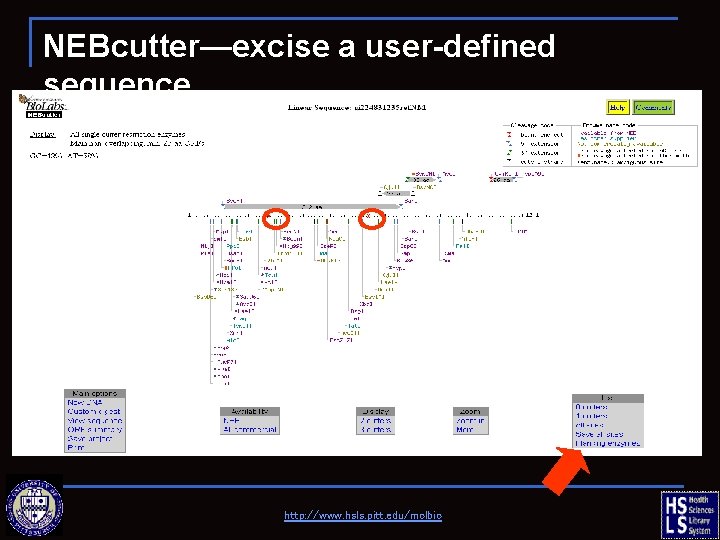
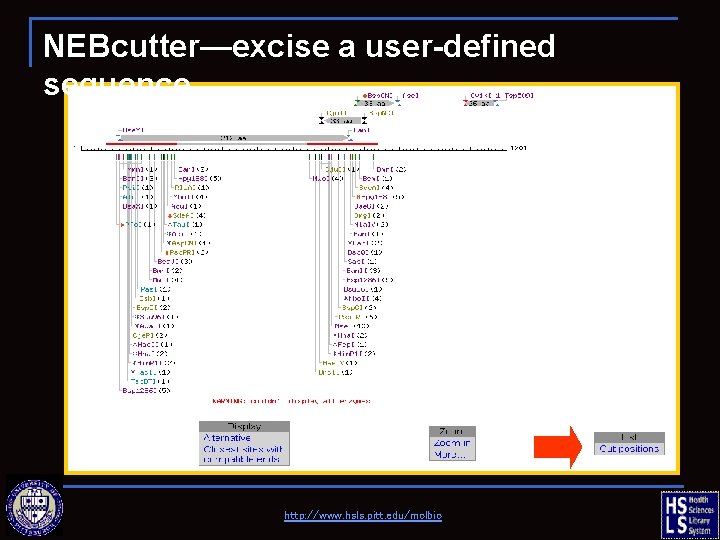
NEBcutter—excise a user-defined sequence http: //www. hsls. pitt. edu/molbio

NEBcutter—excise a user-defined sequence http: //www. hsls. pitt. edu/molbio

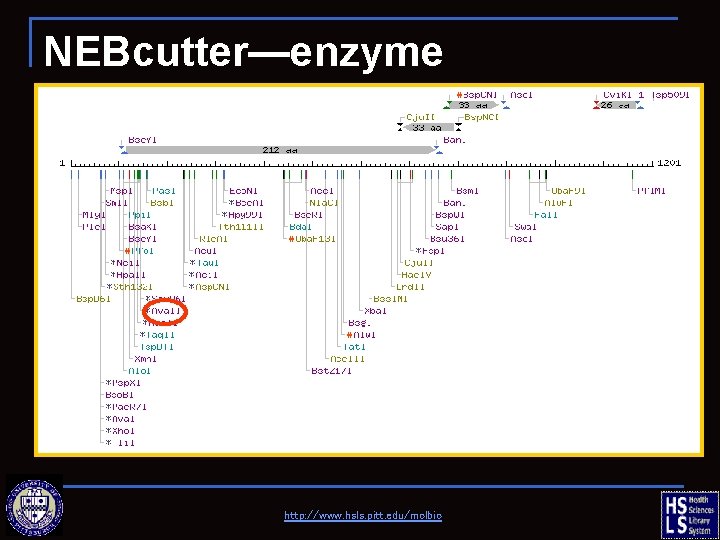
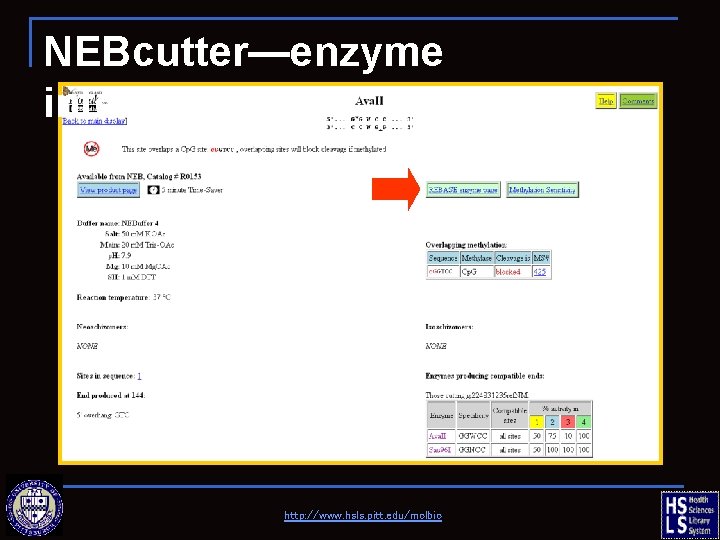
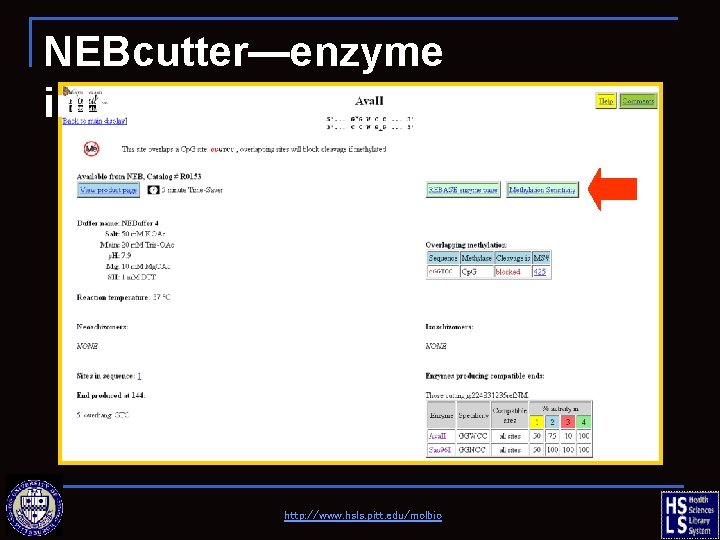
NEBcutter—enzyme information http: //www. hsls. pitt. edu/molbio

NEBcutter—enzyme information http: //www. hsls. pitt. edu/molbio

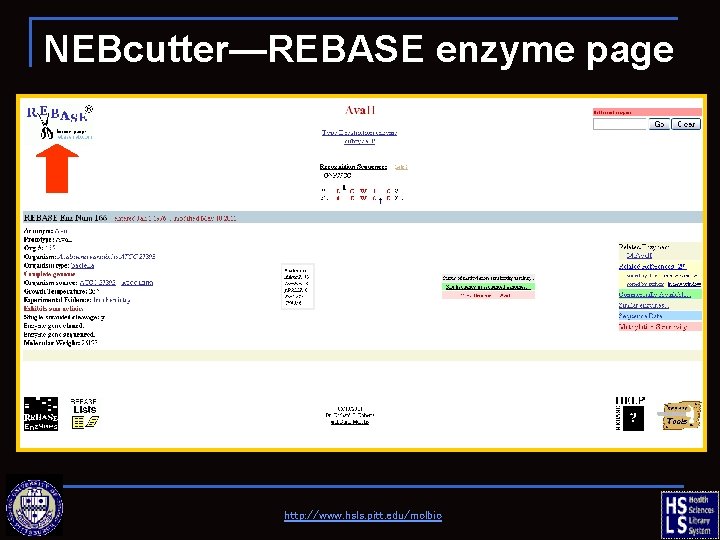
NEBcutter—REBASE enzyme page http: //www. hsls. pitt. edu/molbio

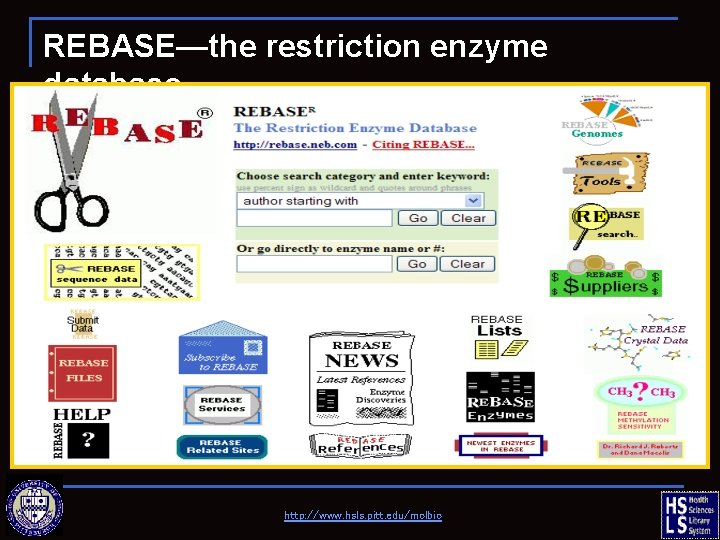
REBASE—the restriction enzyme database http: //www. hsls. pitt. edu/molbio

NEBcutter—enzyme information http: //www. hsls. pitt. edu/molbio

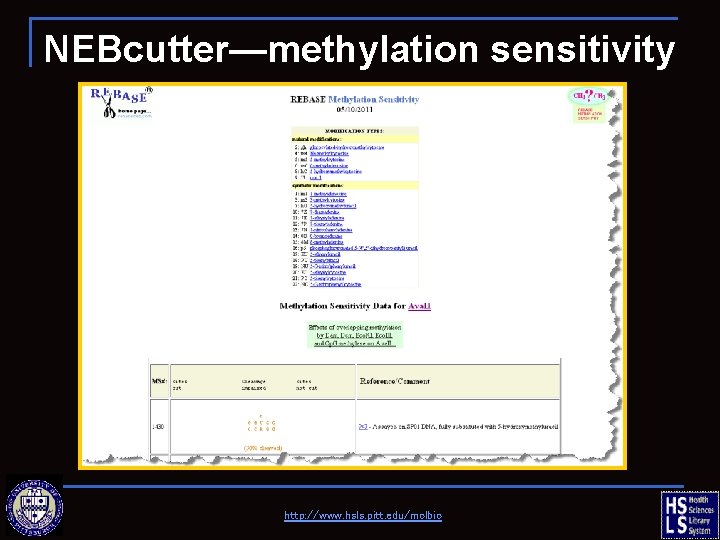
NEBcutter—methylation sensitivity http: //www. hsls. pitt. edu/molbio

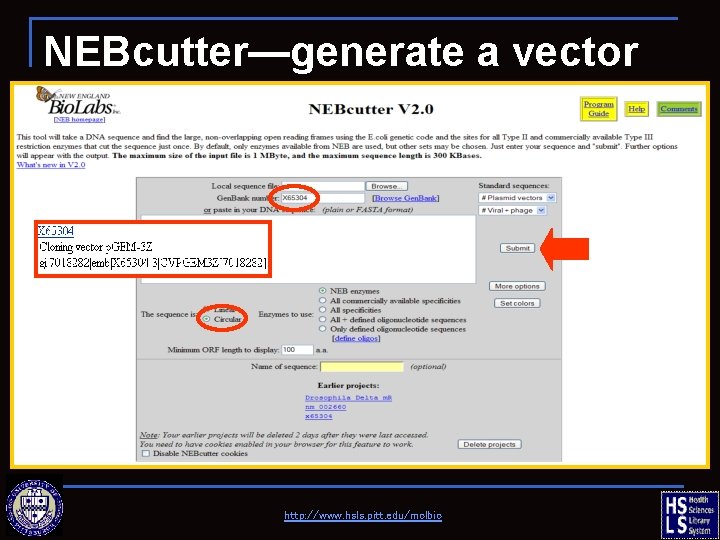
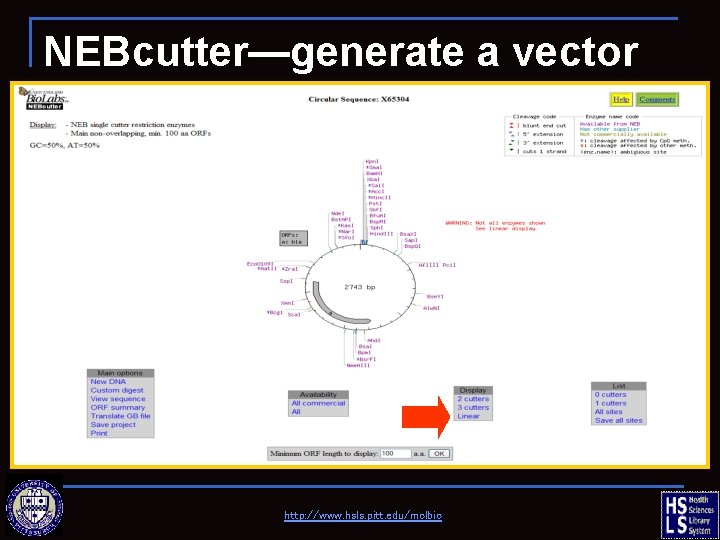
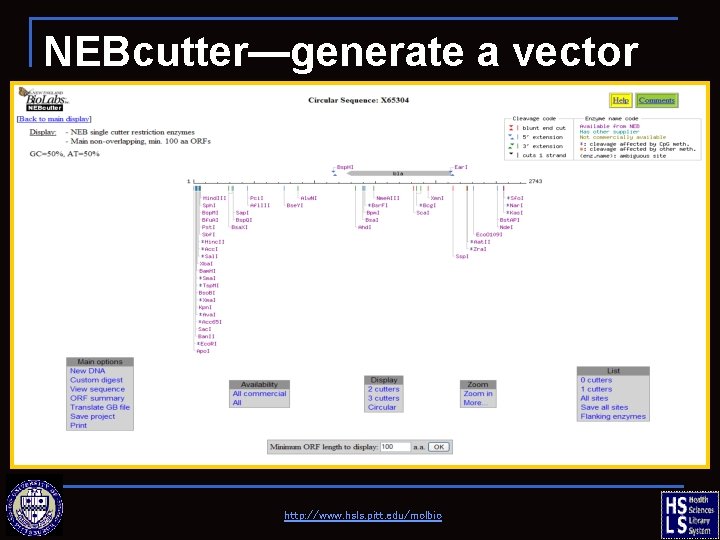
NEBcutter—generate a vector map http: //www. hsls. pitt. edu/molbio

NEBcutter—generate a vector map http: //www. hsls. pitt. edu/molbio

NEBcutter—generate a vector map http: //www. hsls. pitt. edu/molbio

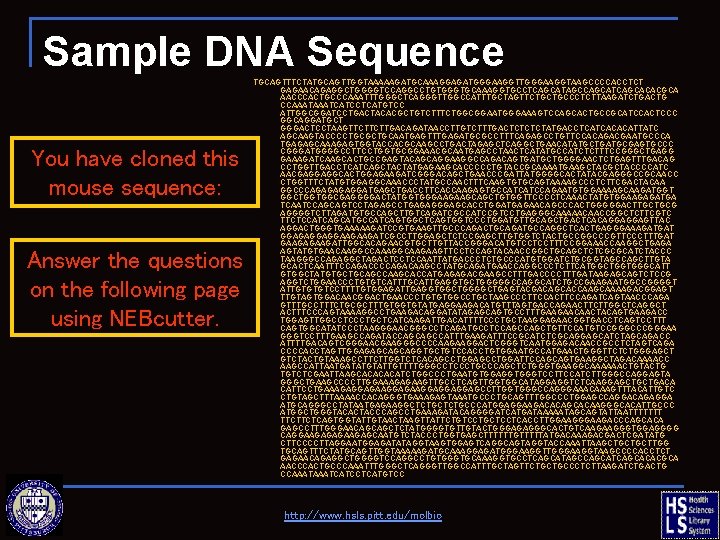
Sample DNA Sequence You have cloned this mouse sequence: Answer the questions on the following page using NEBcutter. TGCAGTTTCTATGCAGTTGGTAAAAAGATGCAAAGGAGATGGGAAGGTAAGCCCCACCTCT GAGAACAGAGGCTGGGGTCCAGGCCTGTGGGTGCAAAGGTGCCTCAGCATAGCCAGCATCAGCACACGCA AACCCACTGCCCAAATTTGGGCTCAGGGTTGGCCATTTGCTAGTTCTGCTGCCCTCTTAAGATCTGACTG CCAAATCATCCTCATGTCC ATTGGCGGATCCTGACTACACGCTGTCTTTCTGGCGGAATGGGAAAGTCCAGCACTGCCGCATCCACTCCC GGCAGGATGCT GGGACTCCTAAGTTCTTCTTGACAGATAACCTTGTCTTTGACTCTATGACCTCATCACACATTATC AGCAAGTACCCCTGCGCTGCAATGAGTTTGAGATGCGCCTTTCAGAGCCTGTTCCACAGACGAATGCCCA TGAGAGCAAAGAGTGGTACCACGCAAGCCTGACTAGAGCTCAGGCTGAACATATGCTGATGCGAGTGCCC CGGGATGGGGCCTTCCTGGTGCGGAAACGCAATGAGCCTAACTCATATGCCATCTCTTTCCGGGCTGAGG GAAAGATCAAGCACTGCCGAGTACAGCAGGAAGGCCAGACAGTGATGCTGGGGAACTCTGAGTTTGACAG CCTGGTTGACCTCATCAGCTACTATGAGAAGCACCCCCTGTACCGCAAAATGAAGCTACCCCATC AACGAGGAGGCACTGGAGAAGATCGGGACAGCTGAACCCGATTATGGGGCACTATACGAGGGCCGCAACC CTGGTTTCTATGTGGAGGCAAACCCTATGCCAACTTTCAAGTGTGCAGTAAAAGCCCTCTTCGACTACAA GGCCCAGAGGATGAGCTGACCTTCACCAAGAGTGCCATCATCCAGAATGTGGAAAAGCAAGATGGT GGCTGGTGGCGAGGGGACTATGGTGGGAAGAAGCAGCTGTGGTTCCCCTCAAACTATGTGGAAGAGATGA TCAATCCAGCAGTCCTAGAGCCTGAGAGGGAGCACCTGGATGAGAACAGCCCACTGGGGGACTTGCTGCG AGGGGTCTTAGATGTGCCAGCTTGTCAGATCGCCATCCGTCCTGAGGGCAAAAACAACCGGCTCTTCGTC TTCTCCATCAGCATGCCATCAGTGGCTCAGTGGTCCCTGGATGTTGCAGCTGACTCACAGGAGGAGTTAC AGGACTGGGTGAAAAAGATCCGTGAAGTTGCCCAGACTGCAGATGCCAGGCTCACTGAGGGAAAGATGAT GGAGAGGAGGAAGAAGATCGCCTTGGAGCTCTCCGAGCTTGTGGTCTACTGCCGGCCCGTTCCCTTTGAT GAAGATTGGCACAGAACGTGCTTGTTACCGGGACATGTCCTCCTTTCCGGAAACCAAGGCTGAGA AGTATGTGAACAAGGCCAAAGGCAAGAAGTTCCTCCAGTACAACCGGCTGCAGCTCTCGCGCATCTACCC TAAGGGCCAGAGGCTAGACTCCTCCAATTATGACCCTCTGCCCATGTGGATCTGCGGTAGCCAGCTTGTA GCACTCAATTTCCAGACCCCAGACAAGCCTATGCAGATGAACCAGGCCCTCTTCATGGCTGGTGGGCATT GTGGCTATGTGCTGCAGCCAAGCACCATGAGAGACGAAGCCTTTGACCCCTTTGATAAGAGCAGTCTCCG AGGTCTGGAACCCTGTGTCATTTGCATTGAGGTGCTGGGGGCCAGGCATCTGCCGAAGAATGGCCGGGGT ATTGTGTGTCCTTTTGTGGAGATTGAGGTGGCTGGGGCTGAGTACGACAGCACCAAGCAAAAGACGGAGT TTGTAGTGGACAACGGACTGAACCCTGTGTGGCCTGCTAAGCCCTTCCAGATCAGTAACCCAGA GTTTGCCTTTCTGCGCTTTGTGGTGTATGAGGAAGACATGTTTAGTGACCAGAACTTCTTGGCTCAGGCT ACTTTCCCAGTAAAAGGCCTGAAGACAGGATATAGAGCAGTGCCTTTGAAGAACAACTACAGTGAAGACC TGGAGTTGGCCTCCCTGCTCATCAAGATTGACATTTTCCCTGCTAAGGAGAACGGTGACCTCAGTCCTTT CAGTGGCATATCCCTAAGGGAACGGGCCTCAGATGCCTCCAGCTGTTCCATGTCCGGGCCCGGGAA GGGTCCTTTGAAGCCAGATACCAGCAGCCATTTGAAGATTTCCGCATCTCGCAGGAGCATCTAGCAGACC ATTTTGACAGTCGGGAACGAAGGGCCCCAAGAAGGACTCGGGTCAATGGAGACAACCGCCTCTAGTCAGA CCCCACCTAGTTGGAGAGCAGCAGGTGCTGTCCACCTGTGGAATGCCATGAACTGGGTTCTCTGGGAGCT GTCTACTGTAAAGCCTTCTTGGTCTCACAGCCTGGATTCCAGCAGTGAAGGCTAGACAAAACC AAGCCATTAATGATATGTATTGTTTTGGGCCTCCCTGCCCAGCTCTGGGTGAAGGCAAAAAACTGTACTG TGTCTCGAATTAAGCACATCTGGCCCTGAATGTGGAGGTGGGTCCTTCCATCTTGGGCCAGGAGTA GGGCTGAAGCCCCTTGGAAAGAGAAGTTGCCTCAGTTGGTGGCATAGGAGGTCTCAAGGAGCTGCTGACA CATTCCTGAAAGAGGAGAAGGAGGAGGAGCCTTGGTGGGCCAGGGAAACAAAGTTTACATTGTC CTGTAGCTTTAAAACCACAGGGTGAAAGAGTAAATGCCCTGCAGTTTGGCCCTGGAGCCAGGACAGAGGA ATGCAGGGCCTATAATGAGAAGGCTCTGCCCATGGAGGAAGACACAGCACAAGGGCACATTGCCC ATGGCTGGGTACACTACCCAGCCTGAAAGATACAGGGGATCATGATAAAAATAGCAGTATTAATTTTTTT TTCTTCTCAGTGGTATTGTAACTAAGTTATTCTGTCCTGCTCCTCACCTTGGAAGACCCAGCACA GAGCCTTTGGGAACAGCAGCTCTATGGGGTGTTGTACTGGGAGAGGGCACTGTCAAGAAGGGTGGAGGGG CAGGAAGAGAGAAGAGCAATGTCTACCCTGGTGAGCTTTTTTGTTTTTATGACAAAGACGACTCGATATG CTTCCCCTTAGGAATGGAGATATAGGTAAGTGGAGTCAGGCAGTAGGTACCAAATTAAGCTGCTGCTTGG TGCAGTTTCTATGCAGTTGGTAAAAAGATGCAAAGGAGATGGGAAGGTAAGCCCCACCTCT GAGAACAGAGGCTGGGGTCCAGGCCTGTGGGTGCAAAGGTGCCTCAGCATAGCCAGCATCAGCACACGCA AACCCACTGCCCAAATTTGGGCTCAGGGTTGGCCATTTGCTAGTTCTGCTGCCCTCTTAAGATCTGACTG CCAAATCATCCTCATGTCC http: //www. hsls. pitt. edu/molbio

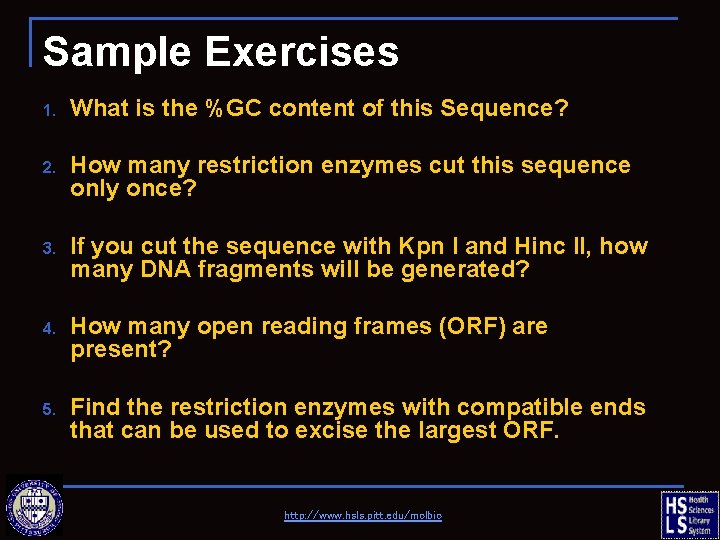
Sample Exercises 1. What is the %GC content of this Sequence? 2. How many restriction enzymes cut this sequence only once? 3. If you cut the sequence with Kpn I and Hinc II, how many DNA fragments will be generated? 4. How many open reading frames (ORF) are present? 5. Find the restriction enzymes with compatible ends that can be used to excise the largest ORF. http: //www. hsls. pitt. edu/molbio

Sample Exercises Hints (NEBcutter) 1. What is the %GC content of this Sequence? q 2. How many restriction enzymes cut this sequence only once? q 3. Select Custom digest, then View gel How many open reading frames (ORF) are present? q 5. Select for single cutters If you cut the sequence with Kpn I and Hinc II, how many DNA fragments will be generated? q 4. See top left of page (after entering sequence info) Select ORF summary Find the restriction enzymes with compatible ends that can be used to excise the largest ORF. q Select the ORF, then locate multiple cutters, cut positions http: //www. hsls. pitt. edu/molbio

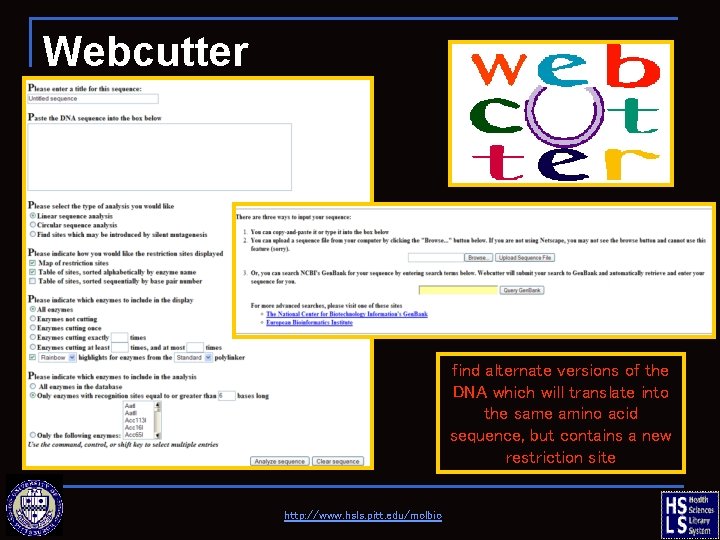
Webcutter 2. 0 n http: //bio. biomedicine. gu. se/cutter 2/ n Free Major features: n q q q Rainbow cutters Highlight your favorite enzymes in color or boldface for easy at-a-glance identification Silent cutters Find sites which may be introduced by silent mutagenesis of your coding sequence Sequence uploads Input sequences directly into Webcutter from a file on your hard drive without needing to cut-and-paste Degenerate sequences Analyze restriction maps of sequences containing ambiguous nucleotides like N, Y, and R. Circular sequences Choose whether to treat your sequence as linear or circular Enzyme info Click into the wealth of references and ordering information at New England Bio. Labs' REBASE, directly from your restriction map results http: //www. hsls. pitt. edu/molbio

Webcutter find alternate versions of the DNA which will translate into the same amino acid sequence, but contains a new restriction site http: //www. hsls. pitt. edu/molbio

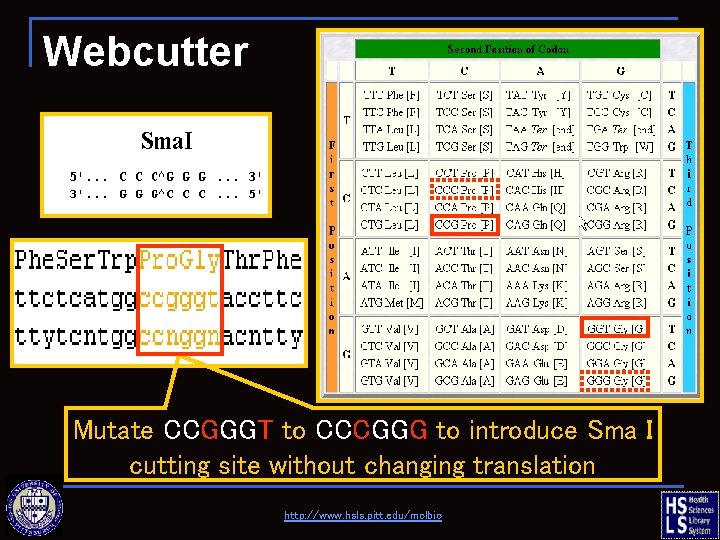
Webcutter Mutate CCGGGT to CCCGGG to introduce Sma I cutting site without changing translation http: //www. hsls. pitt. edu/molbio

Webcutter—silent mutagenesis click here to retrieve sample sequence, then copy/paste into box http: //www. hsls. pitt. edu/molbio

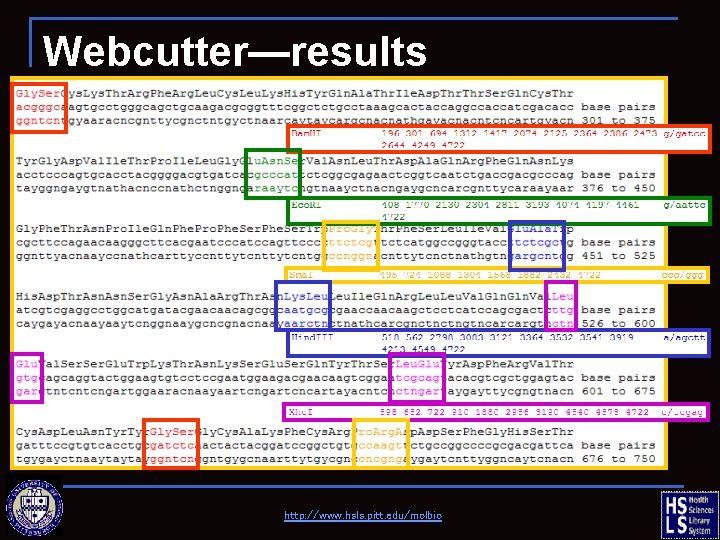
Webcutter—results http: //www. hsls. pitt. edu/molbio

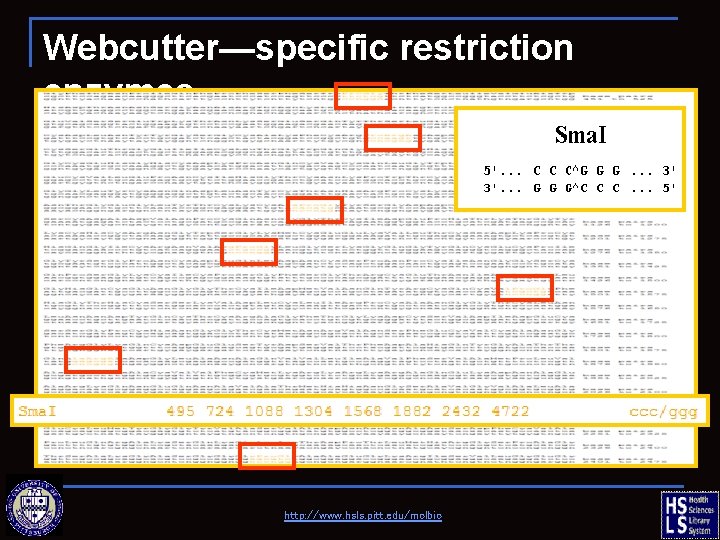
Webcutter—specific restriction enzymes http: //www. hsls. pitt. edu/molbio

Thank you! Any questions? Carrie Iwema iwema@pitt. edu 412 -383 -6887 Ansuman Chattopadhyay ansuman@pitt. edu 412 -648 -1297 http: //www. hsls. pitt. edu/molbio

Sequence Manipulation www. vam. ac. uk/images/image/44010 -large. jpg http: //www. hsls. pitt. edu/molbio

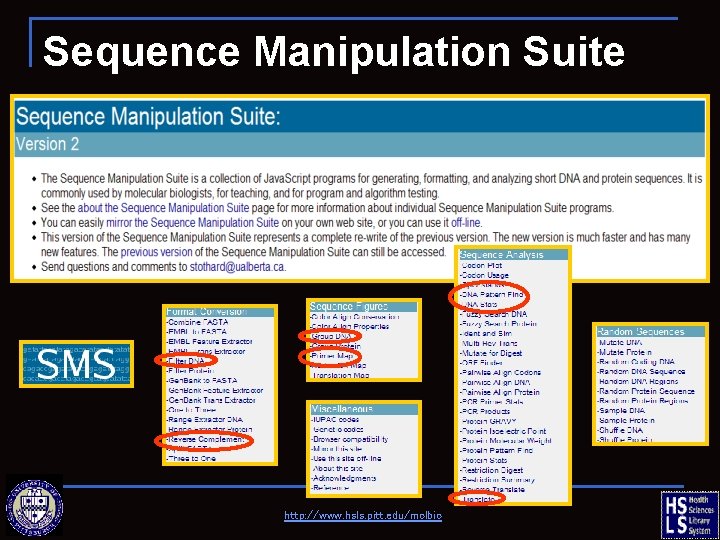
Sequence Manipulation Tools n READSEQ q n http: //www-bimas. cit. nih. gov/molbio/readseq/ Sequence Manipulation Suite q http: //www. bioinformatics. org/sms 2/ http: //www. hsls. pitt. edu/molbio

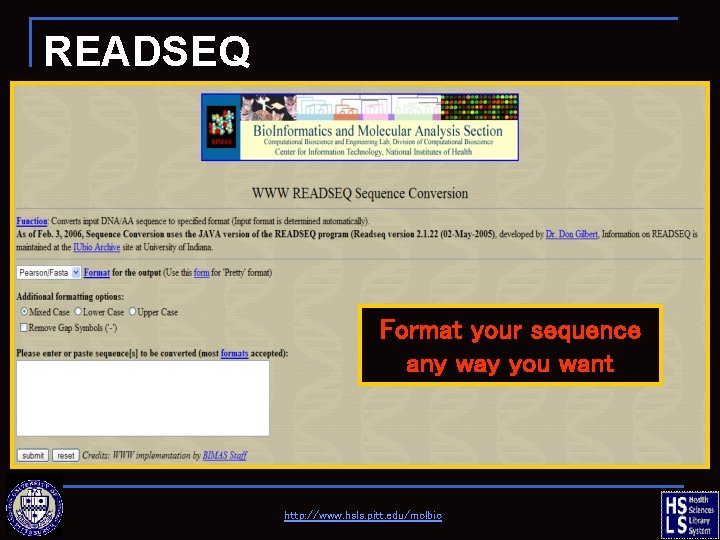
READSEQ Format your sequence any way you want http: //www. hsls. pitt. edu/molbio

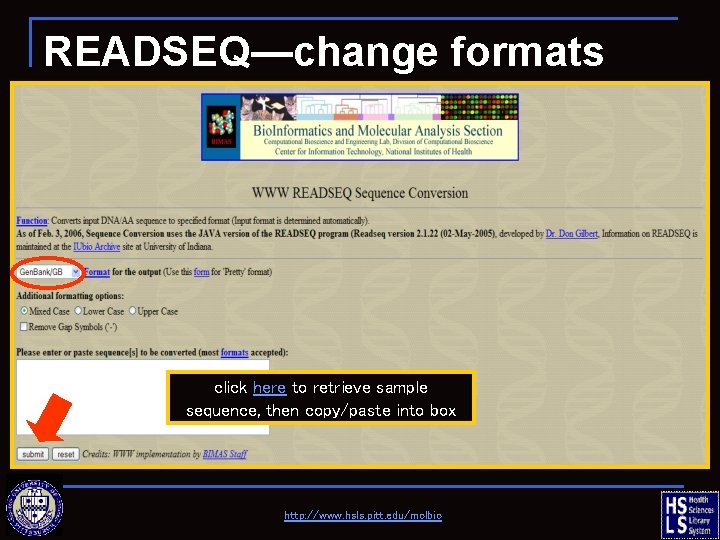
READSEQ—change formats click here to retrieve sample sequence, then copy/paste into box http: //www. hsls. pitt. edu/molbio

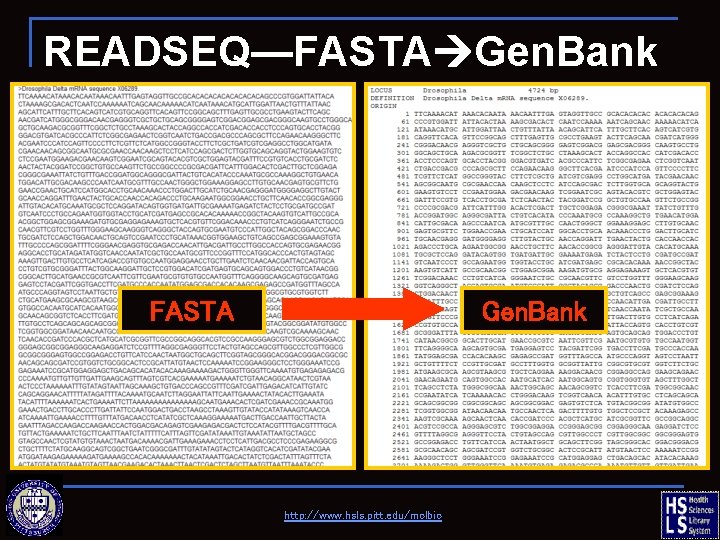
READSEQ—FASTA Gen. Bank http: //www. hsls. pitt. edu/molbio

Sequence Manipulation Suite http: //www. hsls. pitt. edu/molbio

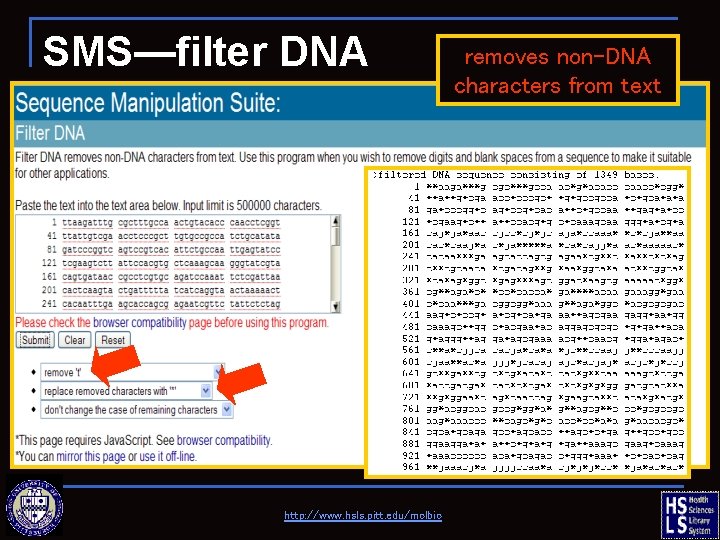
SMS—filter DNA http: //www. hsls. pitt. edu/molbio removes non-DNA characters from text

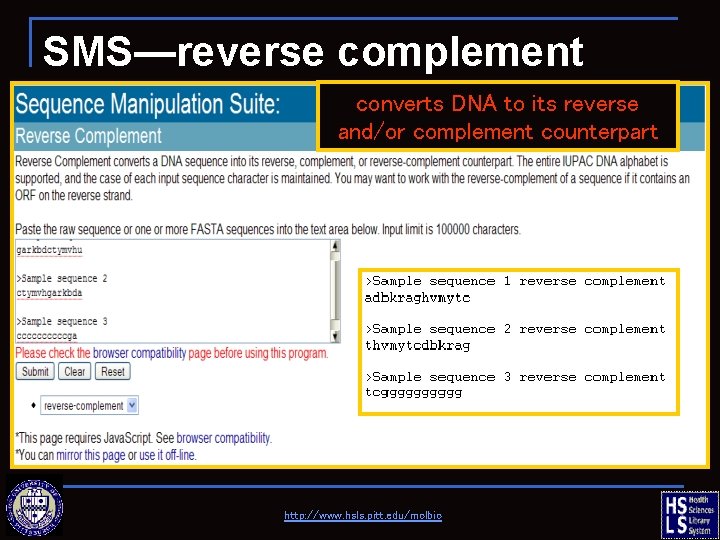
SMS—reverse complement converts DNA to its reverse and/or complement counterpart http: //www. hsls. pitt. edu/molbio

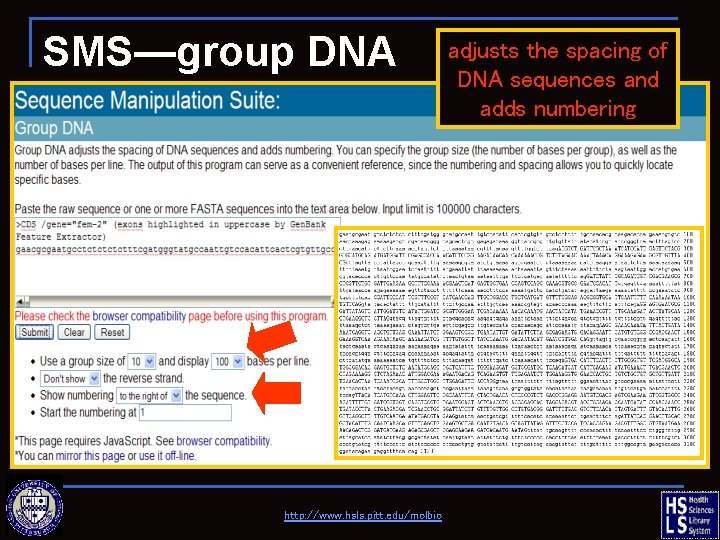
SMS—group DNA http: //www. hsls. pitt. edu/molbio adjusts the spacing of DNA sequences and adds numbering

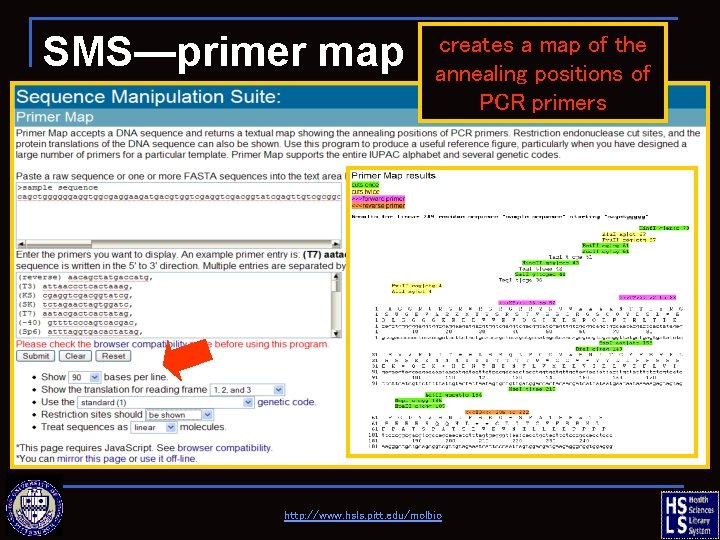
SMS—primer map creates a map of the annealing positions of PCR primers http: //www. hsls. pitt. edu/molbio

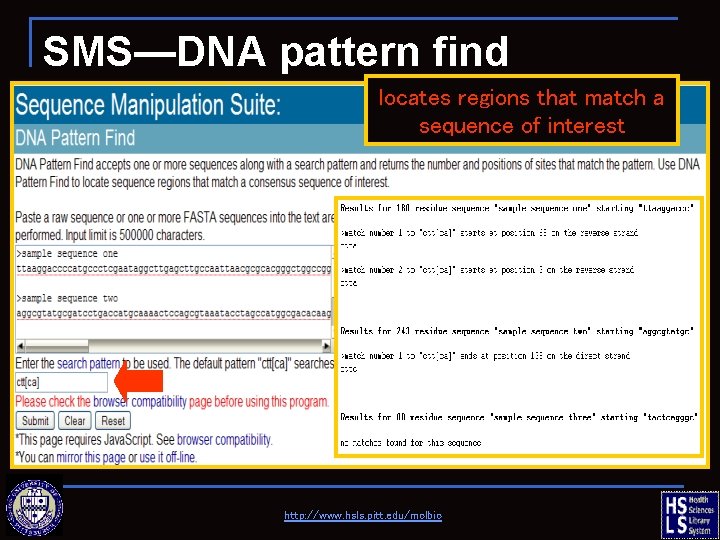
SMS—DNA pattern find locates regions that match a sequence of interest http: //www. hsls. pitt. edu/molbio

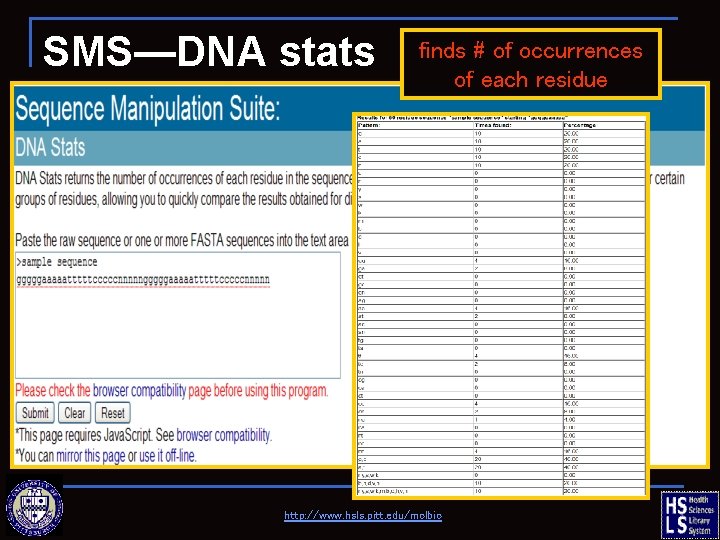
SMS—DNA stats finds # of occurrences of each residue http: //www. hsls. pitt. edu/molbio

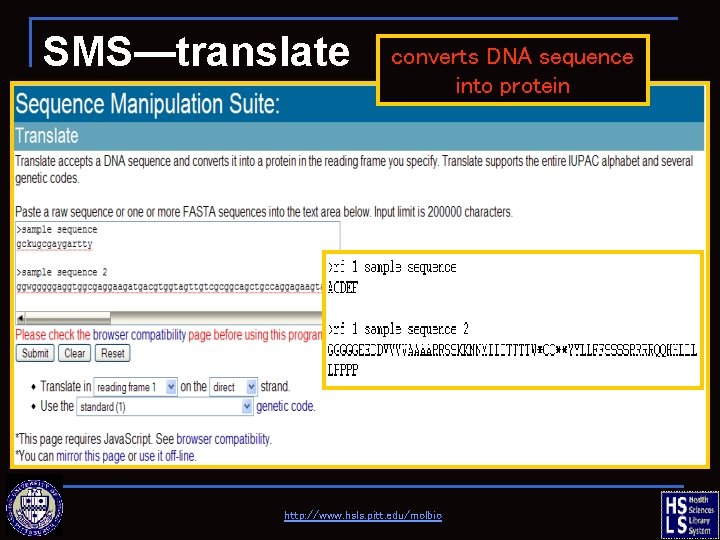
SMS—translate converts DNA sequence into protein http: //www. hsls. pitt. edu/molbio