Restriction mapping Sitespecific restriction endonucleases are used to



- Slides: 13

Restriction mapping Site-specific restriction endonucleases are used to identify DNA molecules

What are restriction endonucleases (REs)? How can REs be used to identify DNA molecules? How can I find RE recognition sites in the MET plasmids?

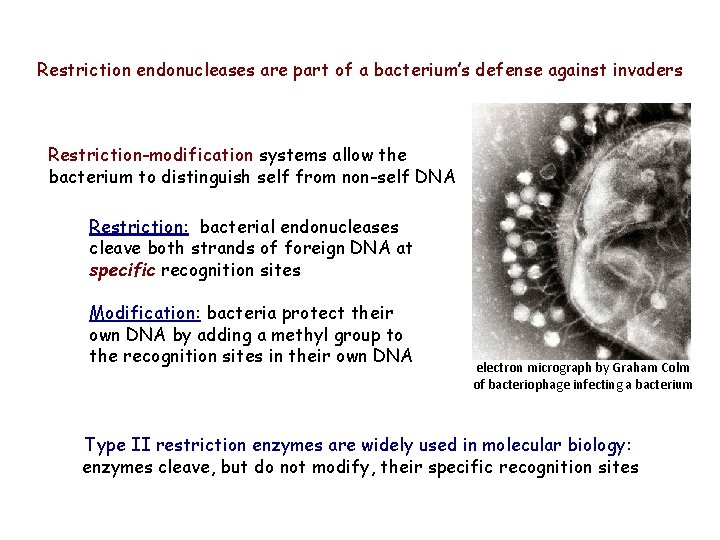
Restriction endonucleases are part of a bacterium’s defense against invaders Restriction-modification systems allow the bacterium to distinguish self from non-self DNA Restriction: bacterial endonucleases cleave both strands of foreign DNA at specific recognition sites Modification: bacteria protect their own DNA by adding a methyl group to the recognition sites in their own DNA electron micrograph by Graham Colm of bacteriophage infecting a bacterium Type II restriction enzymes are widely used in molecular biology: enzymes cleave, but do not modify, their specific recognition sites

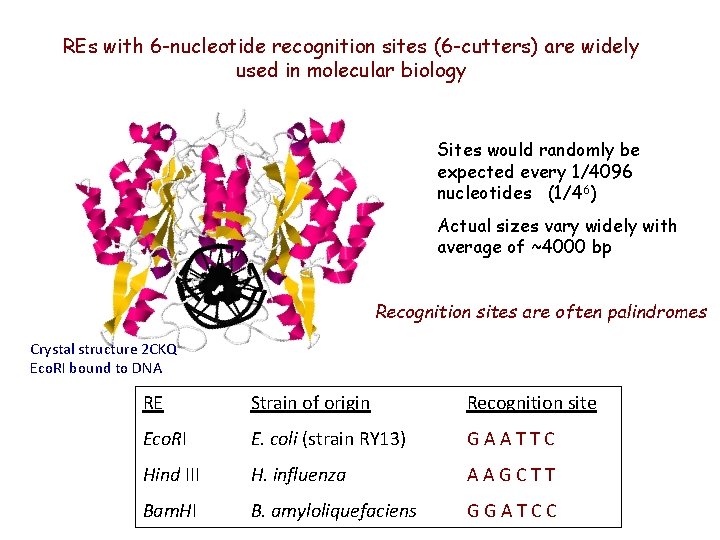
REs with 6 -nucleotide recognition sites (6 -cutters) are widely used in molecular biology Sites would randomly be expected every 1/4096 nucleotides (1/46) Actual sizes vary widely with average of ~4000 bp Recognition sites are often palindromes Crystal structure 2 CKQ Eco. RI bound to DNA RE Strain of origin Recognition site Eco. RI E. coli (strain RY 13) GAATTC Hind III H. influenza AAGCTT Bam. HI B. amyloliquefaciens GGATCC

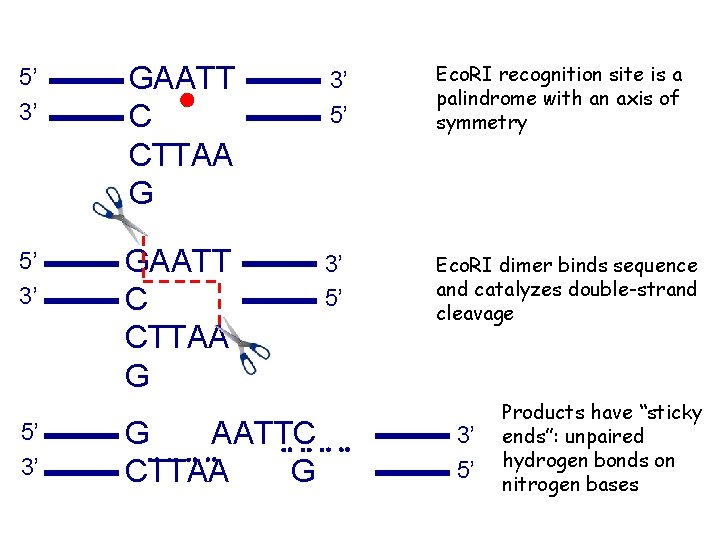
5’ 3’ GAATT C CTTAA G 3’ 5’ Eco. RI recognition site is a palindrome with an axis of symmetry 5’ 3’ GAATT C CTTAA G 3’ 5’ Eco. RI dimer binds sequence and catalyzes double-strand cleavage 5’ 3’ G AATTC CTTAA G 3’ 5’ Products have “sticky ends”: unpaired hydrogen bonds on nitrogen bases

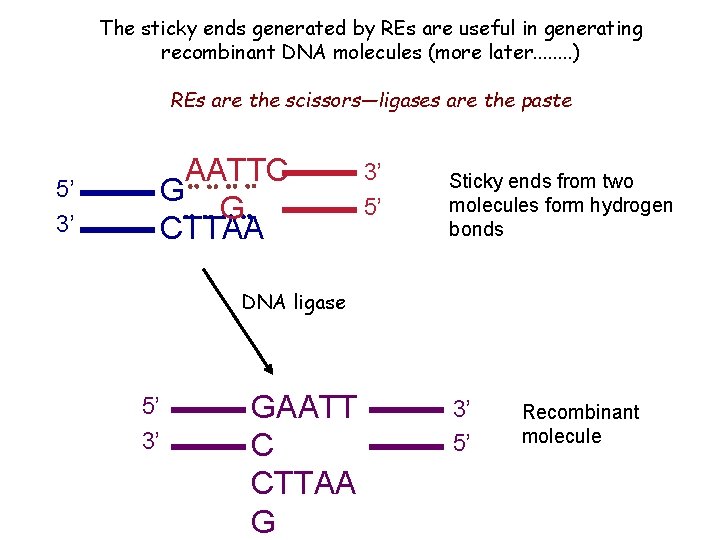
The sticky ends generated by REs are useful in generating recombinant DNA molecules (more later. . . . ) REs are the scissors—ligases are the paste 5’ 3’ AATTC G G CTTAA 3’ 5’ Sticky ends from two molecules form hydrogen bonds DNA ligase 5’ 3’ GAATT C CTTAA G 3’ 5’ Recombinant molecule

What are restriction endonucleases (REs)? How can REs be used to identify DNA molecules? How can I find RE recognition sites in the MET plasmids?

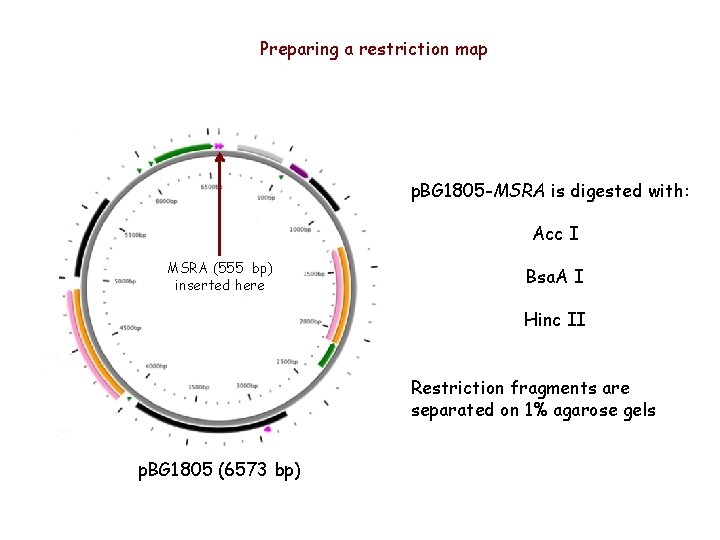
Preparing a restriction map p. BG 1805 -MSRA is digested with: Acc I MSRA (555 bp) inserted here Bsa. A I Hinc II Restriction fragments are separated on 1% agarose gels p. BG 1805 (6573 bp)

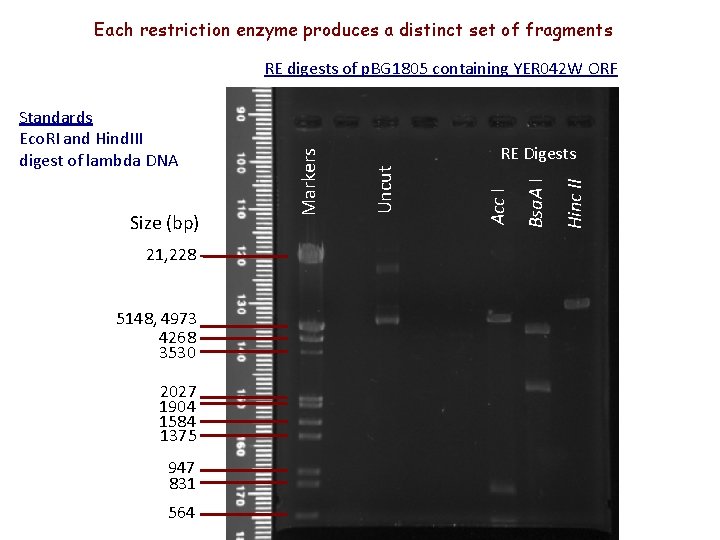
Each restriction enzyme produces a distinct set of fragments 21, 228 5148, 4973 4268 3530 2027 1904 1584 1375 947 831 564 Hinc II Bsa. A I Acc I Size (bp) RE Digests Uncut Standards Eco. RI and Hind. III digest of lambda DNA Markers RE digests of p. BG 1805 containing YER 042 W ORF

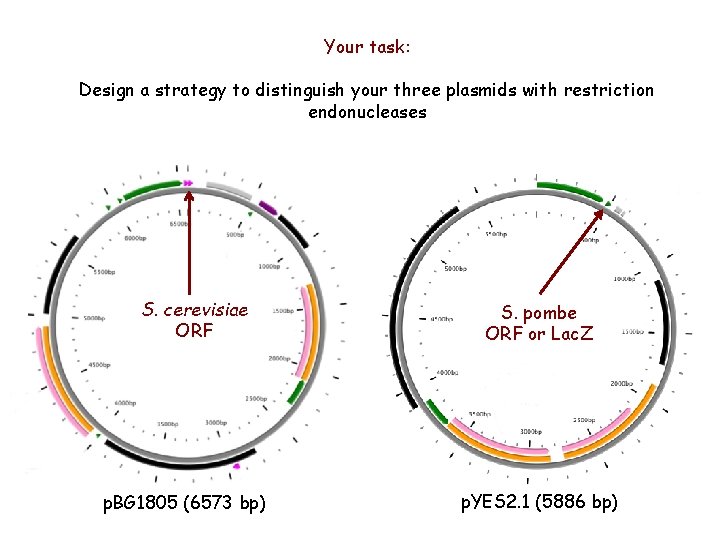
Your task: Design a strategy to distinguish your three plasmids with restriction endonucleases S. cerevisiae ORF p. BG 1805 (6573 bp) S. pombe ORF or Lac. Z p. YES 2. 1 (5886 bp)

What are restriction endonucleases (REs)? How can REs be used to identify DNA molecules? How can I find RE recognition sites in the MET plasmids?

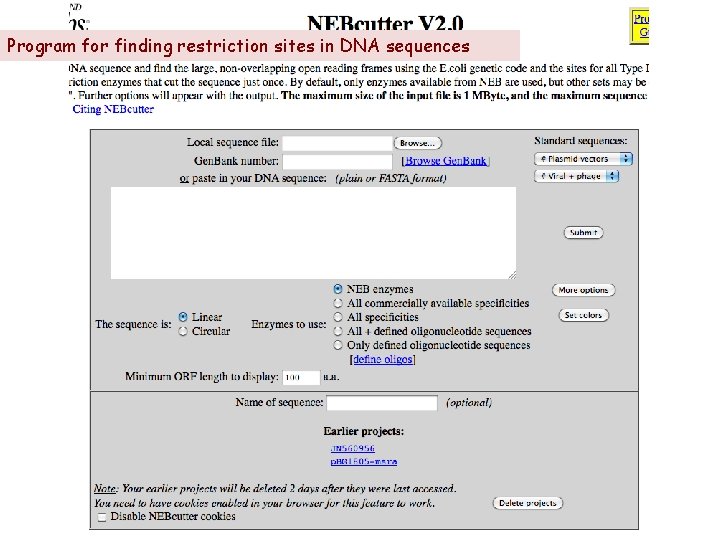
Program for finding restriction sites in DNA sequences

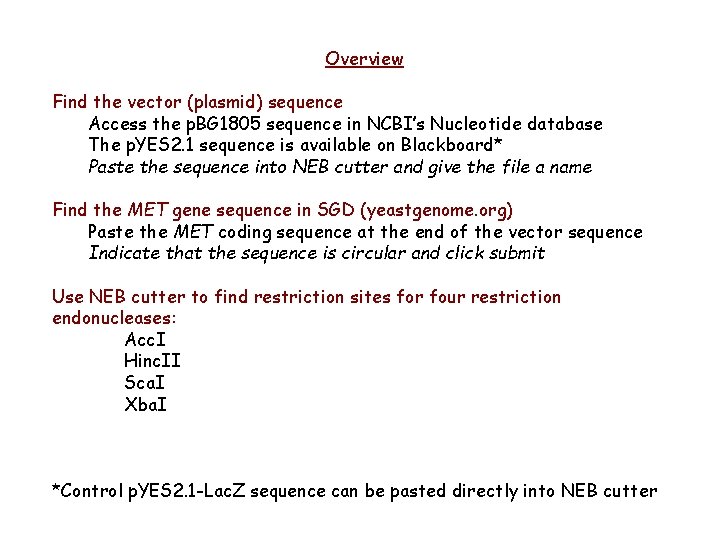
Overview Find the vector (plasmid) sequence Access the p. BG 1805 sequence in NCBI’s Nucleotide database The p. YES 2. 1 sequence is available on Blackboard* Paste the sequence into NEB cutter and give the file a name Find the MET gene sequence in SGD (yeastgenome. org) Paste the MET coding sequence at the end of the vector sequence Indicate that the sequence is circular and click submit Use NEB cutter to find restriction sites for four restriction endonucleases: Acc. I Hinc. II Sca. I Xba. I *Control p. YES 2. 1 -Lac. Z sequence can be pasted directly into NEB cutter