Molecular Scissors Restriction enzymes are molecular scissors Dr





- Slides: 22

Molecular Scissors Restriction enzymes are molecular scissors Dr. A. Avila Jerley, Assistant Professor, PG & Research Department of Zoology, Holy Cross College, Trichy.

What are restriction enzymes? q Restriction enzyme are enzymes that cut double stranded DNA molecules at specific points. q Found naturally in a wide variety of prokaryotes q An important tool for manipulating DNA.

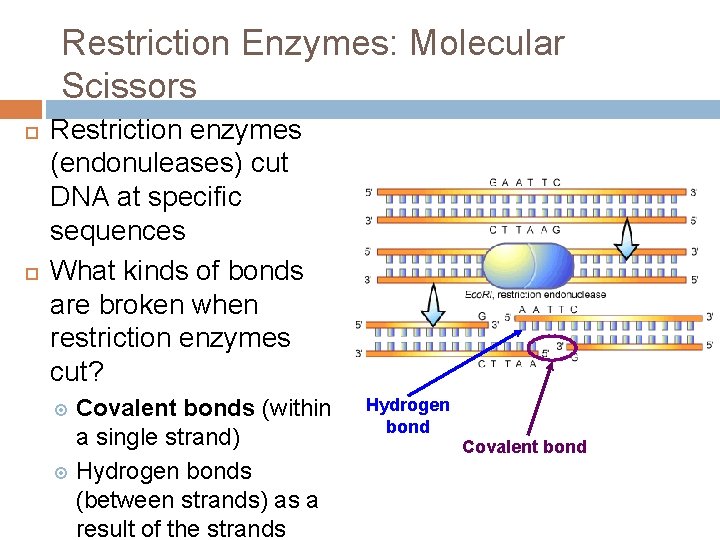
Restriction Enzymes: Molecular Scissors Restriction enzymes (endonuleases) cut DNA at specific sequences What kinds of bonds are broken when restriction enzymes cut? Covalent bonds (within a single strand) Hydrogen bonds (between strands) as a result of the strands Hydrogen bond Covalent bond

Discovery Arbor and Dussoix in 1962 discovered that certain bacteria contain Endonucleases which have the ability to cleave DNA. In 1970 Smith and colleagues purified and characterized the cleavage site of a Restriction Enzyme. Werner Arbor, Hamilton Smith and Daniel Nathans shared the 1978 Nobel prize for Medicine and Physiology for their discovery of Restriction Enzymes.

Biological Role Most bacteria use Restriction Enzymes as a defence against bacteriophages. Restriction enzymes prevent the replication of the phage by cleaving its DNA at specific sites. The host DNA is protected by Methylases which add methyl groups to adenine or cytosine bases within the recognition site thereby modifying the site and protecting the DNA.

Restriction Enzymes scan the DNA code Find a very specific set of nucleotides Make a specific cut Restriction enzymes recognize and make a cut within specific palindromic sequences, known as restriction sites, in the genetic code. This is usually a 4 - or 6 base pair sequence.

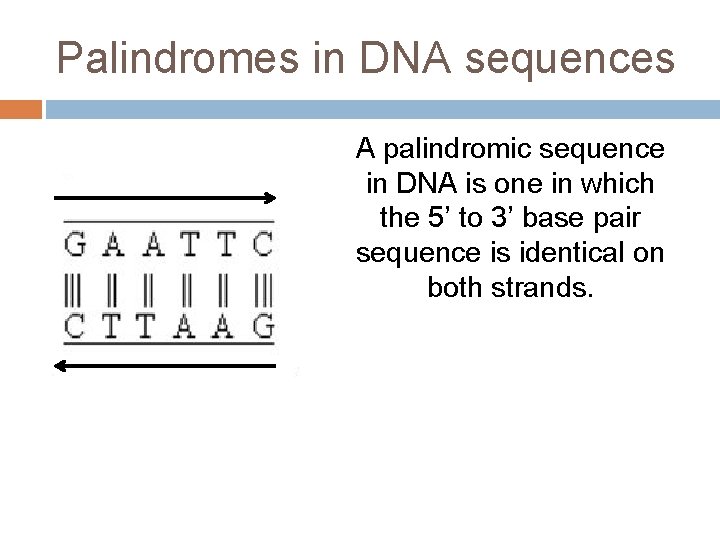
Palindromes in DNA sequences A palindromic sequence in DNA is one in which the 5’ to 3’ base pair sequence is identical on both strands.

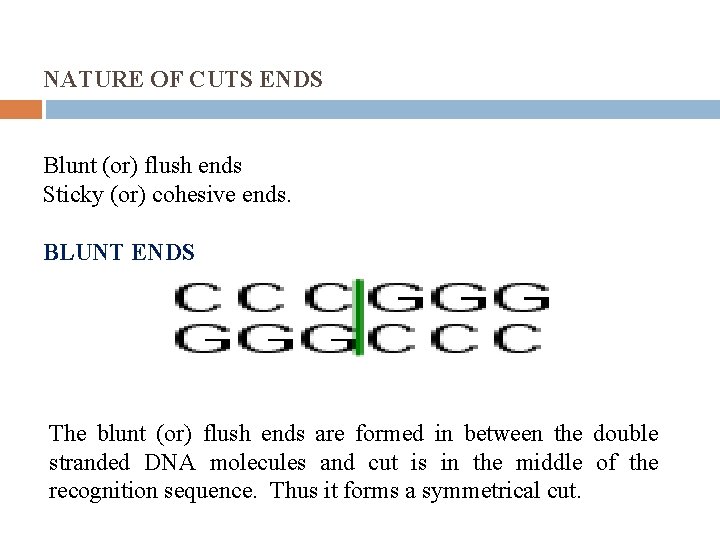
NATURE OF CUTS ENDS Blunt (or) flush ends Sticky (or) cohesive ends. BLUNT ENDS The blunt (or) flush ends are formed in between the double stranded DNA molecules and cut is in the middle of the recognition sequence. Thus it forms a symmetrical cut.

STICKY (OR) COHENSIVE ENDS Several restriction enzymes cut one strand at left side of the axis of symmetry & other strand at the right side axis of the sites. Then they break hydrogen bonds between base pairs lying between the two cut sites. DNA fragment with single strand extension are formed and its called cohesive ends (or) sticky ends.

“sticky ends” are useful DNA fragments with complimentary sticky ends can be combined to create new molecules which allows the creation and manipulation of DNA sequences from different sources.

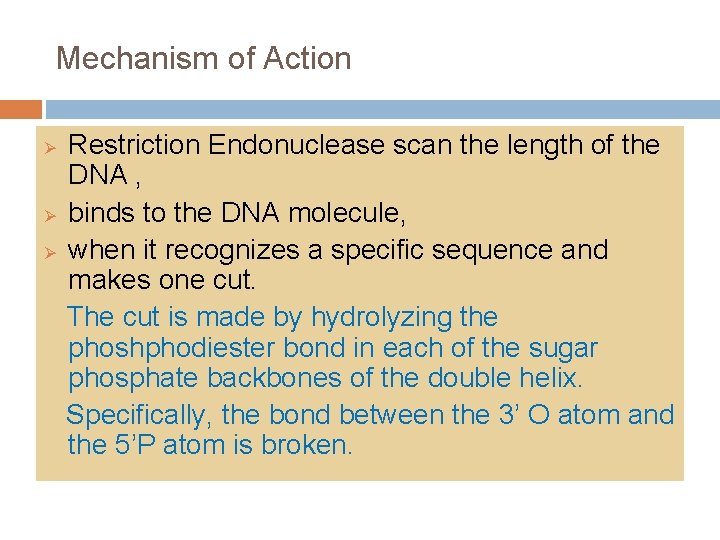
Mechanism of Action Ø Ø Ø Restriction Endonuclease scan the length of the DNA , binds to the DNA molecule, when it recognizes a specific sequence and makes one cut. The cut is made by hydrolyzing the phoshphodiester bond in each of the sugar phosphate backbones of the double helix. Specifically, the bond between the 3’ O atom and the 5’P atom is broken.

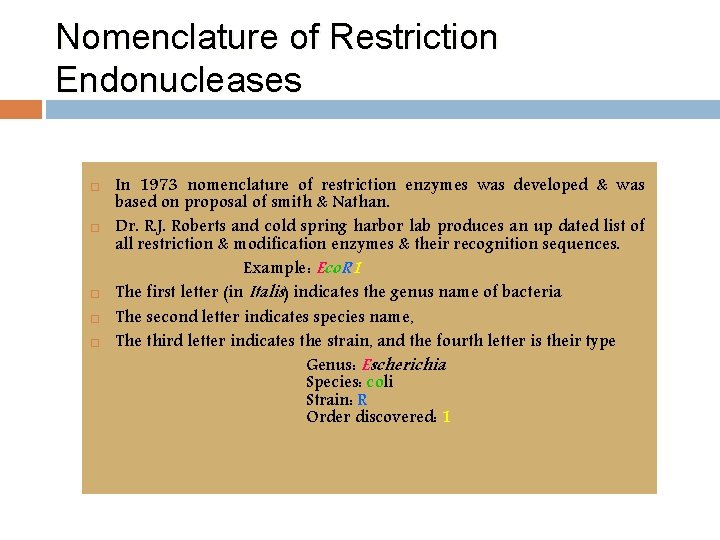
Nomenclature of Restriction Endonucleases In 1973 nomenclature of restriction enzymes was developed & was based on proposal of smith & Nathan. Dr. R. J. Roberts and cold spring harbor lab produces an up dated list of all restriction & modification enzymes & their recognition sequences. Example: Eco. R 1 The first letter (in Italis) indicates the genus name of bacteria The second letter indicates species name, The third letter indicates the strain, and the fourth letter is their type Genus: Escherichia Species: coli Strain: R Order discovered: 1

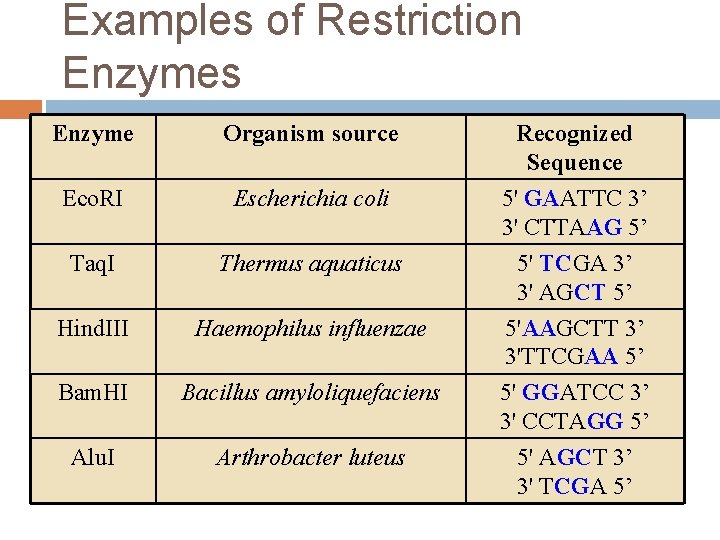
Examples of Restriction Enzymes Enzyme Organism source Eco. RI Escherichia coli Taq. I Thermus aquaticus Hind. III Haemophilus influenzae Bam. HI Bacillus amyloliquefaciens Alu. I Arthrobacter luteus Recognized Sequence 5' GAATTC 3’ 3' CTTAAG 5’ 5' TCGA 3’ 3' AGCT 5’ 5'AAGCTT 3’ 3'TTCGAA 5’ 5' GGATCC 3’ 3' CCTAGG 5’ 5' AGCT 3’ 3' TCGA 5’

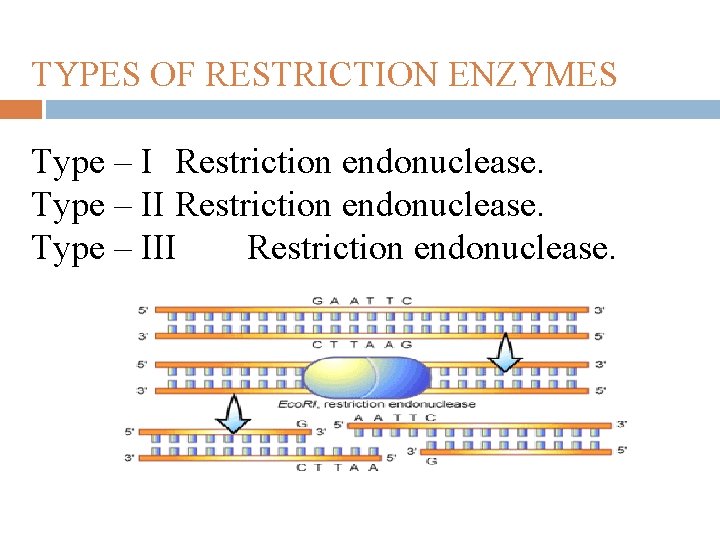
TYPES OF RESTRICTION ENZYMES Type – I Restriction endonuclease. Type – III Restriction endonuclease.

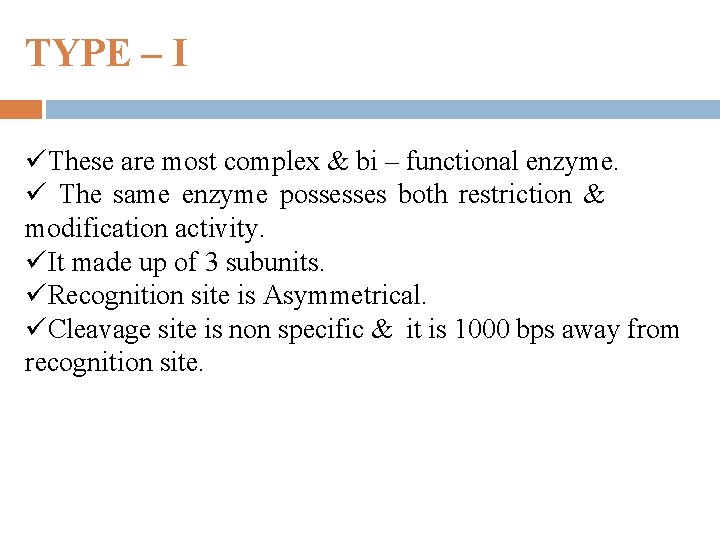
TYPE – I üThese are most complex & bi – functional enzyme. ü The same enzyme possesses both restriction & modification activity. üIt made up of 3 subunits. üRecognition site is Asymmetrical. üCleavage site is non specific & it is 1000 bps away from recognition site.

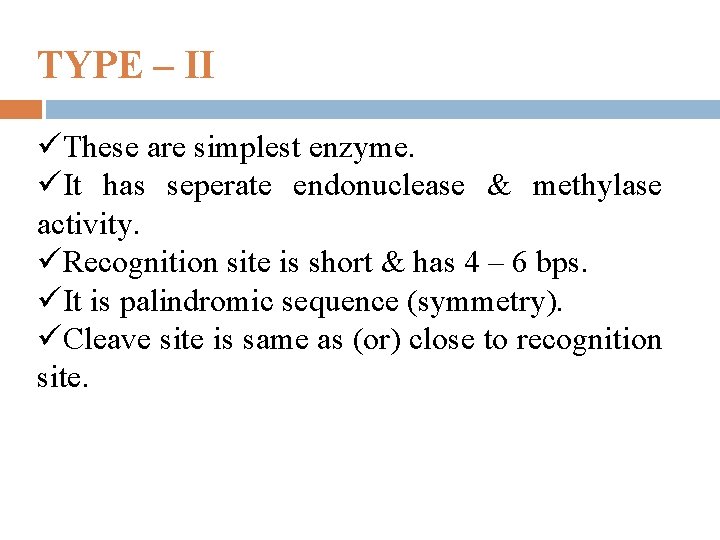
TYPE – II üThese are simplest enzyme. üIt has seperate endonuclease & methylase activity. üRecognition site is short & has 4 – 6 bps. üIt is palindromic sequence (symmetry). üCleave site is same as (or) close to recognition site.

TYPE – III üThese are moderate , complex & bi – functional. ü It has two subunits. ü Recognition site is asymmetrical sequence and has 5 – 7 bps. üCleavage site is 24 -26 bps down stream from recognition site.


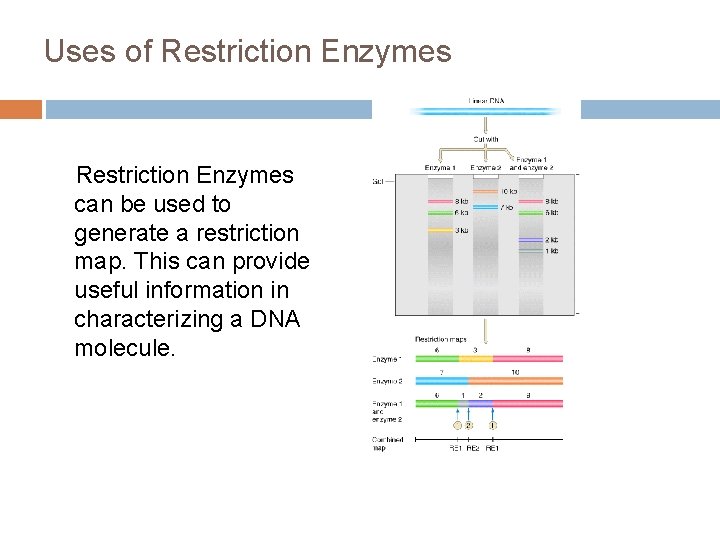
Uses of Restriction Enzymes can be used to generate a restriction map. This can provide useful information in characterizing a DNA molecule.

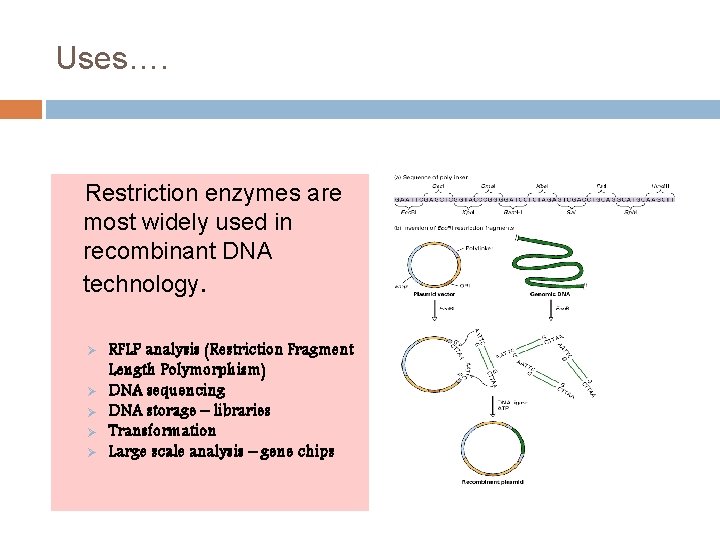
Uses…. Restriction enzymes are most widely used in recombinant DNA technology. Ø Ø Ø RFLP analysis (Restriction Fragment Length Polymorphism) DNA sequencing DNA storage – libraries Transformation Large scale analysis – gene chips

References Biochemistry (1995), Wiley & Sons, Inc. Voet D. and Voet J. G. § Genes to clone- T. A Brown § An Introduction to Genetic Analysis (2000), Freeman & Co. Griffiths, A. , Miller, J. H. , Suzuki, D. T. , Lewontin, R. C. , Gelbart, W. M. § Molecular Cell Biology (2000), Freeman & Co. Lodish, Berk, Zipursky, Matsudaria, Baltimore, Darnell §

THANK YOU