Modelling genome structure and function Ram Samudrala University
- Slides: 31

Modelling genome structure and function Ram Samudrala University of Washington

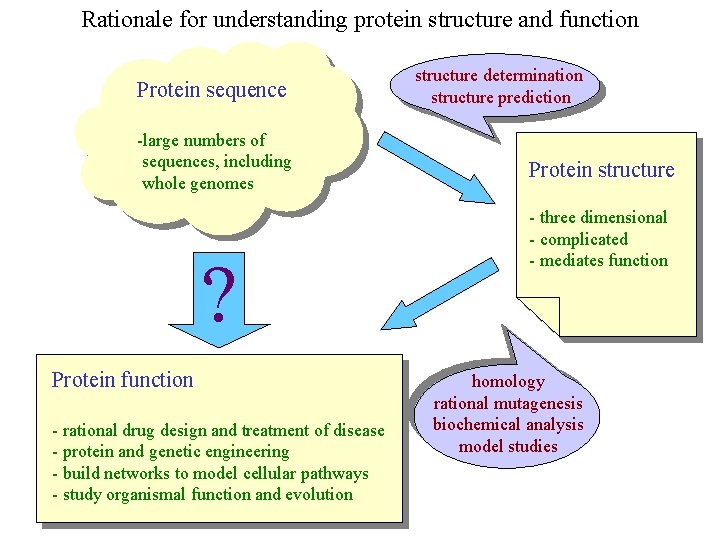
Rationale for understanding protein structure and function Protein sequence -large numbers of sequences, including whole genomes ? Protein function - rational drug design and treatment of disease - protein and genetic engineering - build networks to model cellular pathways - study organismal function and evolution structure determination structure prediction Protein structure - three dimensional - complicated - mediates function homology rational mutagenesis biochemical analysis model studies

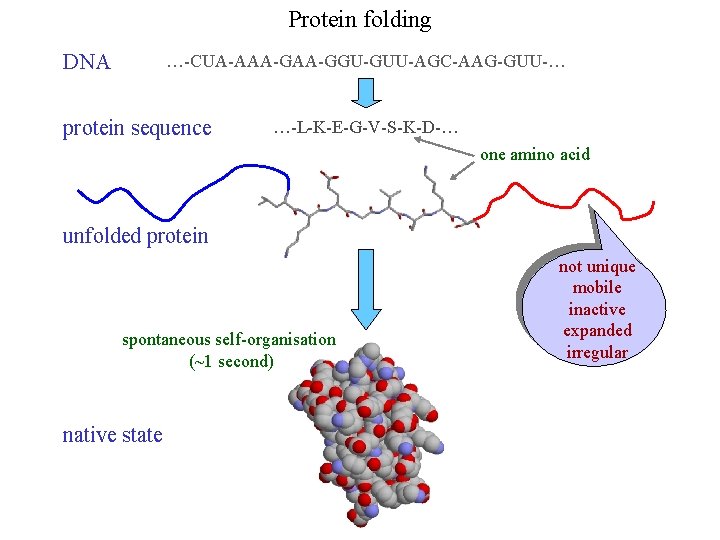
Protein folding DNA …-CUA-AAA-GGU-GUU-AGC-AAG-GUU-… protein sequence …-L-K-E-G-V-S-K-D-… one amino acid unfolded protein spontaneous self-organisation (~1 second) native state not unique mobile inactive expanded irregular

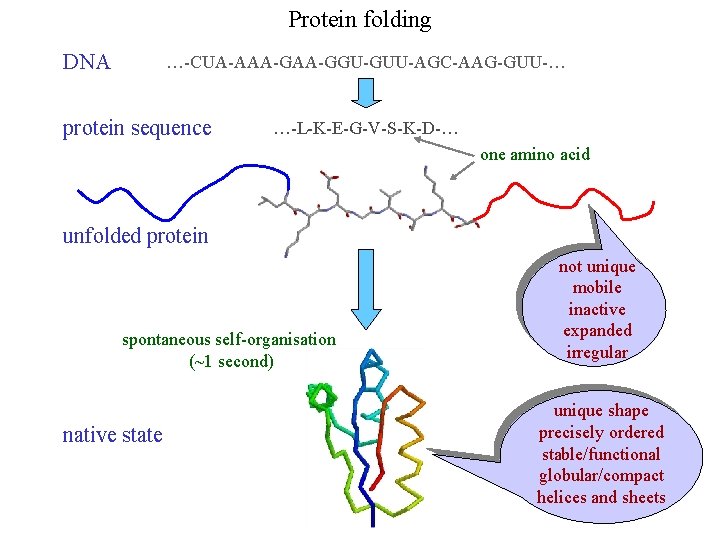
Protein folding DNA …-CUA-AAA-GGU-GUU-AGC-AAG-GUU-… protein sequence …-L-K-E-G-V-S-K-D-… one amino acid unfolded protein spontaneous self-organisation (~1 second) native state not unique mobile inactive expanded irregular unique shape precisely ordered stable/functional globular/compact helices and sheets

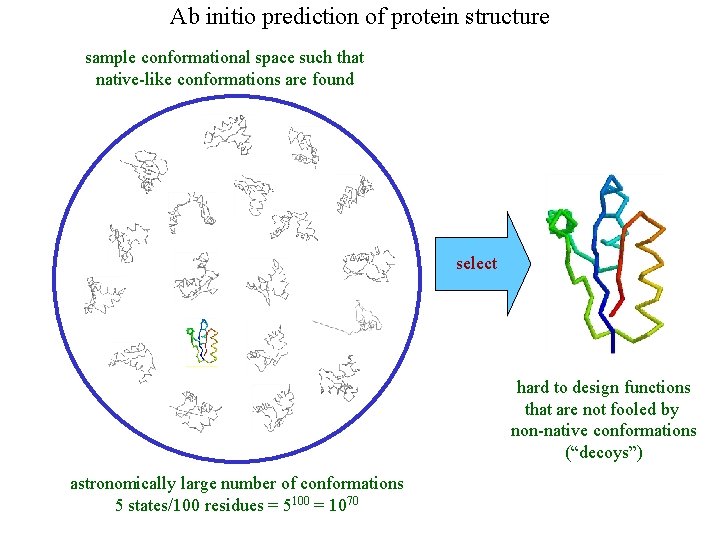
Ab initio prediction of protein structure sample conformational space such that native-like conformations are found select hard to design functions that are not fooled by non-native conformations (“decoys”) astronomically large number of conformations 5 states/100 residues = 5100 = 1070

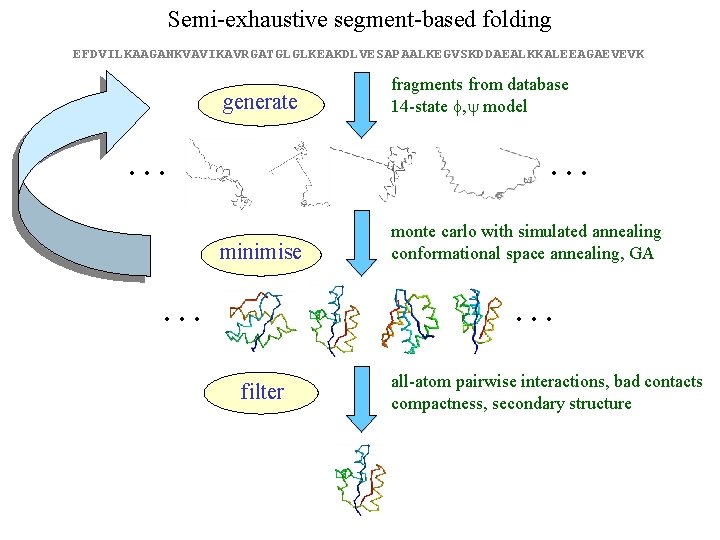
Semi-exhaustive segment-based folding EFDVILKAAGANKVAVIKAVRGATGLGLKEAKDLVESAPAALKEGVSKDDAEALKKALEEAGAEVEVK generate … fragments from database 14 -state f, y model … minimise … monte carlo with simulated annealing conformational space annealing, GA … filter all-atom pairwise interactions, bad contacts compactness, secondary structure

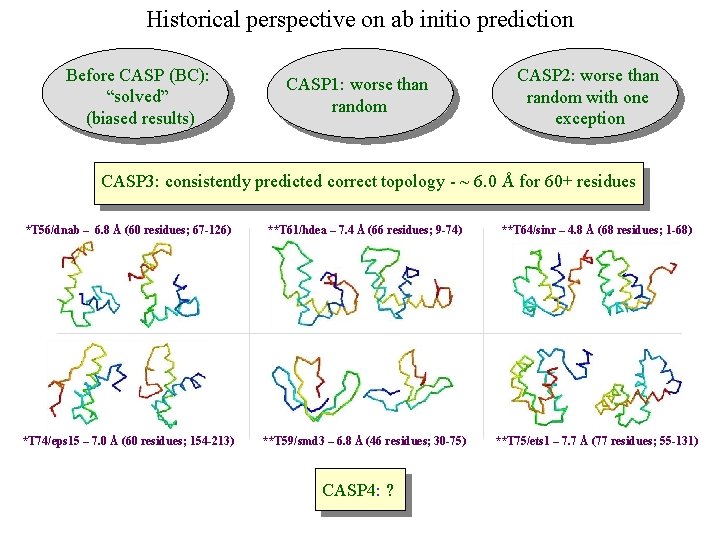
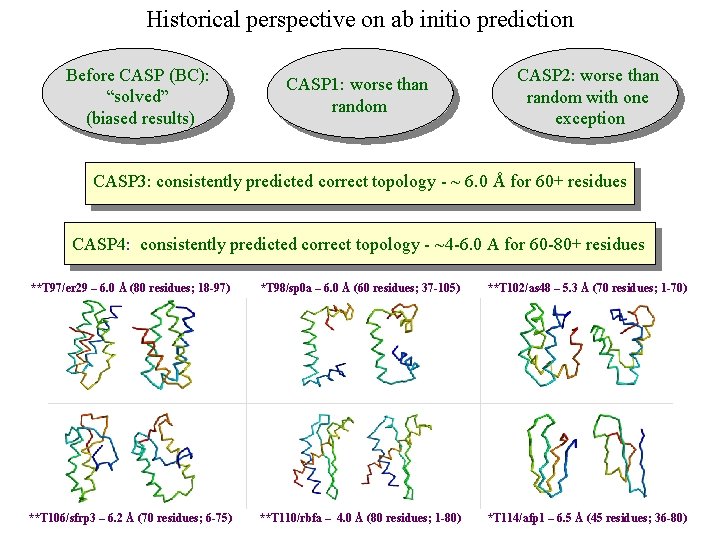
Historical perspective on ab initio prediction Before CASP (BC): “solved” (biased results) CASP 1: worse than random CASP 2: worse than random with one exception CASP 3: consistently predicted correct topology - ~ 6. 0 Å for 60+ residues *T 56/dnab – 6. 8 Å (60 residues; 67 -126) **T 61/hdea – 7. 4 Å (66 residues; 9 -74) **T 64/sinr – 4. 8 Å (68 residues; 1 -68) *T 74/eps 15 – 7. 0 Å (60 residues; 154 -213) **T 59/smd 3 – 6. 8 Å (46 residues; 30 -75) **T 75/ets 1 – 7. 7 Å (77 residues; 55 -131) CASP 4: ?

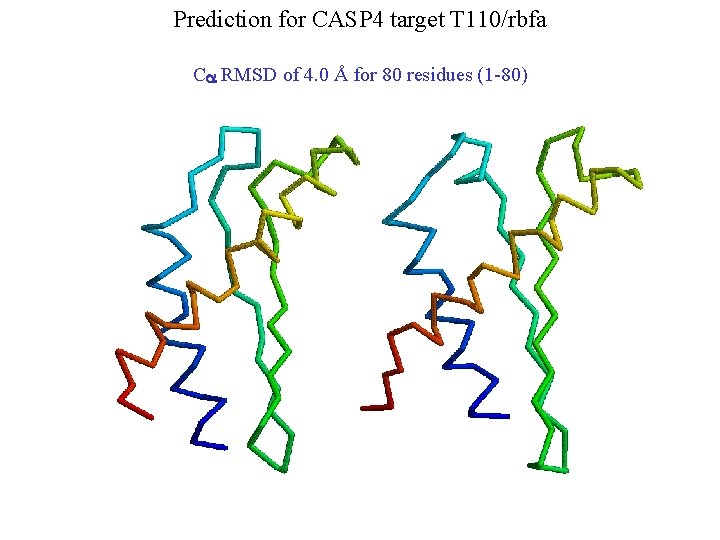
Prediction for CASP 4 target T 110/rbfa Ca RMSD of 4. 0 Å for 80 residues (1 -80)

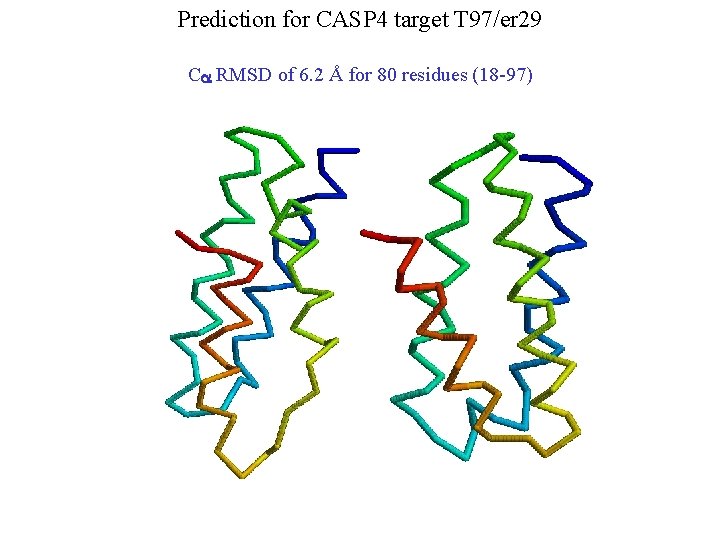
Prediction for CASP 4 target T 97/er 29 Ca RMSD of 6. 2 Å for 80 residues (18 -97)

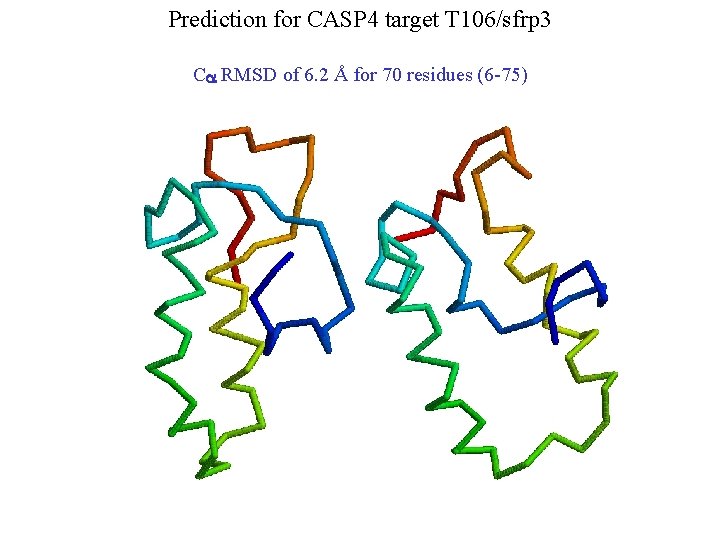
Prediction for CASP 4 target T 106/sfrp 3 Ca RMSD of 6. 2 Å for 70 residues (6 -75)

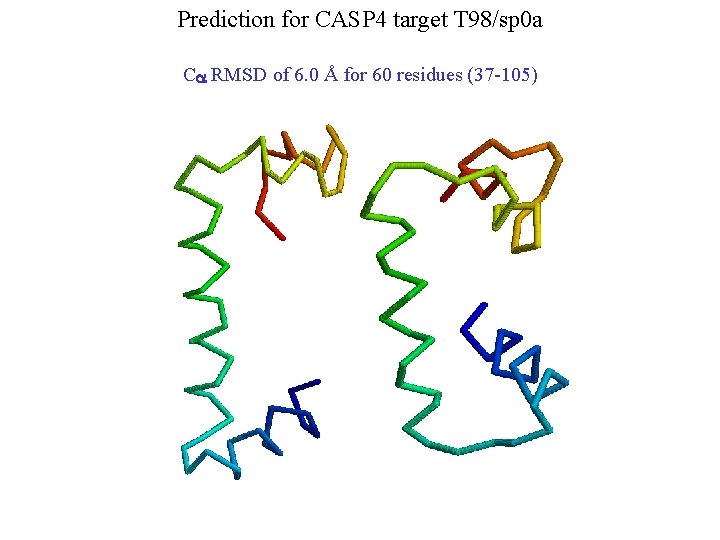
Prediction for CASP 4 target T 98/sp 0 a Ca RMSD of 6. 0 Å for 60 residues (37 -105)

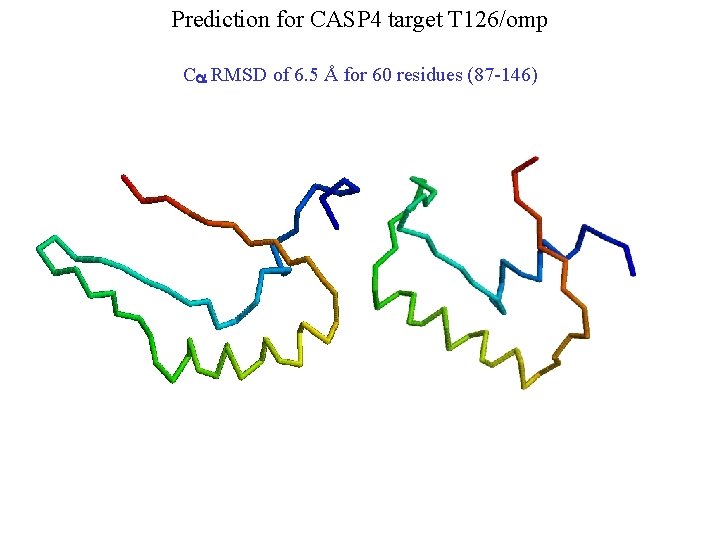
Prediction for CASP 4 target T 126/omp Ca RMSD of 6. 5 Å for 60 residues (87 -146)

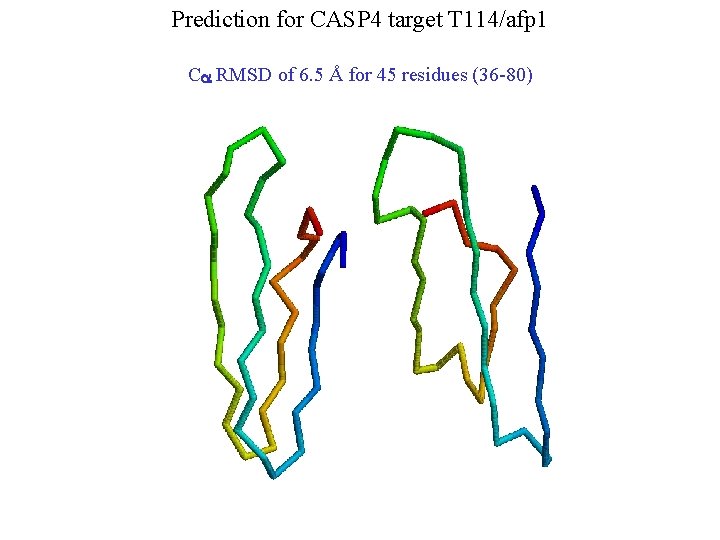
Prediction for CASP 4 target T 114/afp 1 Ca RMSD of 6. 5 Å for 45 residues (36 -80)

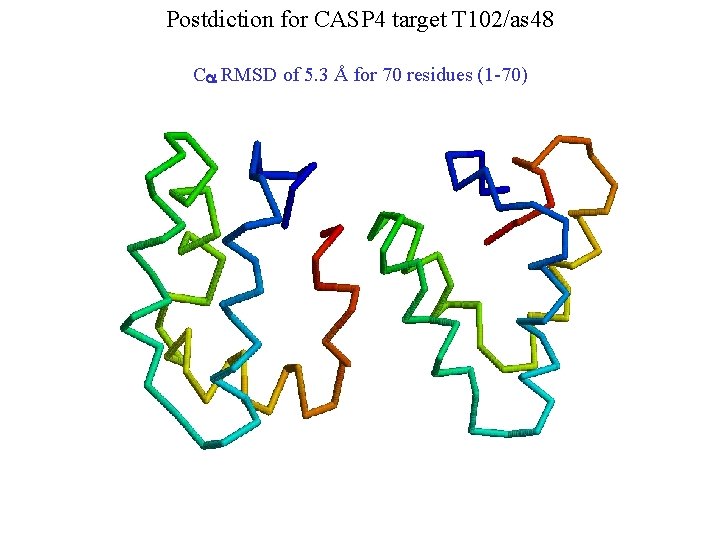
Postdiction for CASP 4 target T 102/as 48 Ca RMSD of 5. 3 Å for 70 residues (1 -70)

Historical perspective on ab initio prediction Before CASP (BC): “solved” (biased results) CASP 1: worse than random CASP 2: worse than random with one exception CASP 3: consistently predicted correct topology - ~ 6. 0 Å for 60+ residues CASP 4: consistently predicted correct topology - ~4 -6. 0 A for 60 -80+ residues **T 97/er 29 – 6. 0 Å (80 residues; 18 -97) *T 98/sp 0 a – 6. 0 Å (60 residues; 37 -105) **T 102/as 48 – 5. 3 Å (70 residues; 1 -70) **T 106/sfrp 3 – 6. 2 Å (70 residues; 6 -75) **T 110/rbfa – 4. 0 Å (80 residues; 1 -80) *T 114/afp 1 – 6. 5 Å (45 residues; 36 -80)

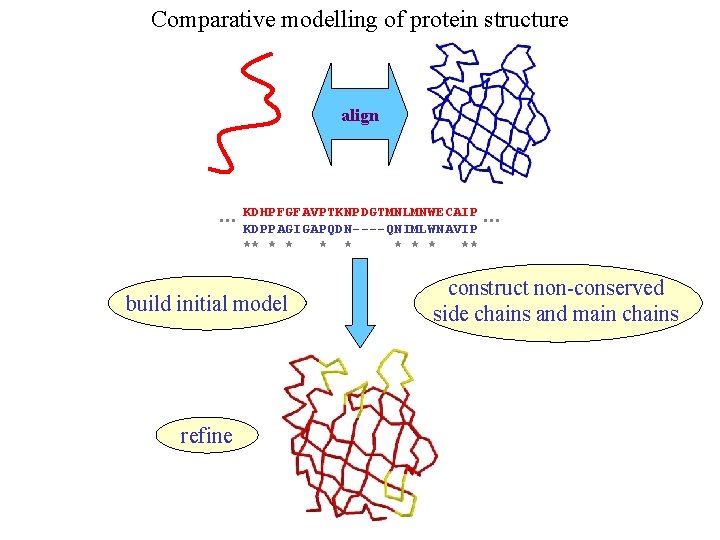
Comparative modelling of protein structure align … KDHPFGFAVPTKNPDGTMNLMNWECAIP KDPPAGIGAPQDN----QNIMLWNAVIP ** * * * ** build initial model refine … construct non-conserved side chains and main chains

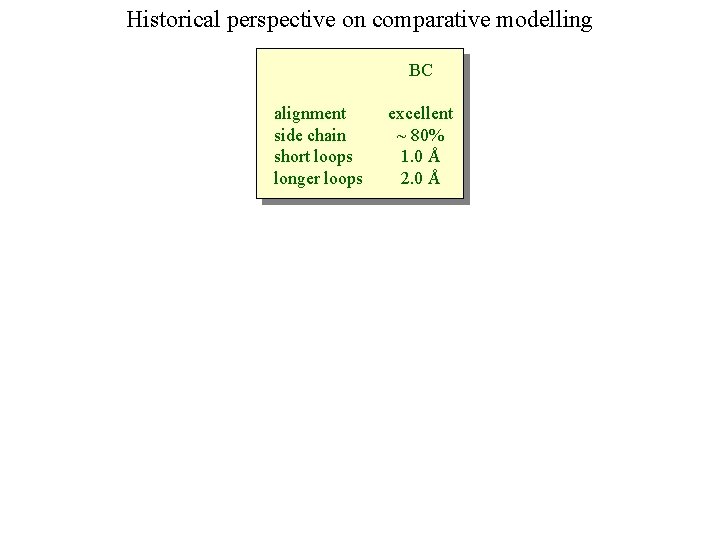
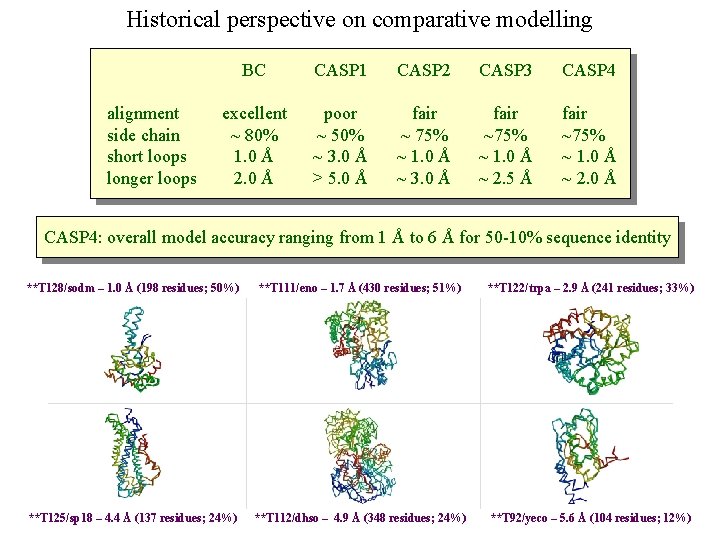
Historical perspective on comparative modelling BC alignment side chain short loops longer loops excellent ~ 80% 1. 0 Å 2. 0 Å

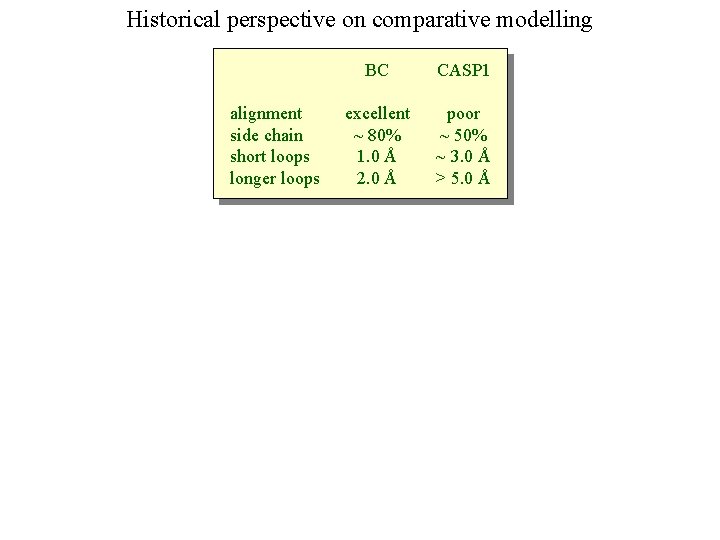
Historical perspective on comparative modelling alignment side chain short loops longer loops BC CASP 1 excellent ~ 80% 1. 0 Å 2. 0 Å poor ~ 50% ~ 3. 0 Å > 5. 0 Å

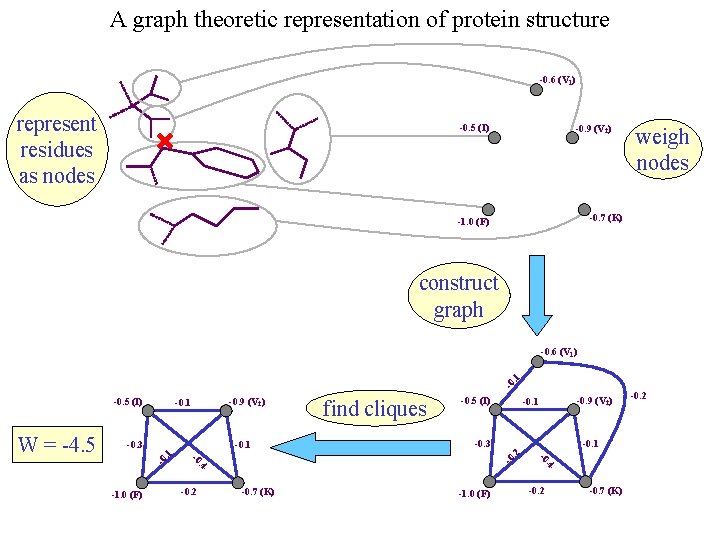
A graph theoretic representation of protein structure -0. 6 (V 1) represent residues as nodes -0. 5 (I) -0. 9 (V 2) weigh nodes -0. 7 (K) -1. 0 (F) construct graph -0 . 1 -0. 6 (V 1) -0. 5 (I) -0. 1 -0 -1. 0 (F) . 4 -0. 7 (K) -0 . 4 -0. 2 -0. 9 (V 2) -0. 1 -0. 3 -0 -1. 0 (F) find cliques -0. 5 (I) . 2 -0. 1. 1 -0. 3 -0 W = -4. 5 -0. 9 (V 2) -0. 1 -0. 2 -0. 7 (K) -0. 2

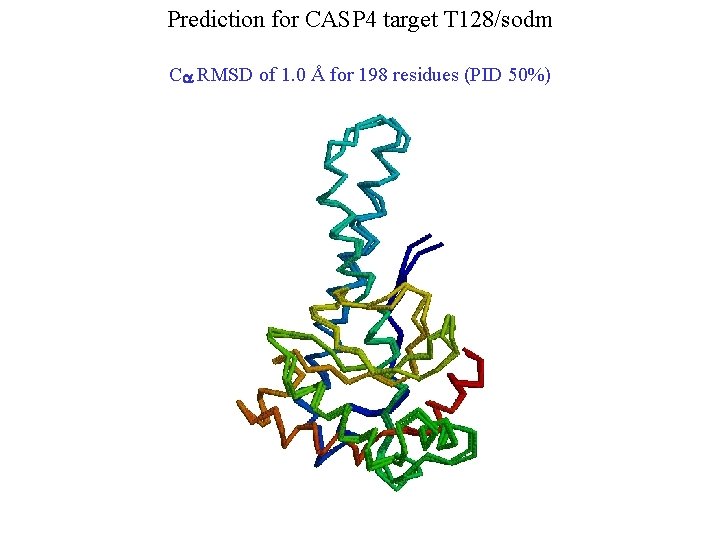
Prediction for CASP 4 target T 128/sodm Ca RMSD of 1. 0 Å for 198 residues (PID 50%)

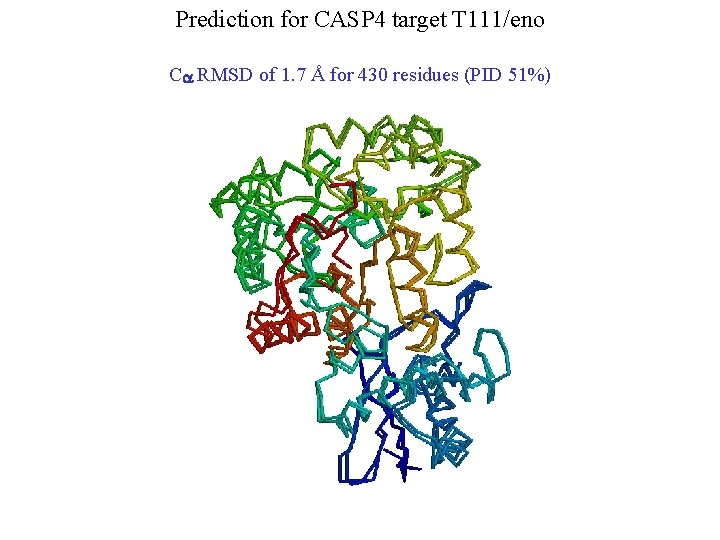
Prediction for CASP 4 target T 111/eno Ca RMSD of 1. 7 Å for 430 residues (PID 51%)

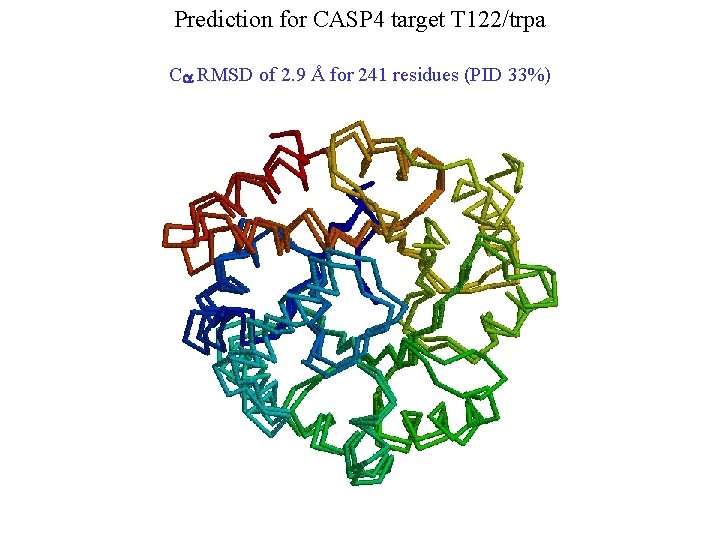
Prediction for CASP 4 target T 122/trpa Ca RMSD of 2. 9 Å for 241 residues (PID 33%)

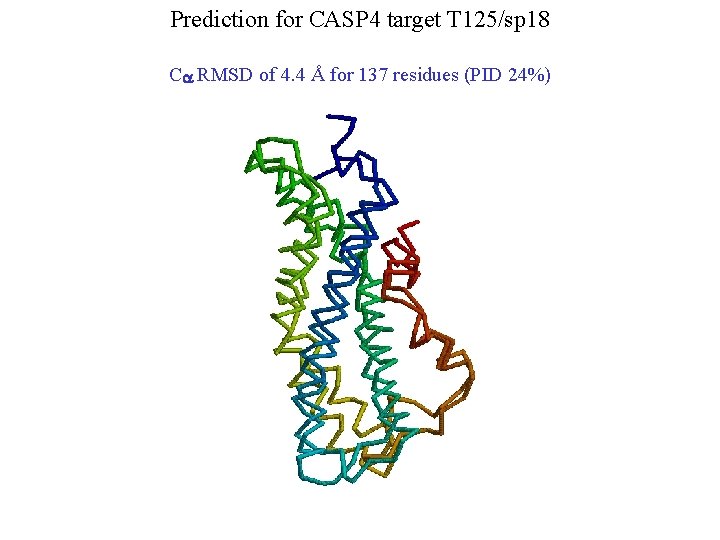
Prediction for CASP 4 target T 125/sp 18 Ca RMSD of 4. 4 Å for 137 residues (PID 24%)

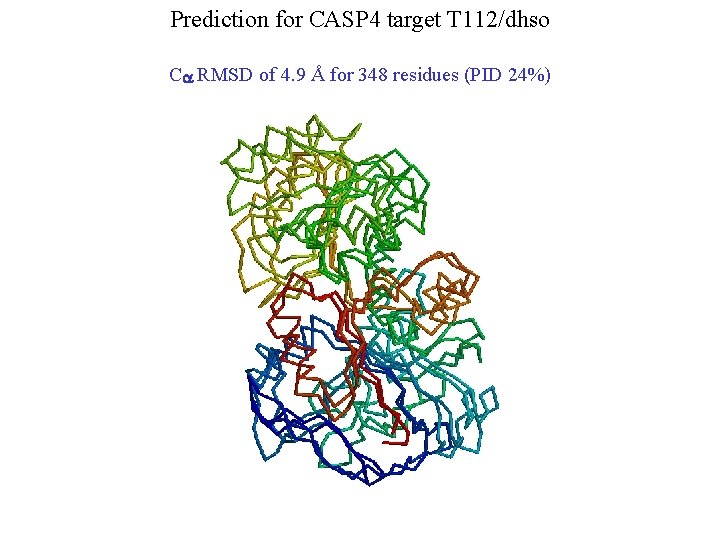
Prediction for CASP 4 target T 112/dhso Ca RMSD of 4. 9 Å for 348 residues (PID 24%)

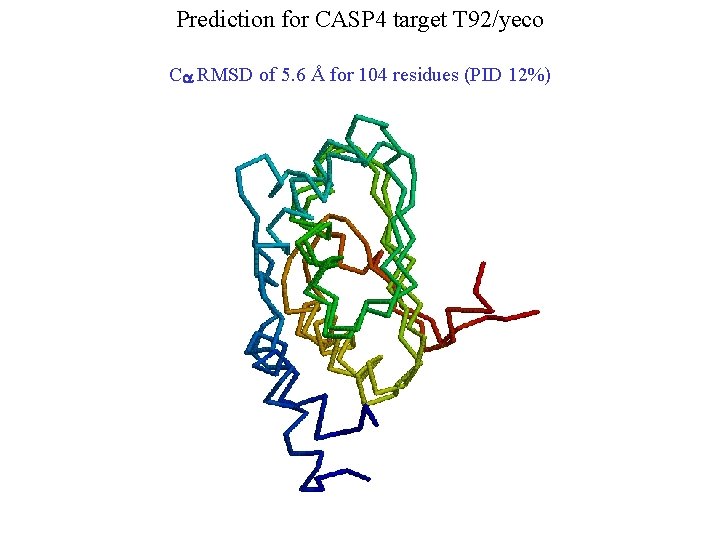
Prediction for CASP 4 target T 92/yeco Ca RMSD of 5. 6 Å for 104 residues (PID 12%)

Historical perspective on comparative modelling alignment side chain short loops longer loops BC CASP 1 CASP 2 CASP 3 CASP 4 excellent ~ 80% 1. 0 Å 2. 0 Å poor ~ 50% ~ 3. 0 Å > 5. 0 Å fair ~ 75% ~ 1. 0 Å ~ 3. 0 Å fair ~75% ~ 1. 0 Å ~ 2. 5 Å fair ~75% ~ 1. 0 Å ~ 2. 0 Å CASP 4: overall model accuracy ranging from 1 Å to 6 Å for 50 -10% sequence identity **T 128/sodm – 1. 0 Å (198 residues; 50%) **T 111/eno – 1. 7 Å (430 residues; 51%) **T 122/trpa – 2. 9 Å (241 residues; 33%) **T 125/sp 18 – 4. 4 Å (137 residues; 24%) **T 112/dhso – 4. 9 Å (348 residues; 24%) **T 92/yeco – 5. 6 Å (104 residues; 12%)

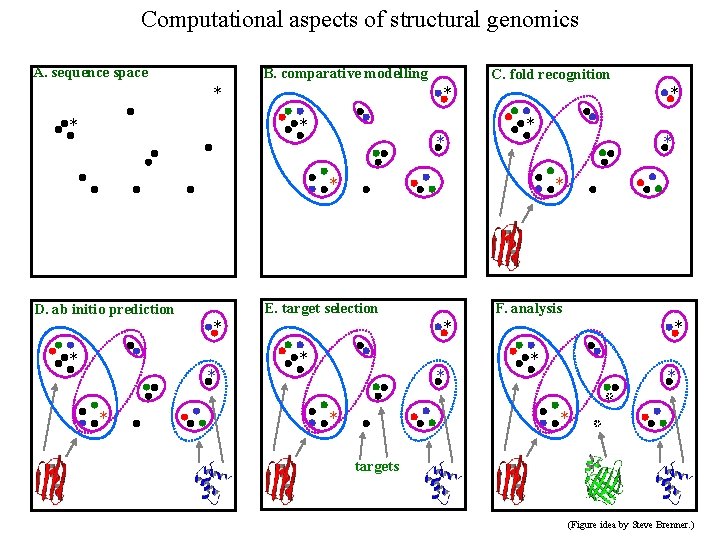
Computational aspects of structural genomics A. sequence space B. comparative modelling * * C. fold recognition * * * * E. target selection D. ab initio prediction * * F. analysis * * * * targets (Figure idea by Steve Brenner. )

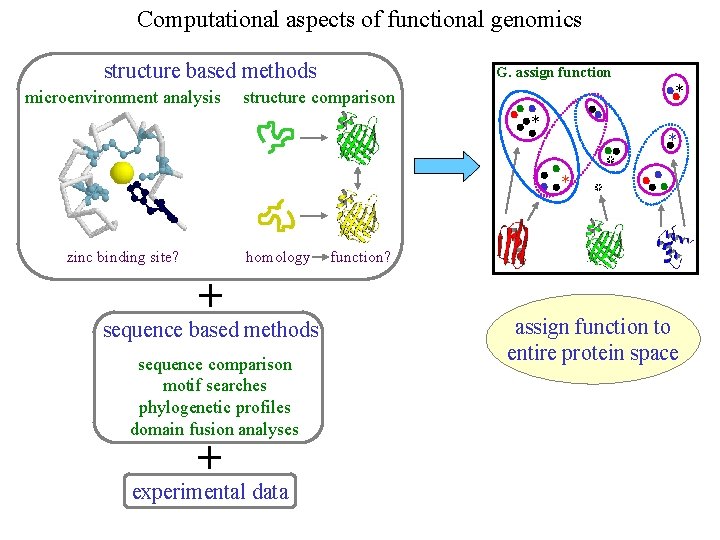
Computational aspects of functional genomics structure based methods microenvironment analysis G. assign function * structure comparison * * * zinc binding site? homology + sequence based methods sequence comparison motif searches phylogenetic profiles domain fusion analyses + experimental data * * function? assign function to entire protein space

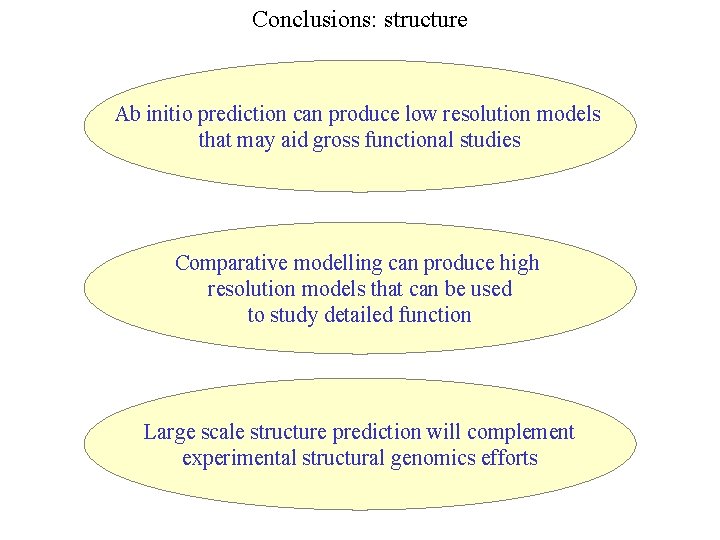
Conclusions: structure Ab initio prediction can produce low resolution models that may aid gross functional studies Comparative modelling can produce high resolution models that can be used to study detailed function Large scale structure prediction will complement experimental structural genomics efforts

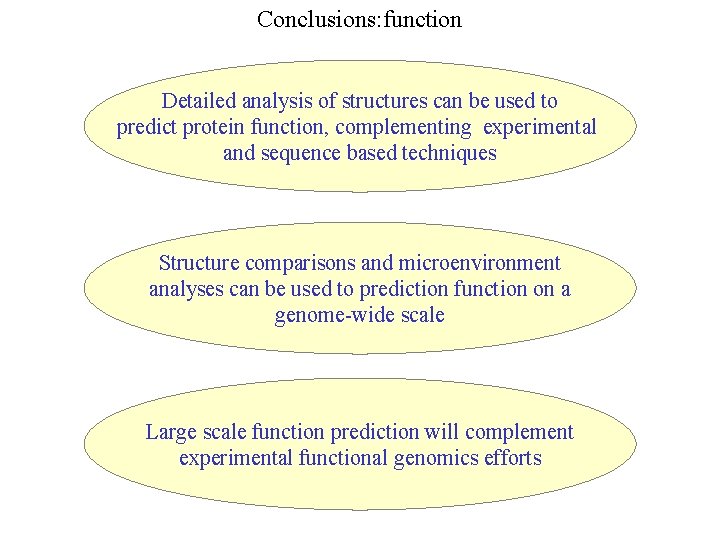
Conclusions: function Detailed analysis of structures can be used to predict protein function, complementing experimental and sequence based techniques Structure comparisons and microenvironment analyses can be used to prediction function on a genome-wide scale Large scale function prediction will complement experimental functional genomics efforts

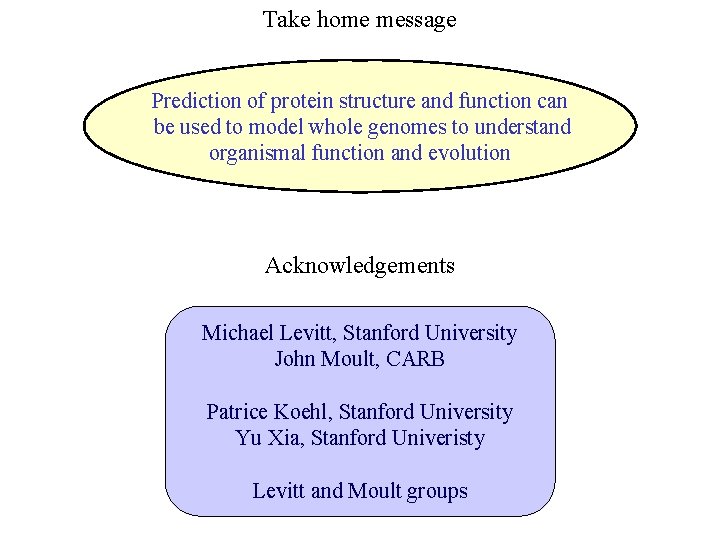
Take home message Prediction of protein structure and function can be used to model whole genomes to understand organismal function and evolution Acknowledgements Michael Levitt, Stanford University John Moult, CARB Patrice Koehl, Stanford University Yu Xia, Stanford Univeristy Levitt and Moult groups