Modelling comparison and analysis of proteomes Ram Samudrala
- Slides: 15

Modelling, comparison, and analysis of proteomes Ram Samudrala University of Washington

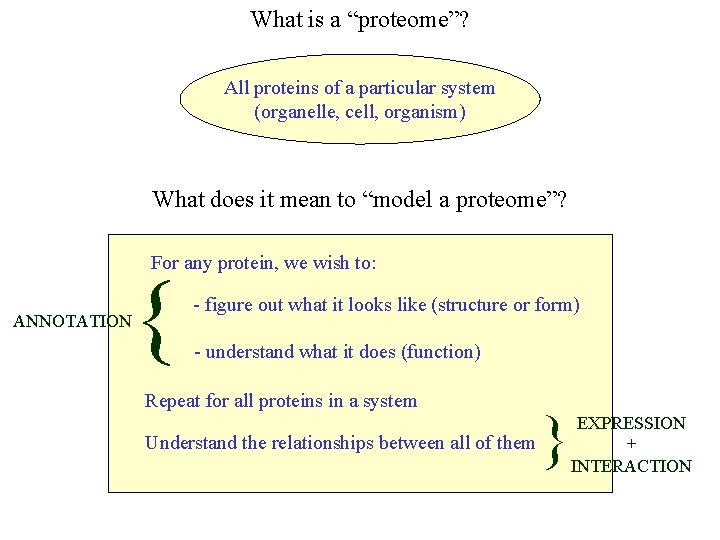
What is a “proteome”? All proteins of a particular system (organelle, cell, organism) What does it mean to “model a proteome”? For any protein, we wish to: ANNOTATION { - figure out what it looks like (structure or form) - understand what it does (function) Repeat for all proteins in a system Understand the relationships between all of them } EXPRESSION + INTERACTION

Why should we model proteomes? Intellectual challenge: Because it’s there! ? Pragmatic reasons: - rational drug design and treatment of disease - protein and genetic engineering - build networks to model cellular pathways - study organismal function and evolution

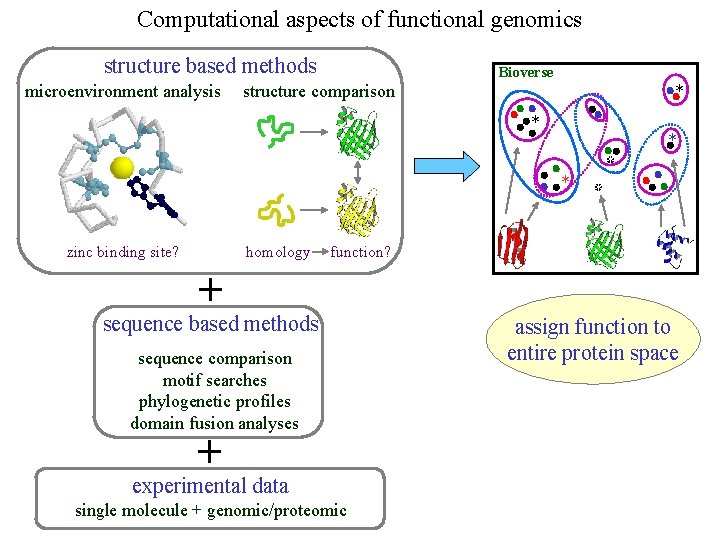
Computational aspects of functional genomics structure based methods microenvironment analysis Bioverse structure comparison * * homology zinc binding site? * * function? + sequence based methods sequence comparison motif searches phylogenetic profiles domain fusion analyses + experimental data single molecule + genomic/proteomic assign function to entire protein space

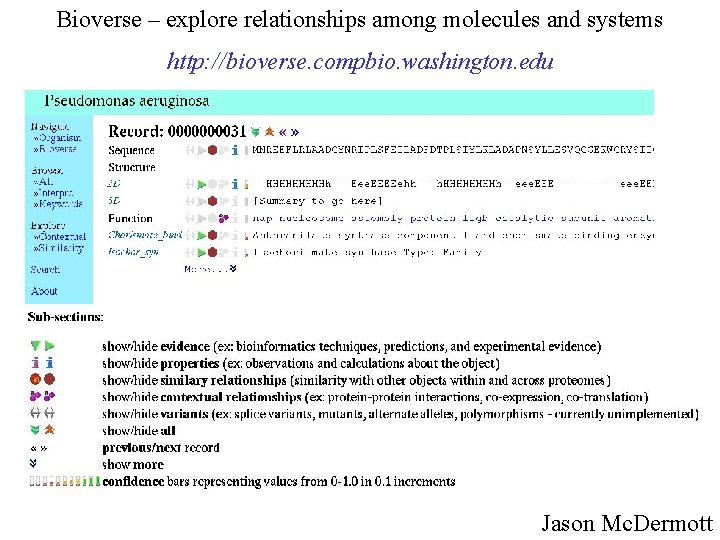
Bioverse – explore relationships among molecules and systems http: //bioverse. compbio. washington. edu Jason Mc. Dermott

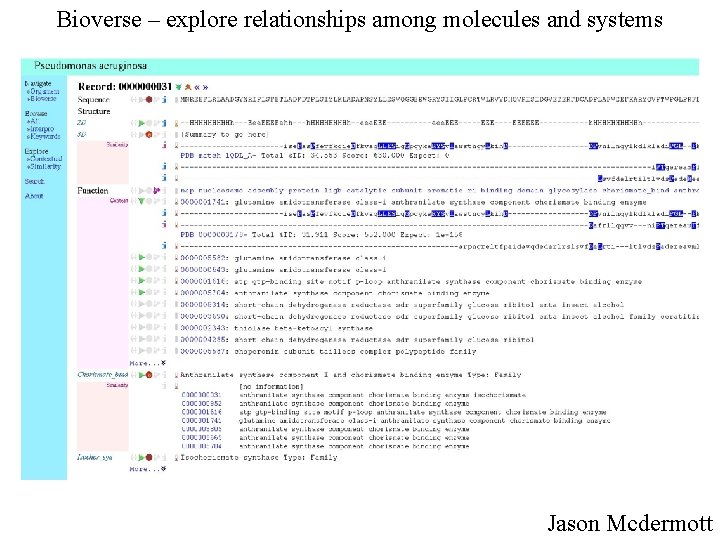
Bioverse – explore relationships among molecules and systems Jason Mcdermott

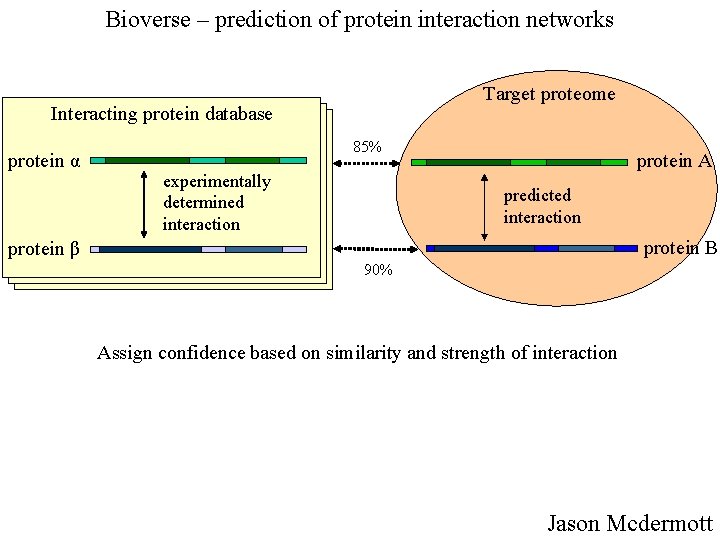
Bioverse – prediction of protein interaction networks Target proteome Interacting protein database protein α 85% experimentally determined interaction protein A predicted interaction protein B protein β 90% Assign confidence based on similarity and strength of interaction Jason Mcdermott

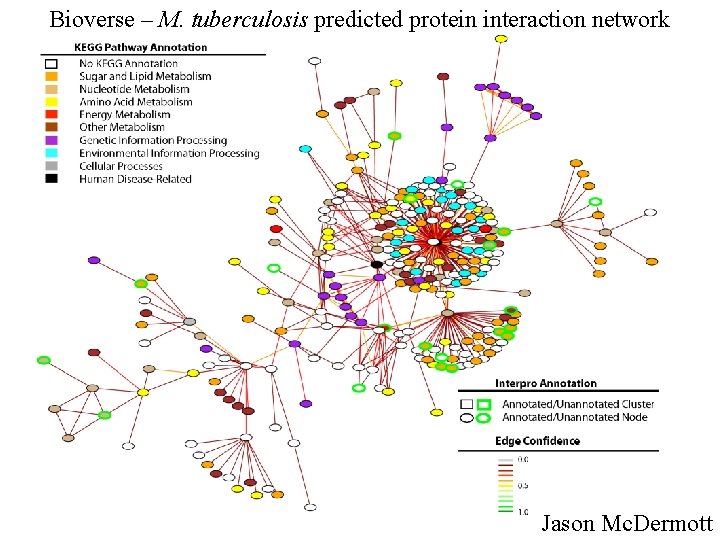
Bioverse – M. tuberculosis predicted protein interaction network Jason Mc. Dermott

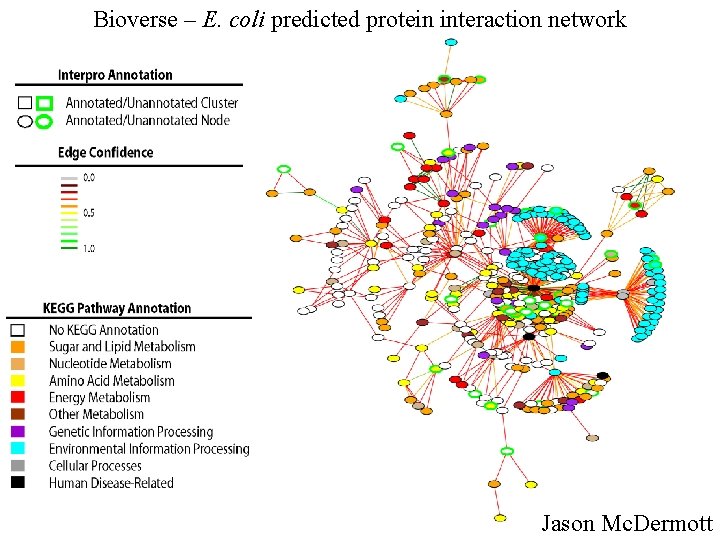
Bioverse – E. coli predicted protein interaction network Jason Mc. Dermott

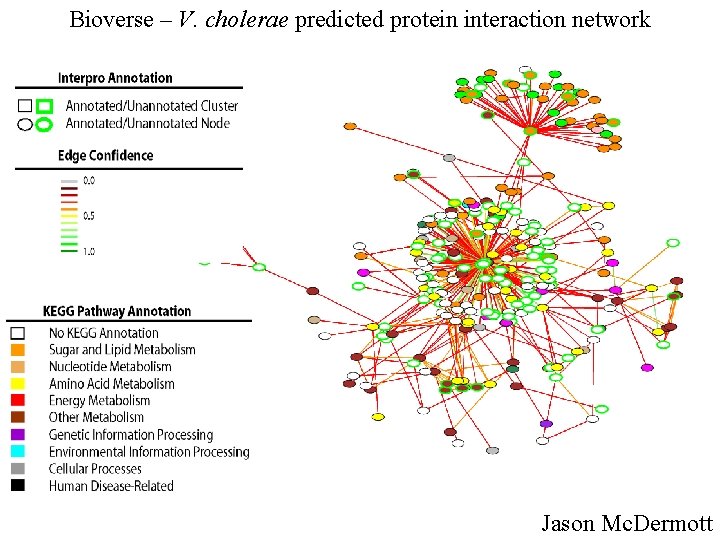
Bioverse – V. cholerae predicted protein interaction network Jason Mc. Dermott

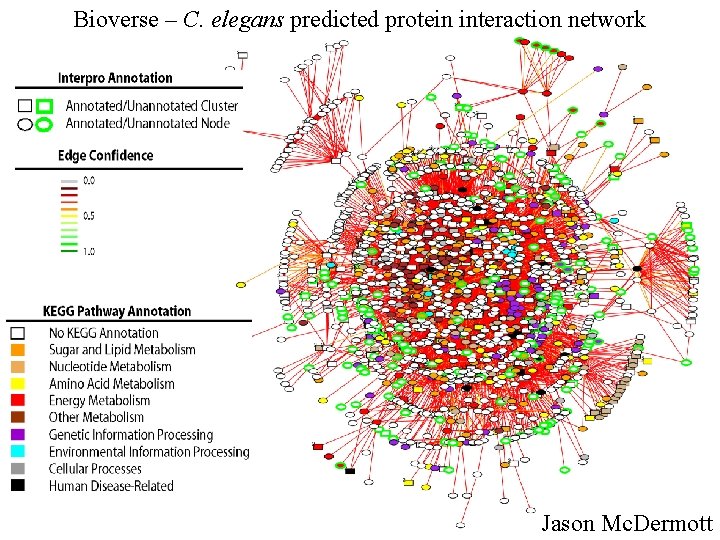
Bioverse – C. elegans predicted protein interaction network Jason Mc. Dermott

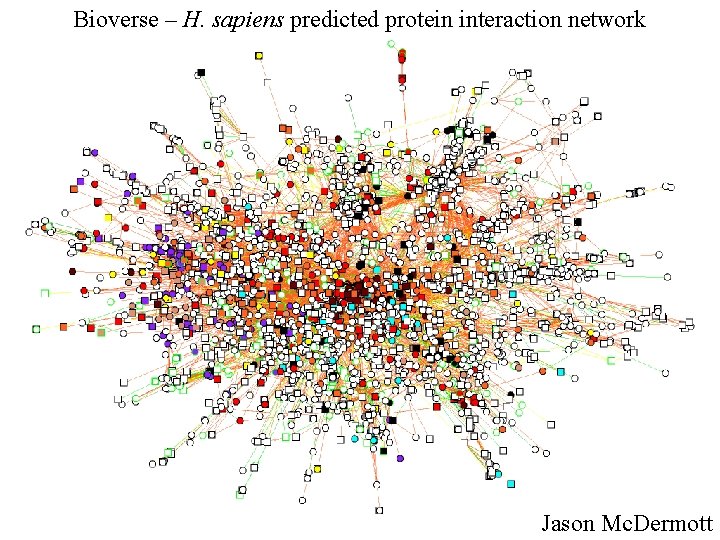
Bioverse – H. sapiens predicted protein interaction network Jason Mc. Dermott

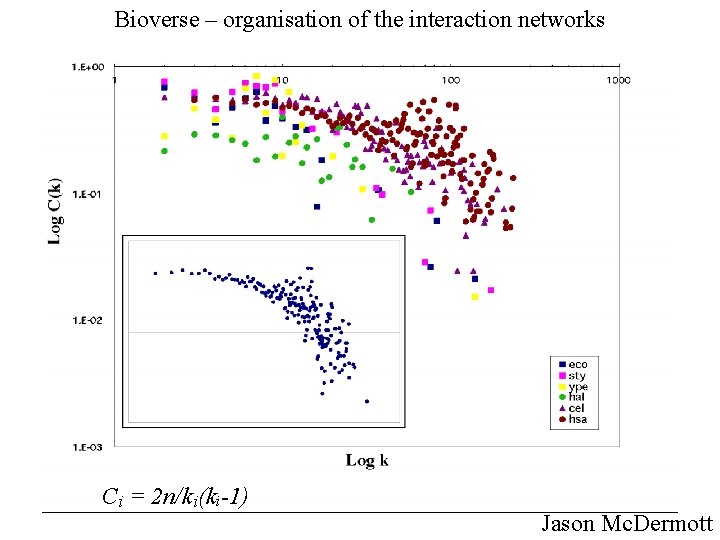
Bioverse – organisation of the interaction networks Ci = 2 n/ki(ki-1) Jason Mc. Dermott

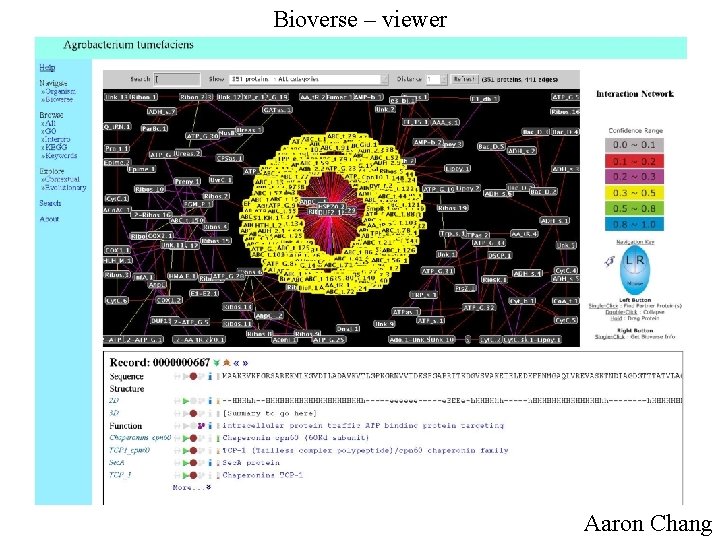
Bioverse – viewer Aaron Chang

Take home message Prediction of protein structure and function can be used to model whole genomes to understand organismal function and evolution Acknowledgements Aaron Chang Ashley Lam Ekachai Jenwitheesuk Gong Cheng Jason Mc. Dermott Kai Wang Ling-Hong Hung Lynne Townsend Marissa La. Madrid Mike Inouye Stewart Moughon Shing-Chung Ngan Yi-Ling Cheng Zach Frazier