MODELLING PROTEOMES RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF


- Slides: 39

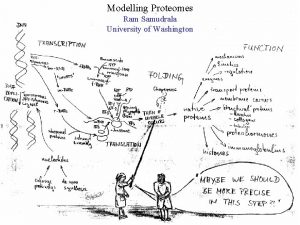
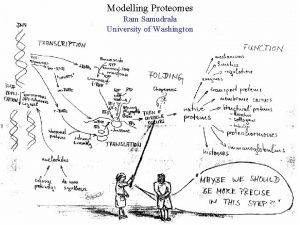
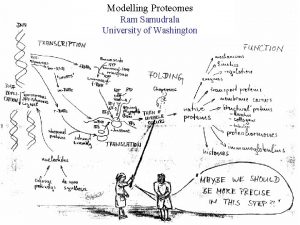
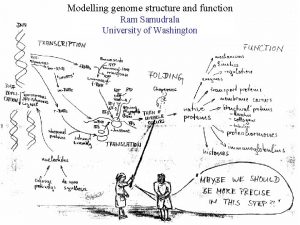
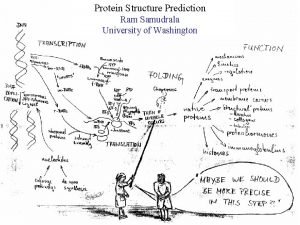
MODELLING PROTEOMES RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF WASHINGTON How does the genome of an organism specify its behaviour and characteristics?

PROTEOME ~60, 000 in human ~60, 000 in rice ~4500 in bacteria Several thousand distinct sequence families

STRUCTURE A few thousand distinct structural folds

FUNCTION Tens of thousands of functions

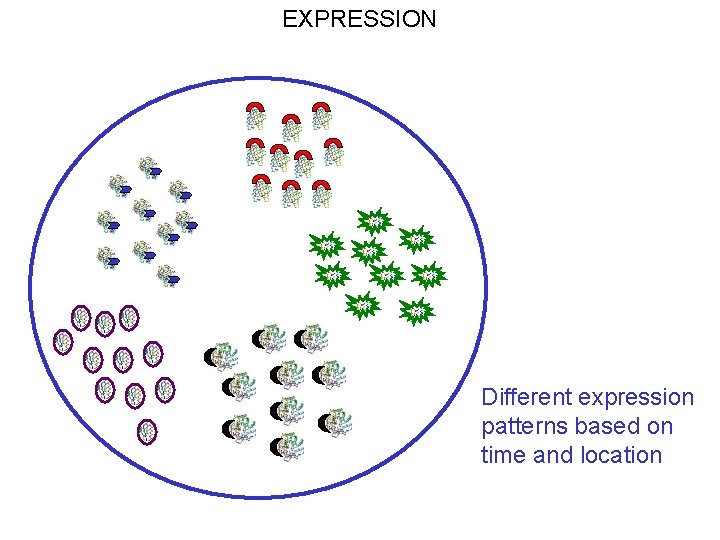
EXPRESSION Different expression patterns based on time and location

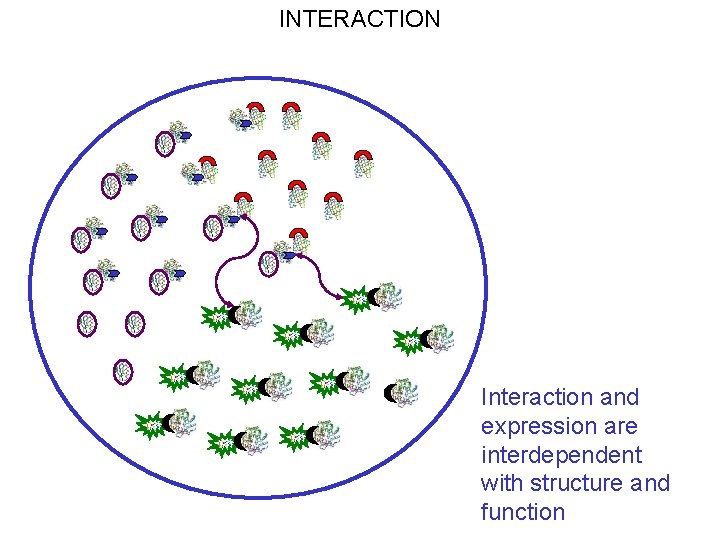
INTERACTION Interaction and expression are interdependent with structure and function

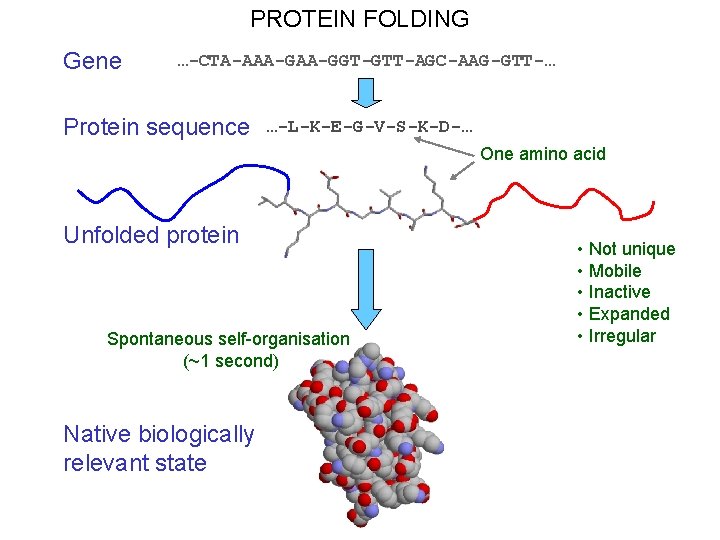
PROTEIN FOLDING Gene …-CTA-AAA-GGT-GTT-AGC-AAG-GTT-… Protein sequence …-L-K-E-G-V-S-K-D-… One amino acid Unfolded protein Spontaneous self-organisation (~1 second) Native biologically relevant state • Not unique • Mobile • Inactive • Expanded • Irregular

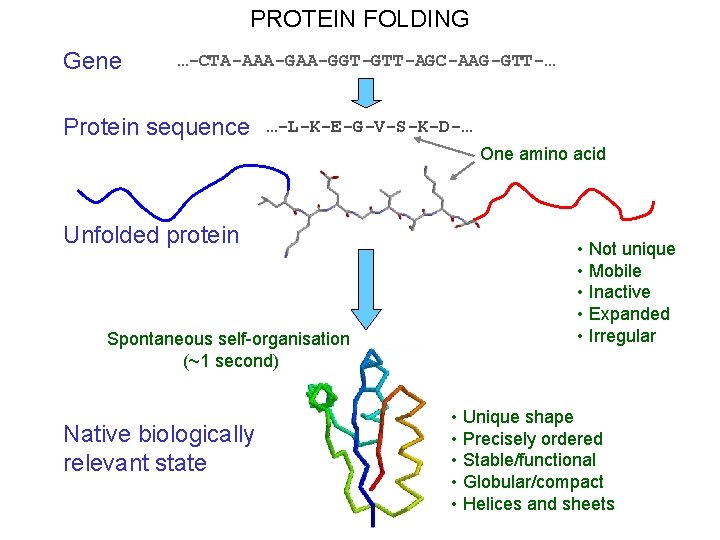
PROTEIN FOLDING Gene …-CTA-AAA-GGT-GTT-AGC-AAG-GTT-… Protein sequence …-L-K-E-G-V-S-K-D-… One amino acid Unfolded protein Spontaneous self-organisation (~1 second) Native biologically relevant state • Not unique • Mobile • Inactive • Expanded • Irregular • Unique shape • Precisely ordered • Stable/functional • Globular/compact • Helices and sheets

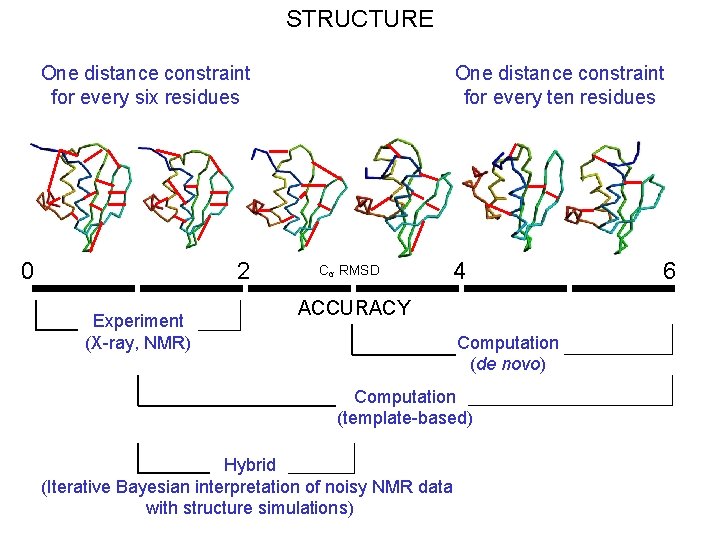
STRUCTURE One distance constraint for every six residues 0 2 Experiment (X-ray, NMR) One distance constraint for every ten residues Cα RMSD 4 ACCURACY Computation (de novo) Computation (template-based) Hybrid (Iterative Bayesian interpretation of noisy NMR data with structure simulations) 6

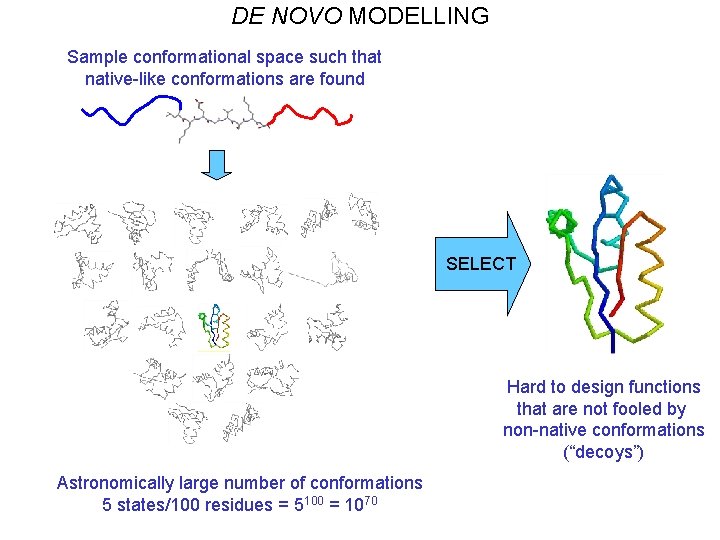
DE NOVO MODELLING Sample conformational space such that native-like conformations are found SELECT Hard to design functions that are not fooled by non-native conformations (“decoys”) Astronomically large number of conformations 5 states/100 residues = 5100 = 1070

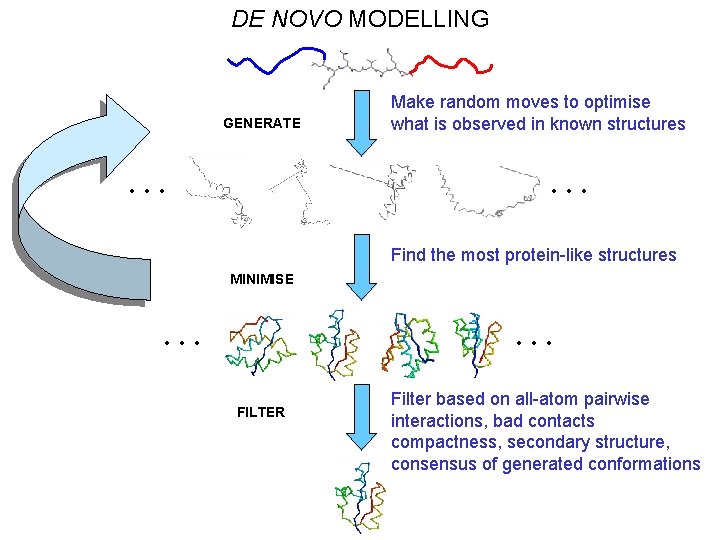
DE NOVO MODELLING GENERATE … Make random moves to optimise what is observed in known structures … Find the most protein-like structures MINIMISE … … FILTER Filter based on all-atom pairwise interactions, bad contacts compactness, secondary structure, consensus of generated conformations

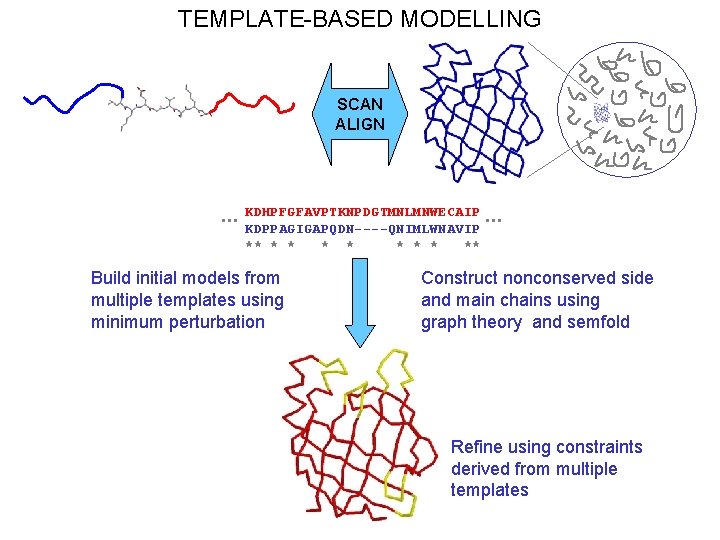
TEMPLATE-BASED MODELLING SCAN ALIGN … KDHPFGFAVPTKNPDGTMNLMNWECAIP KDPPAGIGAPQDN----QNIMLWNAVIP ** * * * ** Build initial models from multiple templates using minimum perturbation … Construct nonconserved side and main chains using graph theory and semfold Refine using constraints derived from multiple templates

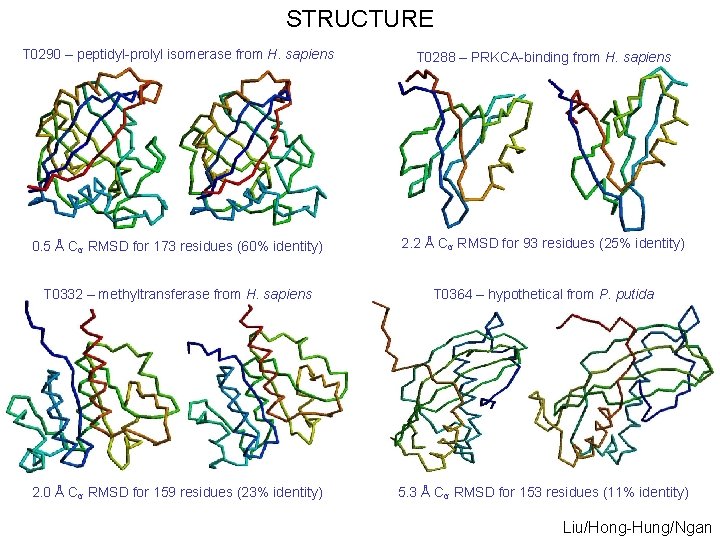
STRUCTURE T 0290 – peptidyl-prolyl isomerase from H. sapiens T 0288 – PRKCA-binding from H. sapiens 0. 5 Å Cα RMSD for 173 residues (60% identity) 2. 2 Å Cα RMSD for 93 residues (25% identity) T 0332 – methyltransferase from H. sapiens T 0364 – hypothetical from P. putida 2. 0 Å Cα RMSD for 159 residues (23% identity) 5. 3 Å Cα RMSD for 153 residues (11% identity) Liu/Hong-Hung/Ngan

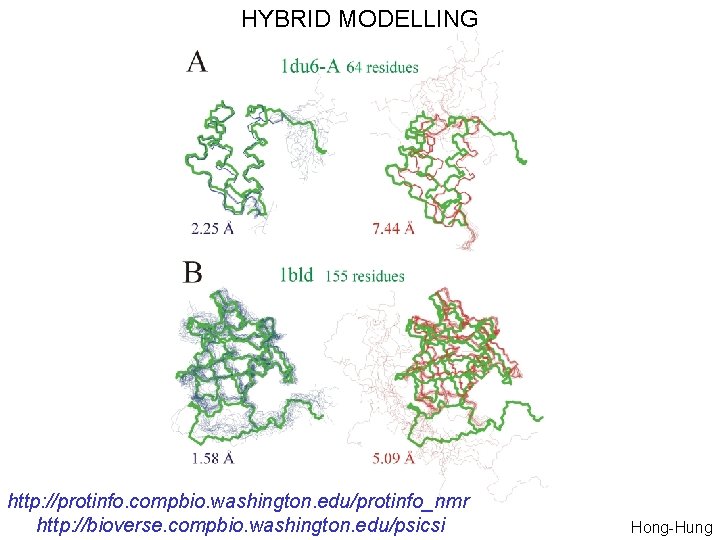
HYBRID MODELLING http: //protinfo. compbio. washington. edu/protinfo_nmr http: //bioverse. compbio. washington. edu/psicsi Hong-Hung

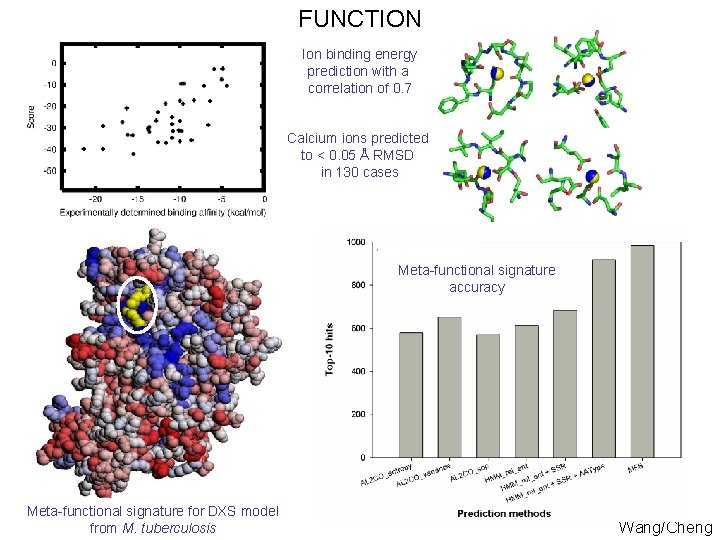
FUNCTION Ion binding energy prediction with a correlation of 0. 7 Calcium ions predicted to < 0. 05 Å RMSD in 130 cases Meta-functional signature accuracy Meta-functional signature for DXS model from M. tuberculosis Wang/Cheng

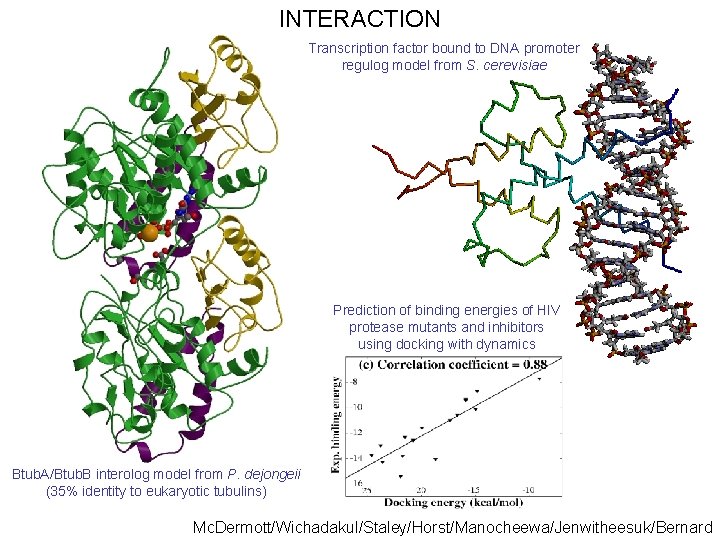
INTERACTION Transcription factor bound to DNA promoter regulog model from S. cerevisiae Prediction of binding energies of HIV protease mutants and inhibitors using docking with dynamics Btub. A/Btub. B interolog model from P. dejongeii (35% identity to eukaryotic tubulins) Mc. Dermott/Wichadakul/Staley/Horst/Manocheewa/Jenwitheesuk/Bernard

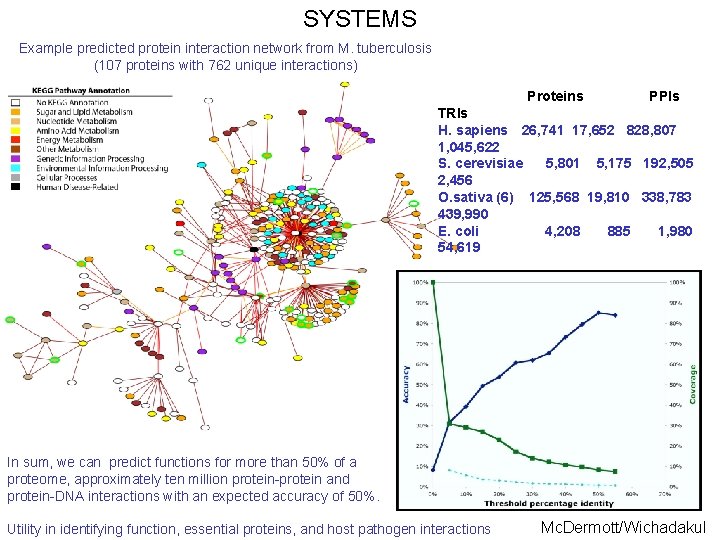
SYSTEMS Example predicted protein interaction network from M. tuberculosis (107 proteins with 762 unique interactions) Proteins PPIs TRIs H. sapiens 26, 741 17, 652 828, 807 1, 045, 622 S. cerevisiae 5, 801 5, 175 192, 505 2, 456 O. sativa (6) 125, 568 19, 810 338, 783 439, 990 E. coli 4, 208 885 1, 980 54, 619 In sum, we can predict functions for more than 50% of a proteome, approximately ten million protein-protein and protein-DNA interactions with an expected accuracy of 50%. Utility in identifying function, essential proteins, and host pathogen interactions Mc. Dermott/Wichadakul

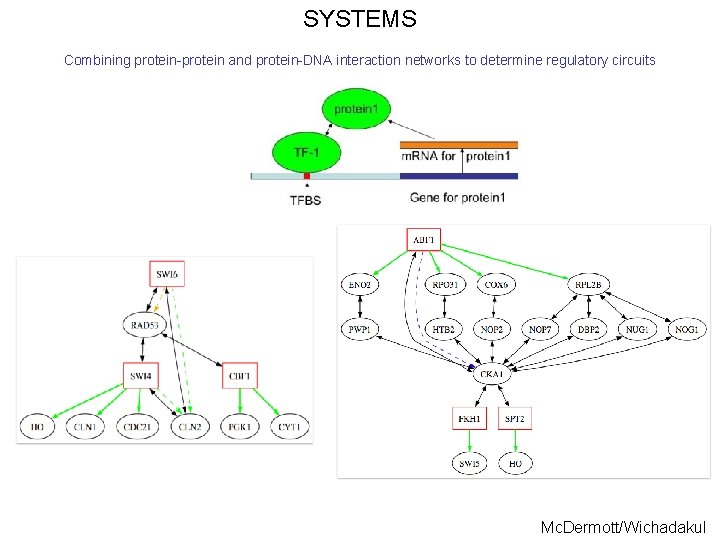
SYSTEMS Combining protein-protein and protein-DNA interaction networks to determine regulatory circuits Mc. Dermott/Wichadakul

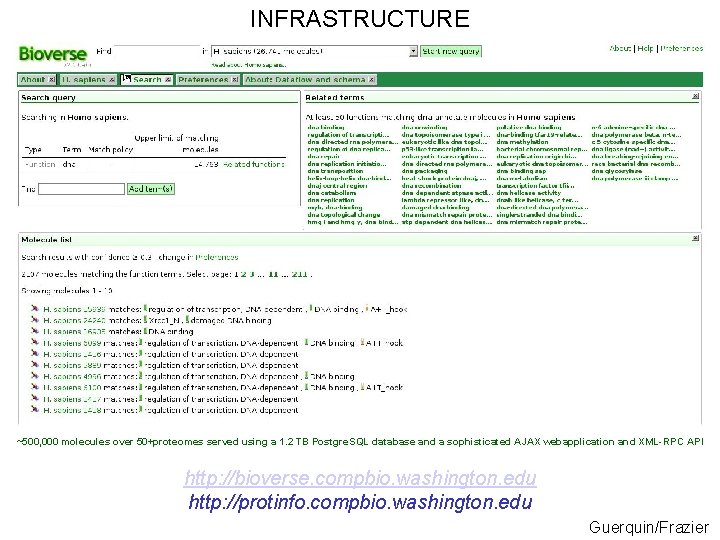
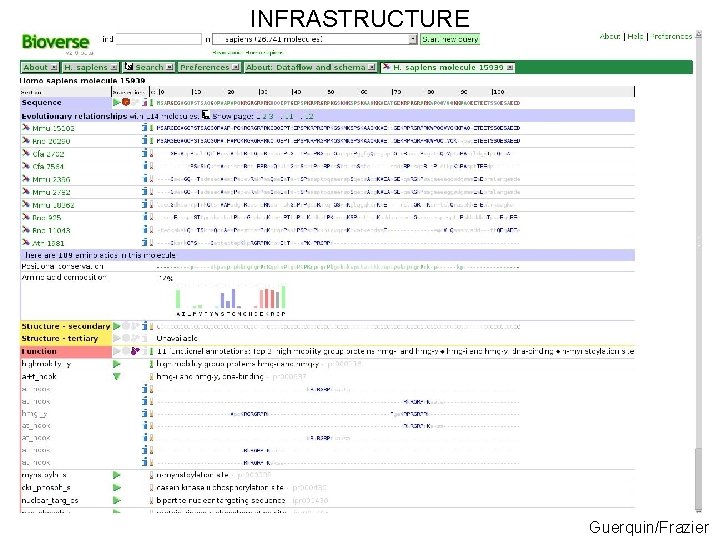
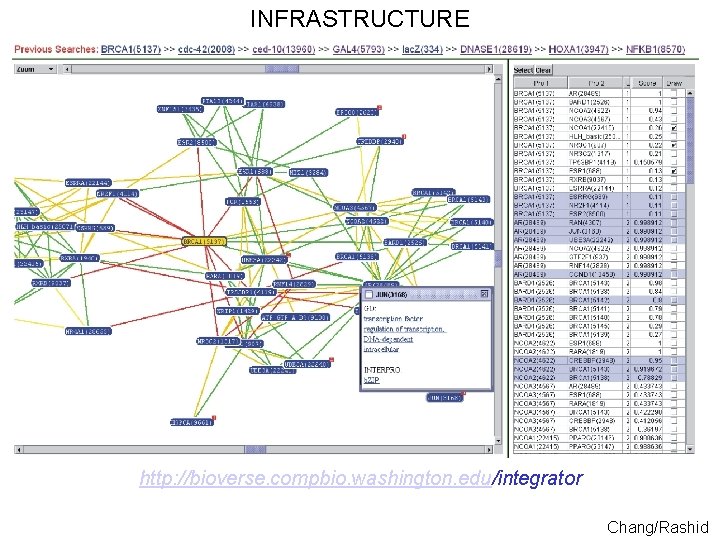
INFRASTRUCTURE ~500, 000 molecules over 50+proteomes served using a 1. 2 TB Postgre. SQL database and a sophisticated AJAX webapplication and XML-RPC API http: //bioverse. compbio. washington. edu http: //protinfo. compbio. washington. edu Guerquin/Frazier

INFRASTRUCTURE Guerquin/Frazier

INFRASTRUCTURE http: //bioverse. compbio. washington. edu/integrator Chang/Rashid

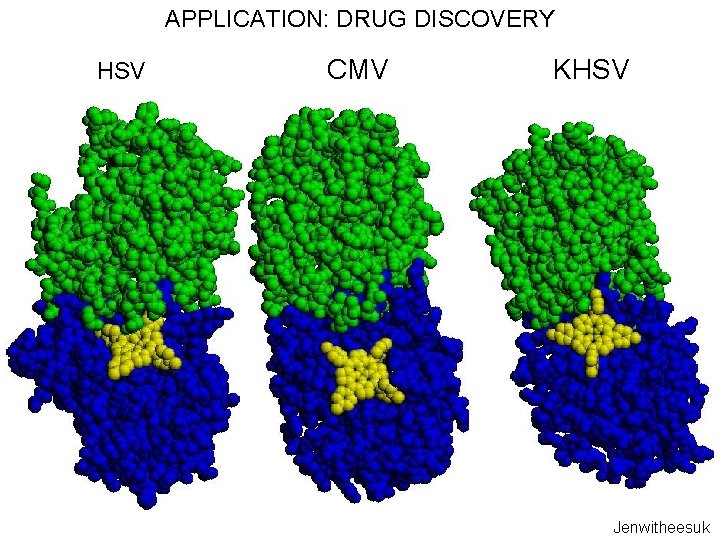
APPLICATION: DRUG DISCOVERY HSV CMV KHSV Jenwitheesuk

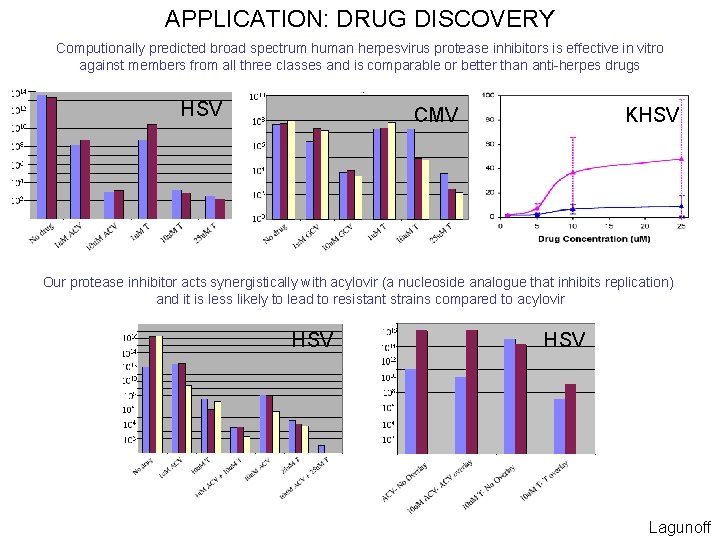
APPLICATION: DRUG DISCOVERY Computionally predicted broad spectrum human herpesvirus protease inhibitors is effective in vitro against members from all three classes and is comparable or better than anti-herpes drugs HSV KHSV CMV Our protease inhibitor acts synergistically with acylovir (a nucleoside analogue that inhibits replication) and it is less likely to lead to resistant strains compared to acylovir HSV Lagunoff

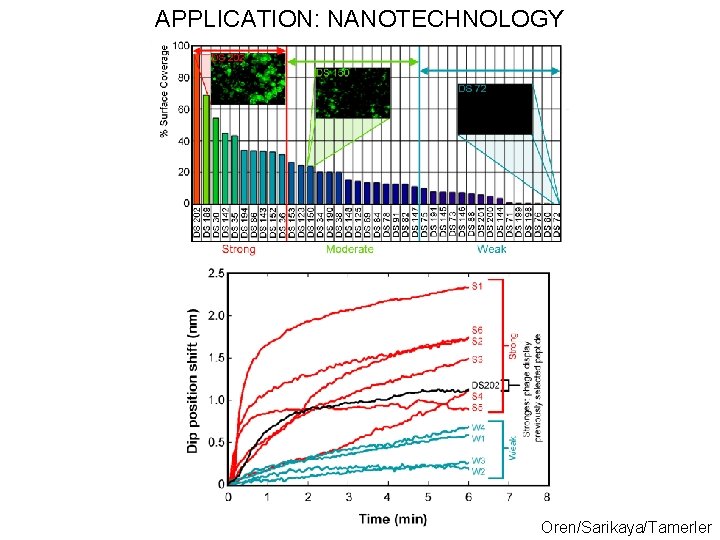
APPLICATION: NANOTECHNOLOGY Oren/Sarikaya/Tamerler

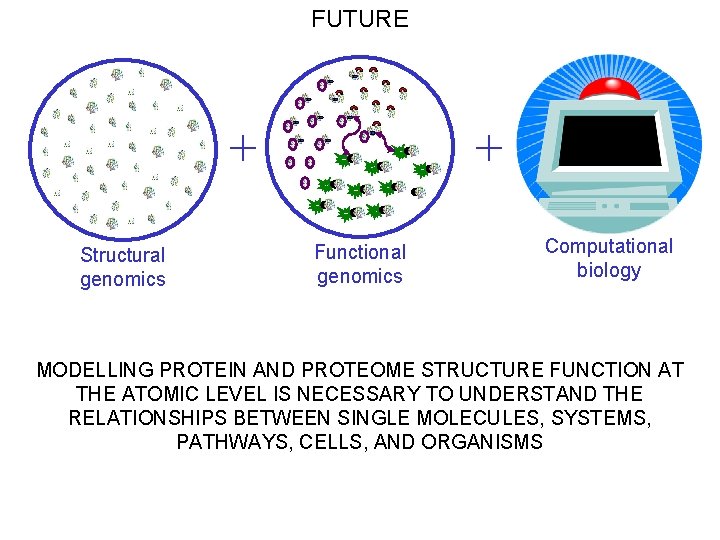
FUTURE + Structural genomics + Functional genomics Computational biology MODELLING PROTEIN AND PROTEOME STRUCTURE FUNCTION AT THE ATOMIC LEVEL IS NECESSARY TO UNDERSTAND THE RELATIONSHIPS BETWEEN SINGLE MOLECULES, SYSTEMS, PATHWAYS, CELLS, AND ORGANISMS

ACKNOWLEDGEMENTS Current group members: Past group members: • Baishali Chanda • Brady Bernard • Chuck Mader • David Nickle • Ersin Emre Oren • Ekachai Jenwitheesuk • Gong Cheng • Imran Rashid • Jeremy Horst • Ling-Hong Hung • Michal Guerquin • Rob Brasier • Rosalia Tungaraza • Shing-Chung Ngan • Siriphan Manocheewa • Somsak Phattarasukol • Stewart Moughon • Tianyun Liu • Vania Wang • Weerayuth Kittichotirat • Zach Frazier • Kristina Montgomery, Program Manager • Aaron Chang • Duncan Milburn • Jason Mc. Dermott • Kai Wang • Marissa La. Madrid Collaborators: • James Staley • Mehmet Sarikaya/Candan Tamerler • Michael Lagunoff • Roger Bumgarner • Wesley Van Voorhis Funding agencies: • National Institutes of Health • National Science Foundation • Searle Scholars Program • Puget Sound Partners in Global Health • UW Advanced Technology Initiative • Washington Research Foundation • UW TGIF


MOTIVATION FOR DETERMINING PROTEIN STRUCTURE The functions necessary for life are undertaken by proteins. Protein function is mediated by protein three-dimensional structure. Knowing protein structure at high resolution will enable us to: Determine and understand molecular function. Understand substrate and ligand binding. Devise intelligent mutagenesis and biochemical experiments to understand biological function. Design therapeutics rationally. Design novel proteins. Knowing the structures of all proteins encoded by an organism’s genome will enable us to understand complex pathways and systems, and ultimately organismal behaviour and evolution. Applications in the area of medicine, nanotechnology, and biological computing.

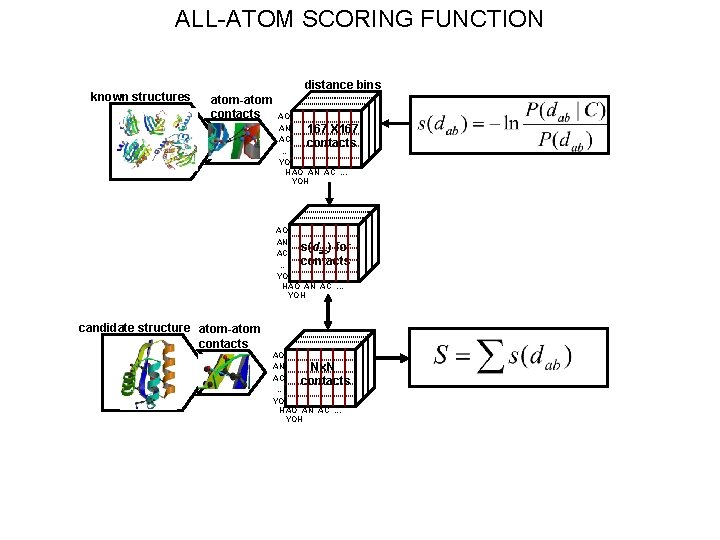
ALL-ATOM SCORING FUNCTION known structures distance bins atom-atom contacts AO AN 167 X 167 AC contacts … YO H AO AN AC. . . YOH AO AN s(dab) for AC contacts … YO HAO AN AC. . . YOH candidate structure atom-atom contacts AO AN Nx. N AC contacts … YO H AO AN AC. . . YOH

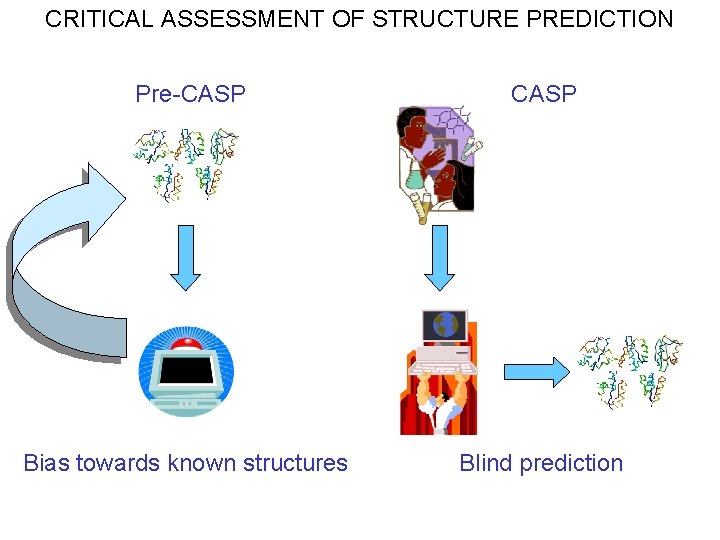
CRITICAL ASSESSMENT OF STRUCTURE PREDICTION Pre-CASP Bias towards known structures Blind prediction

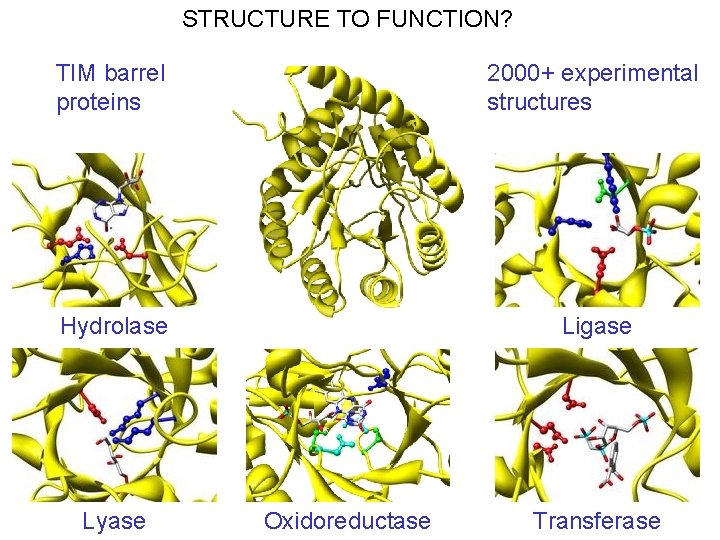
STRUCTURE TO FUNCTION? TIM barrel proteins 2000+ experimental structures Hydrolase Ligase Lyase Oxidoreductase Transferase

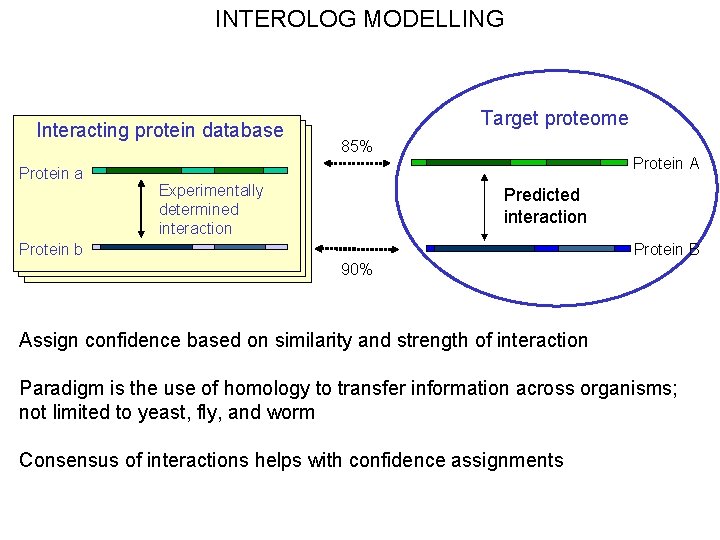
INTEROLOG MODELLING Interacting protein database Protein a Target proteome 85% Experimentally determined interaction Protein A Predicted interaction Protein b Protein B 90% Assign confidence based on similarity and strength of interaction Paradigm is the use of homology to transfer information across organisms; not limited to yeast, fly, and worm Consensus of interactions helps with confidence assignments

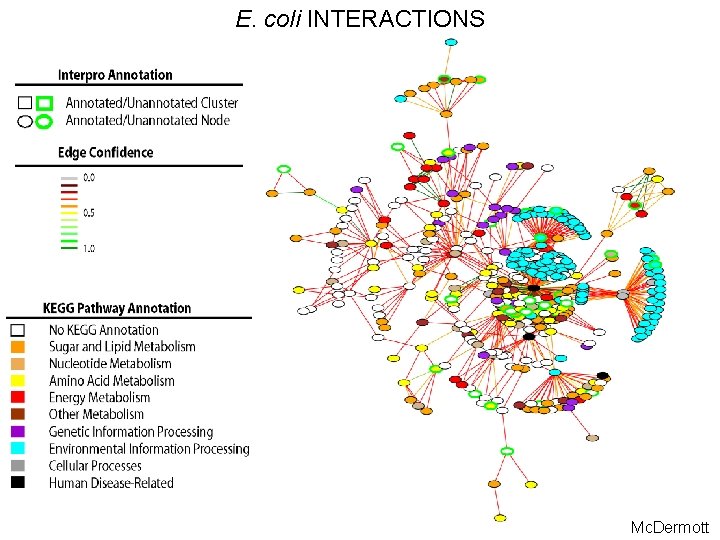
E. coli INTERACTIONS Mc. Dermott

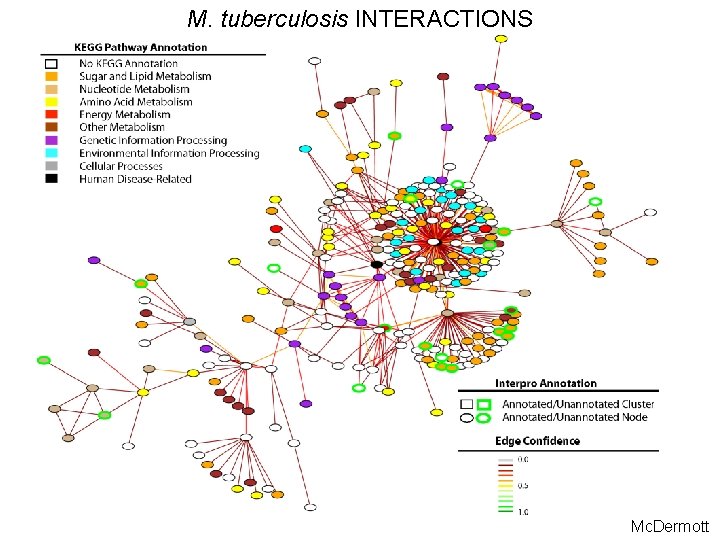
M. tuberculosis INTERACTIONS Mc. Dermott

C. elegans INTERACTIONS Mc. Dermott

H. sapiens INTERACTIONS Mc. Dermott

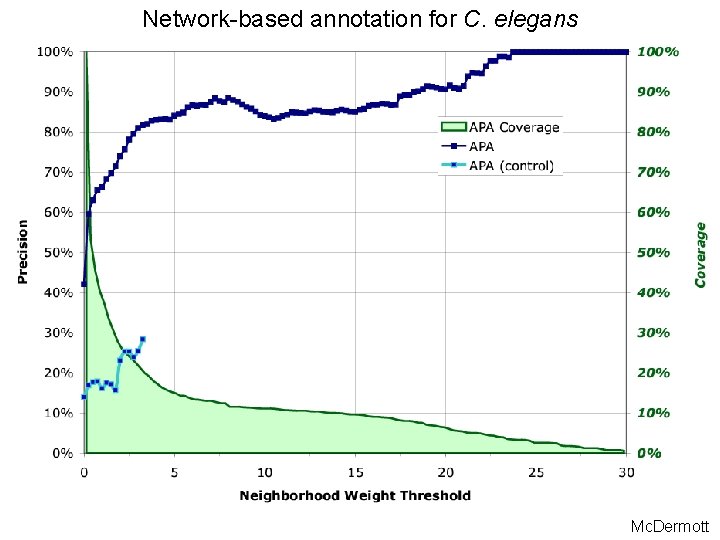
Network-based annotation for C. elegans Mc. Dermott

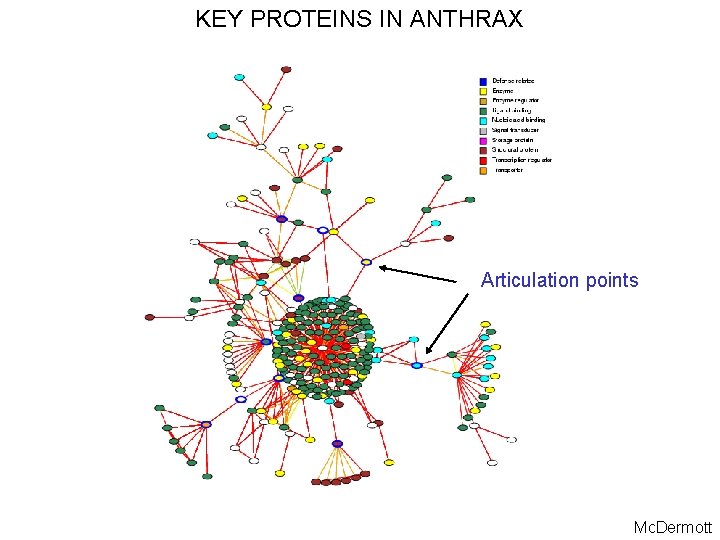
KEY PROTEINS IN ANTHRAX Articulation points Mc. Dermott

HOST PATHOGEN INTERACTIONS Mc. Dermott
 Ram samudrala
Ram samudrala Ram samudrala
Ram samudrala Promotion from assistant to associate professor
Promotion from assistant to associate professor Sruthi samudrala
Sruthi samudrala Ram nam me lin hai dekhat sabme ram
Ram nam me lin hai dekhat sabme ram Bcs membership benefits
Bcs membership benefits Tecniche associate al pensiero computazionale:
Tecniche associate al pensiero computazionale: What does this drawing indicate about the inca civilization
What does this drawing indicate about the inca civilization Lone star college nursing program requirements
Lone star college nursing program requirements Certified system engineering professional
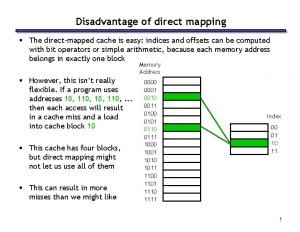
Certified system engineering professional Direct mapped cache
Direct mapped cache Lore taillieu
Lore taillieu Associate degree in the netherlands
Associate degree in the netherlands Berstoff gearbox repair
Berstoff gearbox repair Jeannie watkins
Jeannie watkins Rcog cpd portfolio
Rcog cpd portfolio Associate degree duo gift
Associate degree duo gift Analyst hierarchy
Analyst hierarchy Harper college
Harper college Iter project associate
Iter project associate Critical aad
Critical aad Harbor city community college
Harbor city community college Critical reading
Critical reading برنامهxx
برنامهxx Associate degree rmit
Associate degree rmit Cumsa certificate
Cumsa certificate Cincinnati state associate degrees
Cincinnati state associate degrees Safety associate
Safety associate Associate warden
Associate warden Sales profit chain
Sales profit chain Imeche associate membership
Imeche associate membership To associate
To associate Hea associate fellowship
Hea associate fellowship Associate consultant in capgemini
Associate consultant in capgemini Associate program
Associate program Associate consultant in capgemini
Associate consultant in capgemini Mhp associate partner gehalt
Mhp associate partner gehalt Ruckus certification training
Ruckus certification training What is word association
What is word association Marine casual uniform
Marine casual uniform