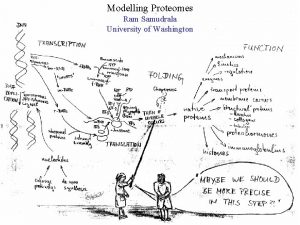
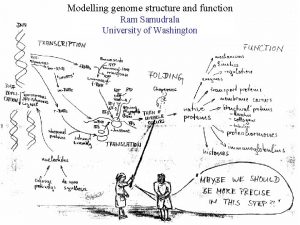
Modelling the rice proteome Ram Samudrala University of

- Slides: 28

Modelling the rice proteome Ram Samudrala University of Washington

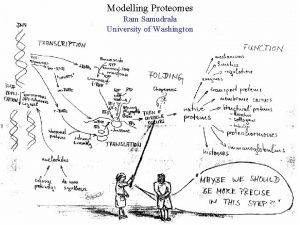
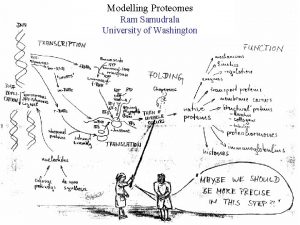
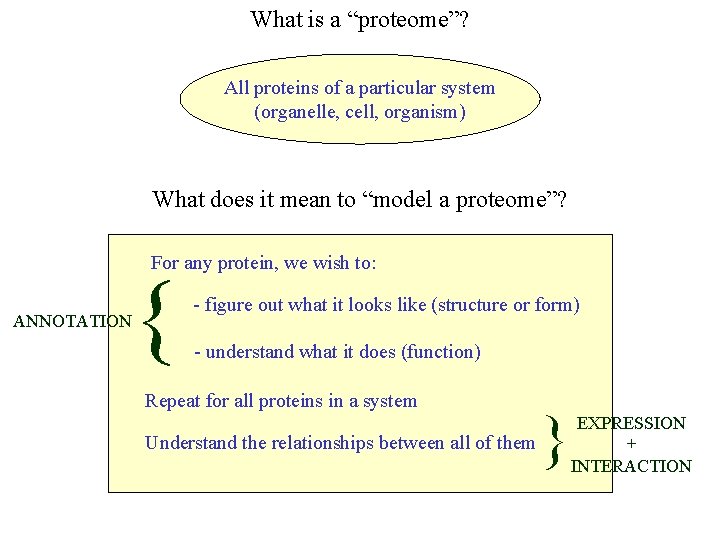
What is a “proteome”? All proteins of a particular system (organelle, cell, organism) What does it mean to “model a proteome”? For any protein, we wish to: ANNOTATION { - figure out what it looks like (structure or form) - understand what it does (function) Repeat for all proteins in a system Understand the relationships between all of them } EXPRESSION + INTERACTION

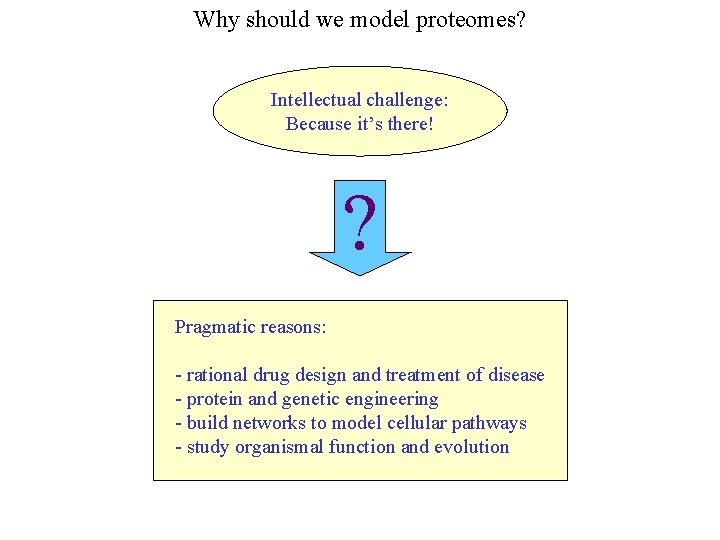
Why should we model proteomes? Intellectual challenge: Because it’s there! ? Pragmatic reasons: - rational drug design and treatment of disease - protein and genetic engineering - build networks to model cellular pathways - study organismal function and evolution

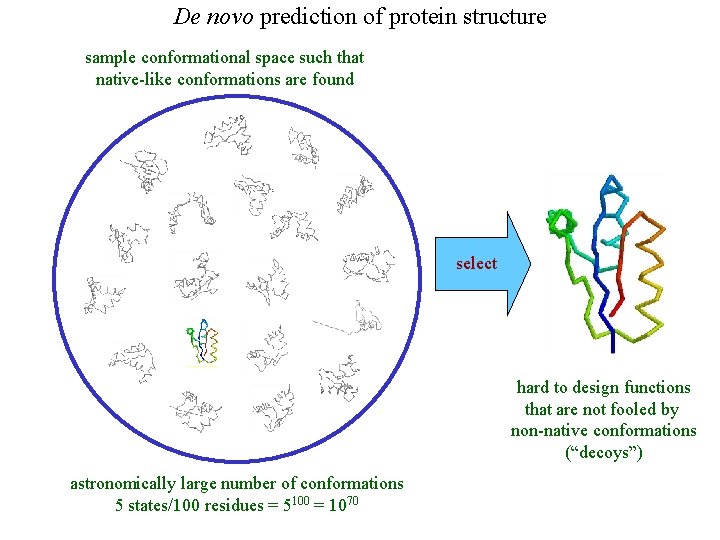
De novo prediction of protein structure sample conformational space such that native-like conformations are found select hard to design functions that are not fooled by non-native conformations (“decoys”) astronomically large number of conformations 5 states/100 residues = 5100 = 1070

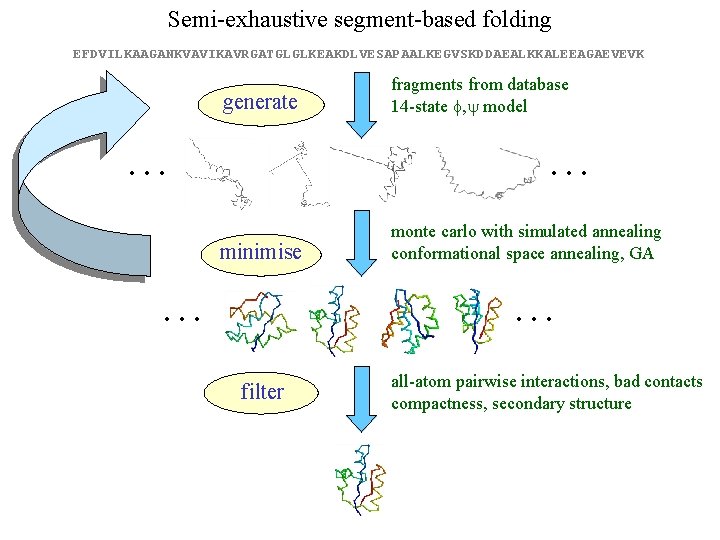
Semi-exhaustive segment-based folding EFDVILKAAGANKVAVIKAVRGATGLGLKEAKDLVESAPAALKEGVSKDDAEALKKALEEAGAEVEVK generate … fragments from database 14 -state f, y model … minimise … monte carlo with simulated annealing conformational space annealing, GA … filter all-atom pairwise interactions, bad contacts compactness, secondary structure

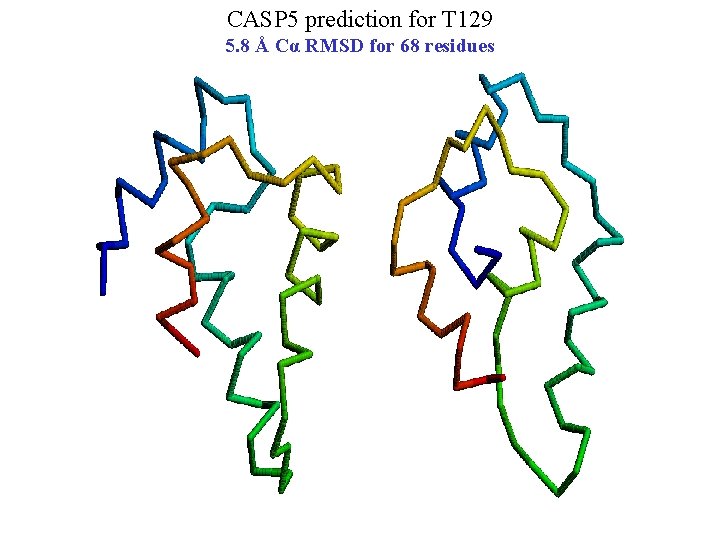
CASP 5 prediction for T 129 5. 8 Å Cα RMSD for 68 residues

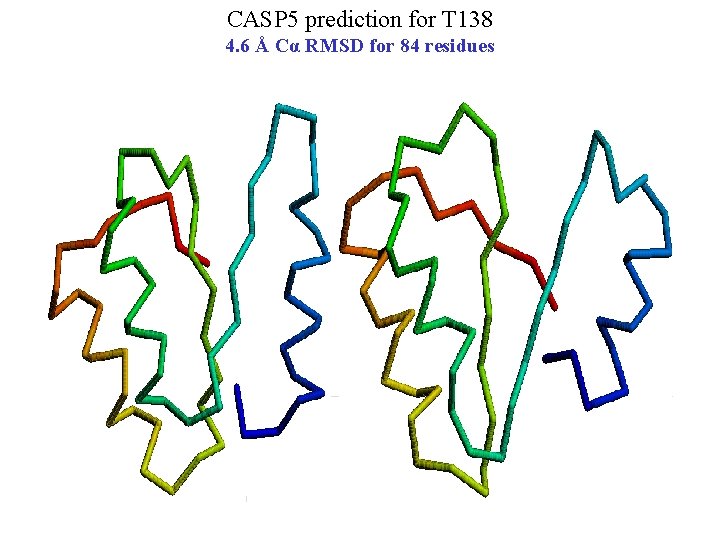
CASP 5 prediction for T 138 4. 6 Å Cα RMSD for 84 residues

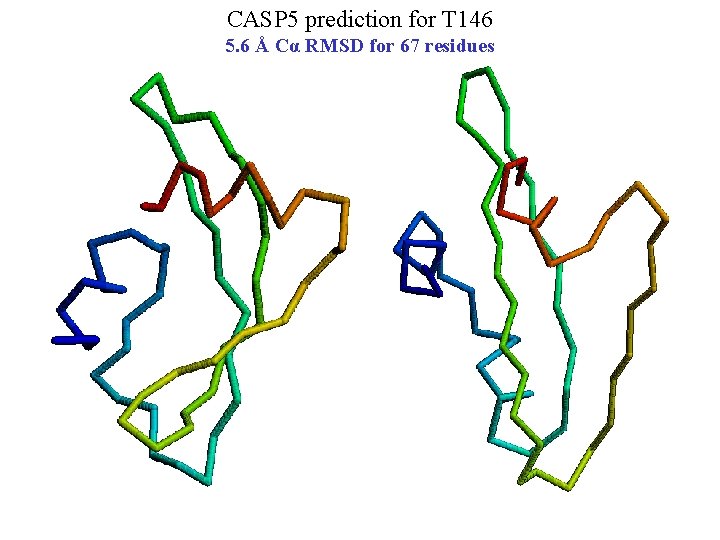
CASP 5 prediction for T 146 5. 6 Å Cα RMSD for 67 residues

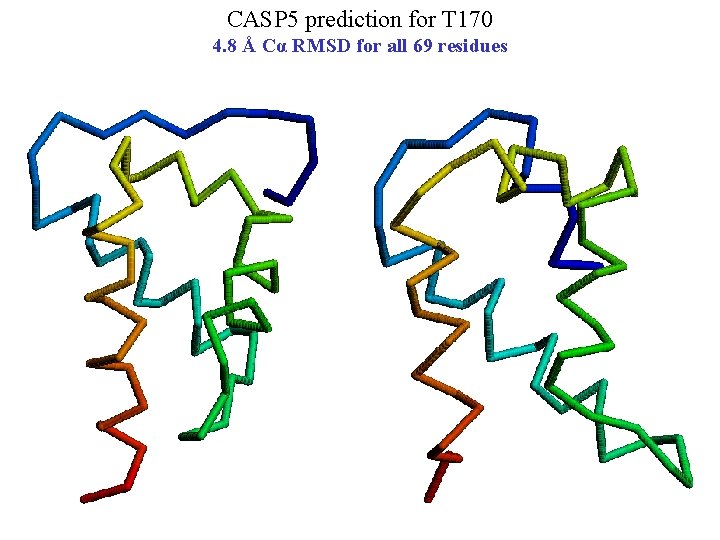
CASP 5 prediction for T 170 4. 8 Å Cα RMSD for all 69 residues

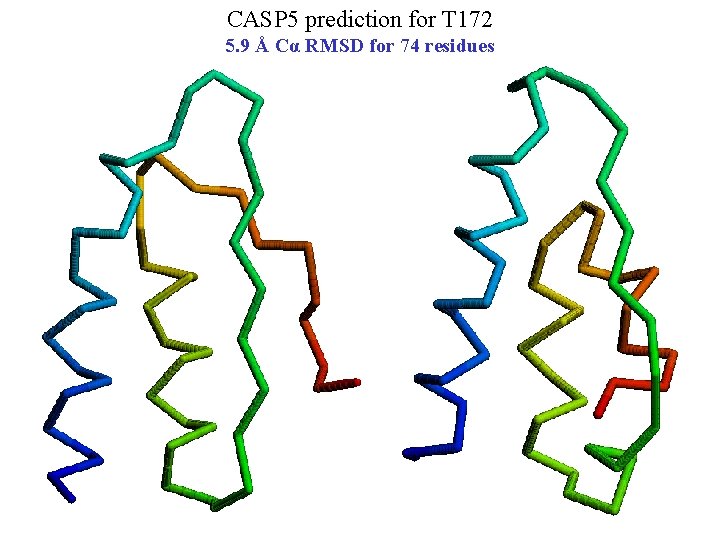
CASP 5 prediction for T 172 5. 9 Å Cα RMSD for 74 residues

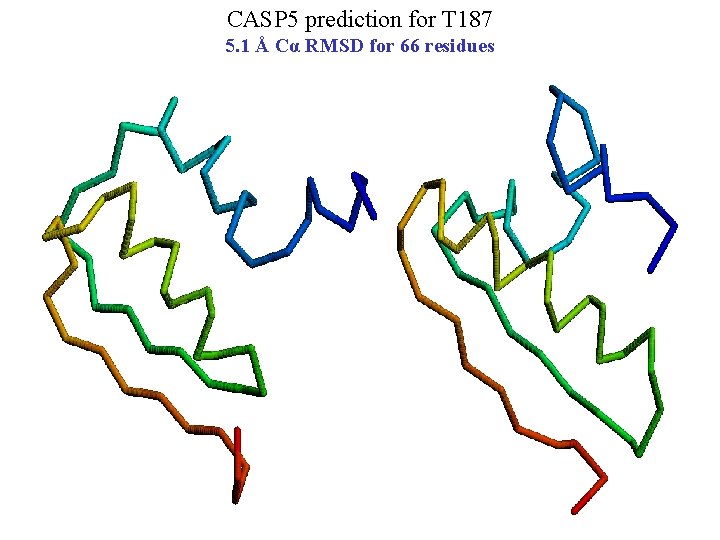
CASP 5 prediction for T 187 5. 1 Å Cα RMSD for 66 residues

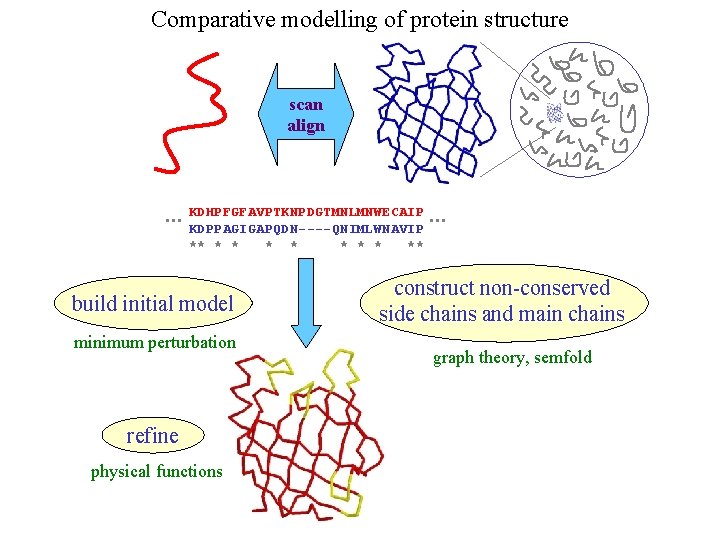
Comparative modelling of protein structure scan align … KDHPFGFAVPTKNPDGTMNLMNWECAIP KDPPAGIGAPQDN----QNIMLWNAVIP ** * * * ** build initial model minimum perturbation refine physical functions … construct non-conserved side chains and main chains graph theory, semfold

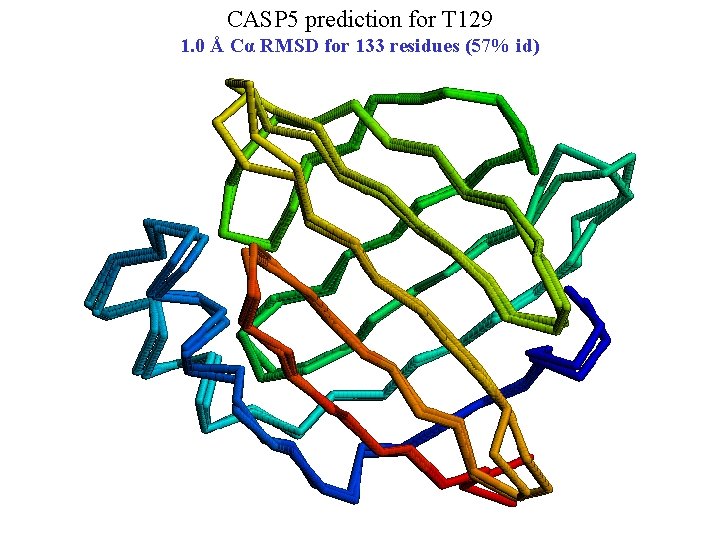
CASP 5 prediction for T 129 1. 0 Å Cα RMSD for 133 residues (57% id)

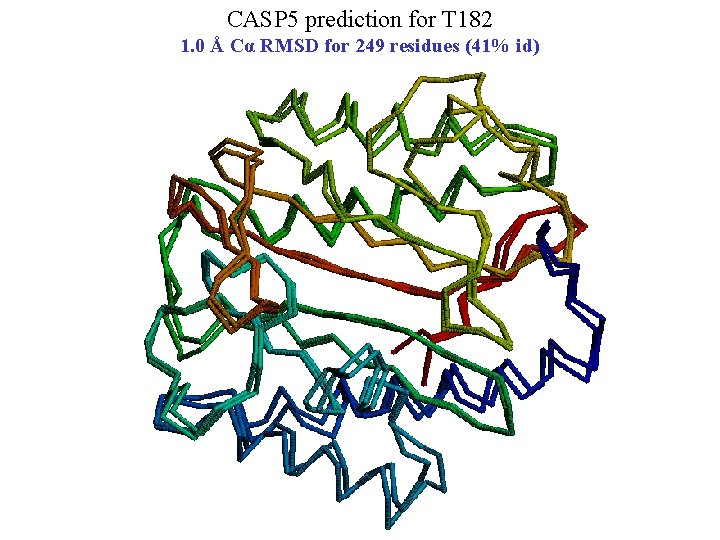
CASP 5 prediction for T 182 1. 0 Å Cα RMSD for 249 residues (41% id)

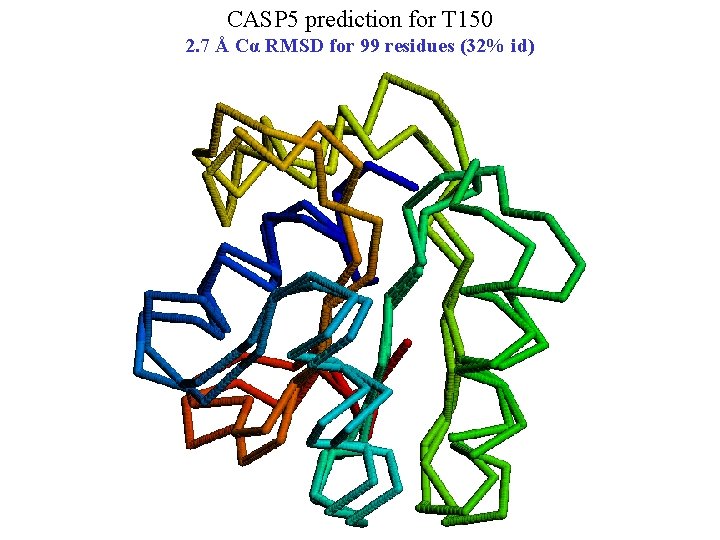
CASP 5 prediction for T 150 2. 7 Å Cα RMSD for 99 residues (32% id)

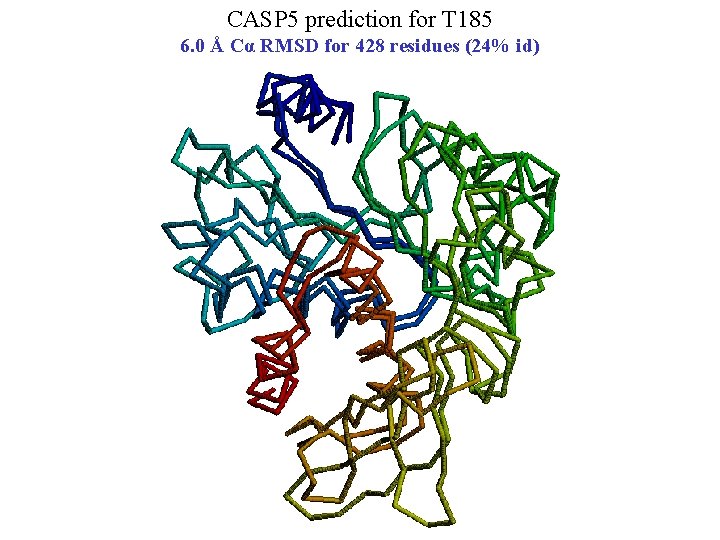
CASP 5 prediction for T 185 6. 0 Å Cα RMSD for 428 residues (24% id)

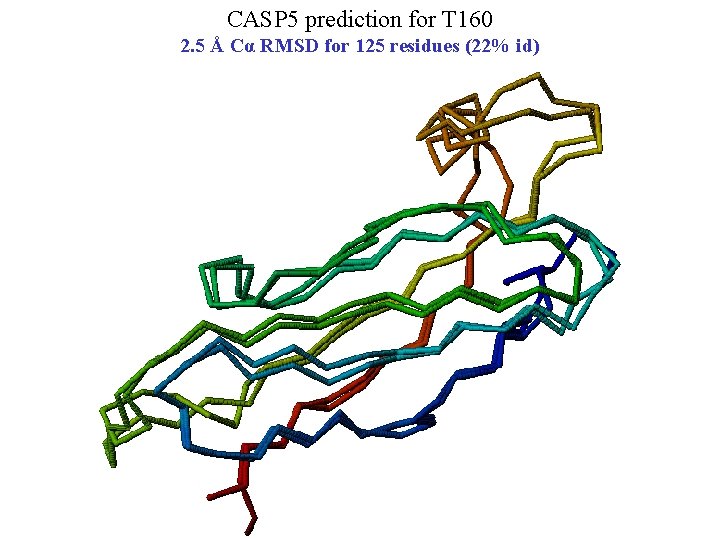
CASP 5 prediction for T 160 2. 5 Å Cα RMSD for 125 residues (22% id)

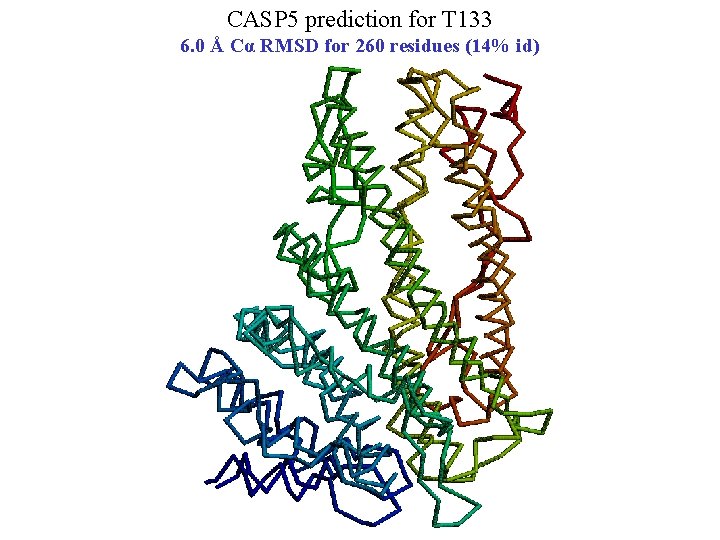
CASP 5 prediction for T 133 6. 0 Å Cα RMSD for 260 residues (14% id)

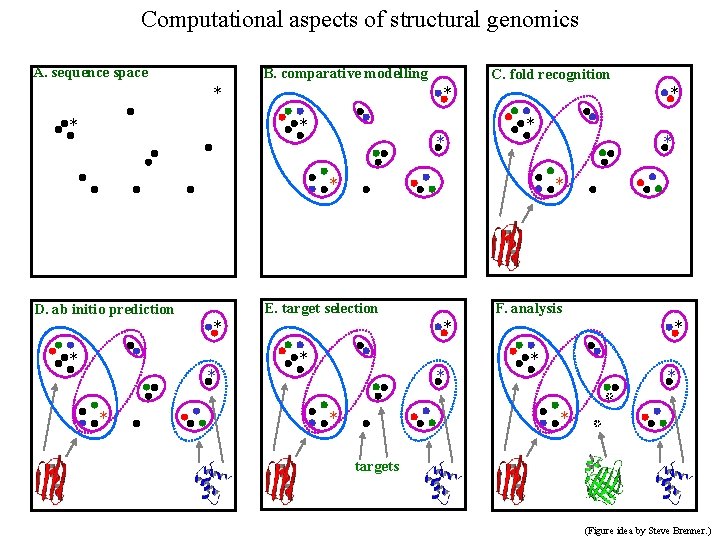
Computational aspects of structural genomics A. sequence space B. comparative modelling * * C. fold recognition * * * * E. target selection D. ab initio prediction * * F. analysis * * * * targets (Figure idea by Steve Brenner. )

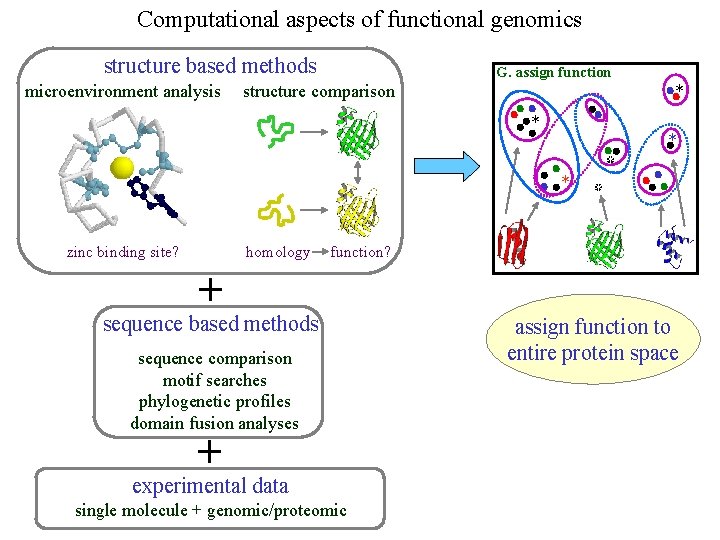
Computational aspects of functional genomics structure based methods microenvironment analysis G. assign function structure comparison * * homology zinc binding site? * * function? + sequence based methods sequence comparison motif searches phylogenetic profiles domain fusion analyses + experimental data single molecule + genomic/proteomic assign function to entire protein space

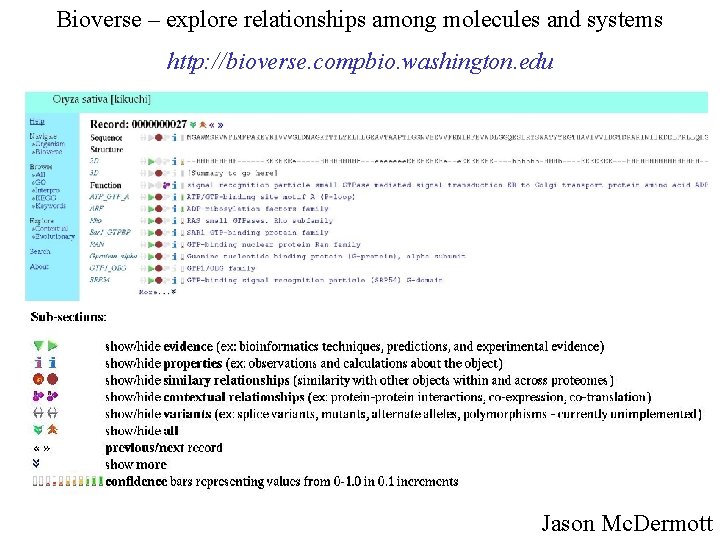
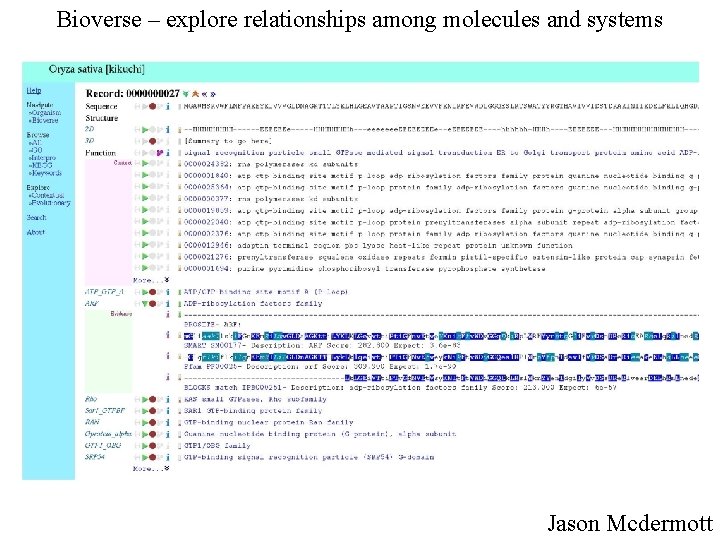
Bioverse – explore relationships among molecules and systems http: //bioverse. compbio. washington. edu Jason Mc. Dermott

Bioverse – explore relationships among molecules and systems Jason Mcdermott

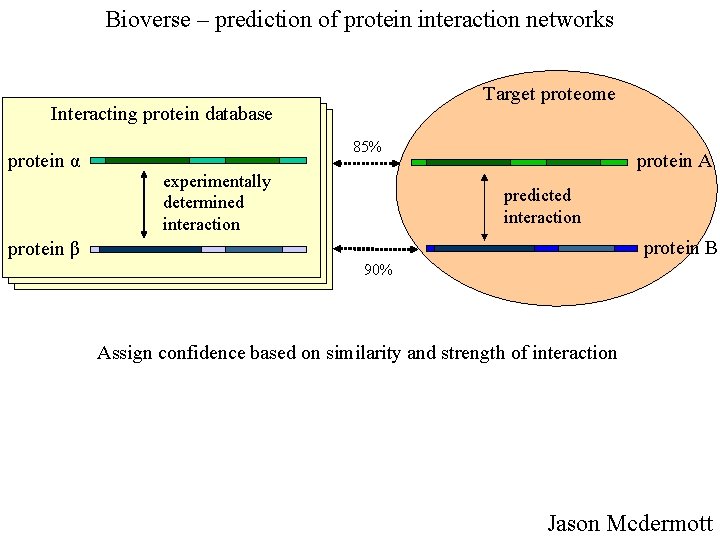
Bioverse – prediction of protein interaction networks Target proteome Interacting protein database protein α 85% experimentally determined interaction protein A predicted interaction protein B protein β 90% Assign confidence based on similarity and strength of interaction Jason Mcdermott

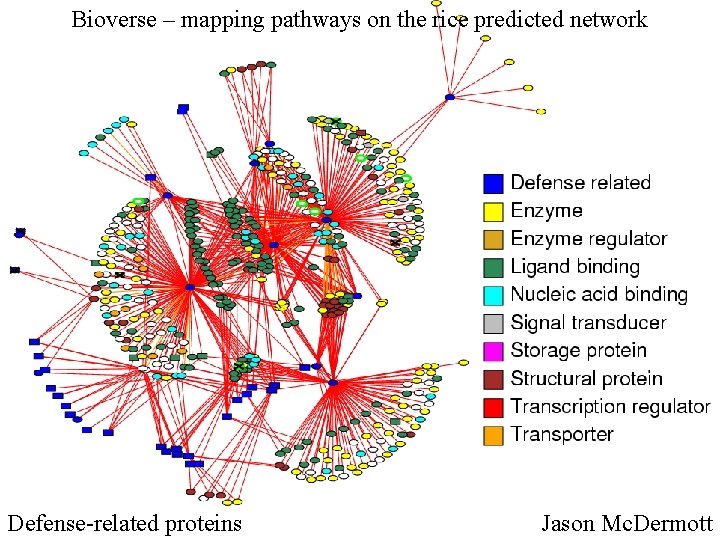
Bioverse – mapping pathways on the rice predicted network Defense-related proteins Jason Mc. Dermott

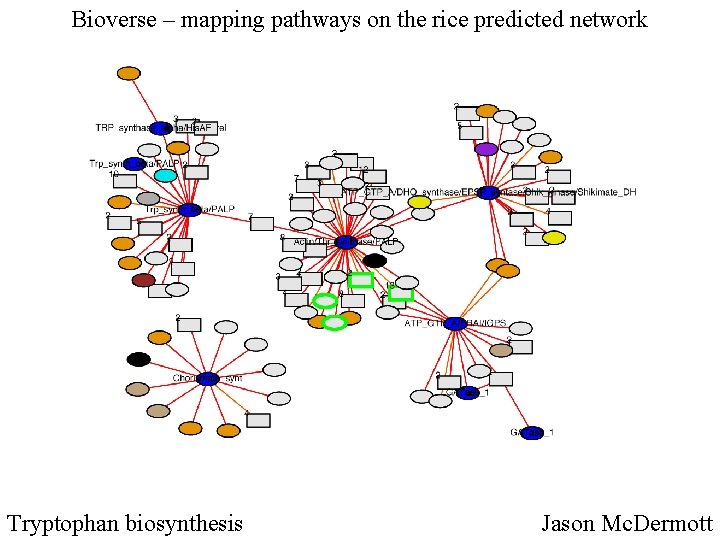
Bioverse – mapping pathways on the rice predicted network Tryptophan biosynthesis Jason Mc. Dermott

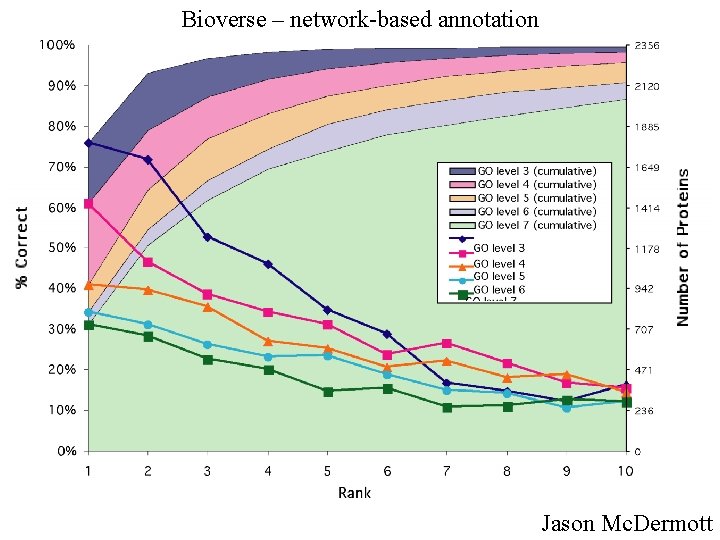
Bioverse – network-based annotation Jason Mc. Dermott

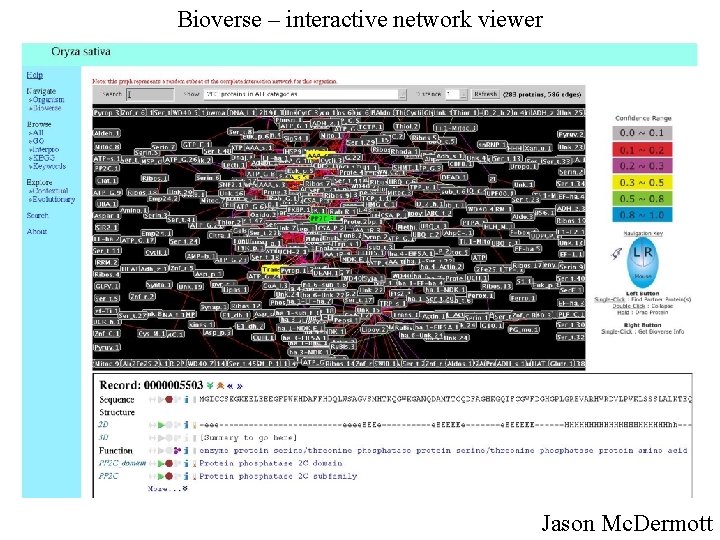
Bioverse – interactive network viewer Jason Mc. Dermott

Take home message Prediction of protein structure and function can be used to model whole genomes to understand organismal function and evolution Acknowledgements Aaron Chang Ashley Lam Ekachai Jenwitheesuk Gong Cheng Jason Mc. Dermott Kai Wang Ling-Hong Hung Lynne Townsend Marissa La. Madrid Mike Inouye Stewart Moughon Shing-Chung Ngan Yi-Ling Cheng Zach Frazier
 Ram samudrala
Ram samudrala Ram samudrala
Ram samudrala Sruthi samudrala
Sruthi samudrala Ram nam me lin hai dekhat sabme ram
Ram nam me lin hai dekhat sabme ram Daniel cohan rice university
Daniel cohan rice university Environmental awareness presentation
Environmental awareness presentation What is technological modelling
What is technological modelling Homology modelling steps
Homology modelling steps Jsimgraph
Jsimgraph Molecular modelling laboratory
Molecular modelling laboratory Modelling madness what's new
Modelling madness what's new Richer interaction in hci
Richer interaction in hci Lbo case study
Lbo case study Wire frame modeling advantages
Wire frame modeling advantages Homology modelling steps
Homology modelling steps Hair modelling
Hair modelling What is edlc in embedded system
What is edlc in embedded system Homology modelling steps
Homology modelling steps Exercise 4
Exercise 4 Mathematical modelling of electrical systems
Mathematical modelling of electrical systems Modelling rich interaction
Modelling rich interaction Unit 5 data modelling assignment 2
Unit 5 data modelling assignment 2 Is graphical modelling tool for structured analysis
Is graphical modelling tool for structured analysis Algorithmic cost modelling
Algorithmic cost modelling Class responsibility collaborator modelling
Class responsibility collaborator modelling State modelling
State modelling Object oriented modelling
Object oriented modelling Multijet modeling
Multijet modeling Cognitive modelling
Cognitive modelling