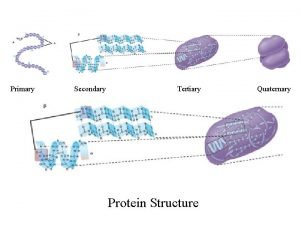
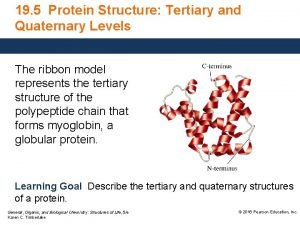
Modelling protein tertiary structure Ram Samudrala University of
- Slides: 7

Modelling protein tertiary structure Ram Samudrala University of Washington

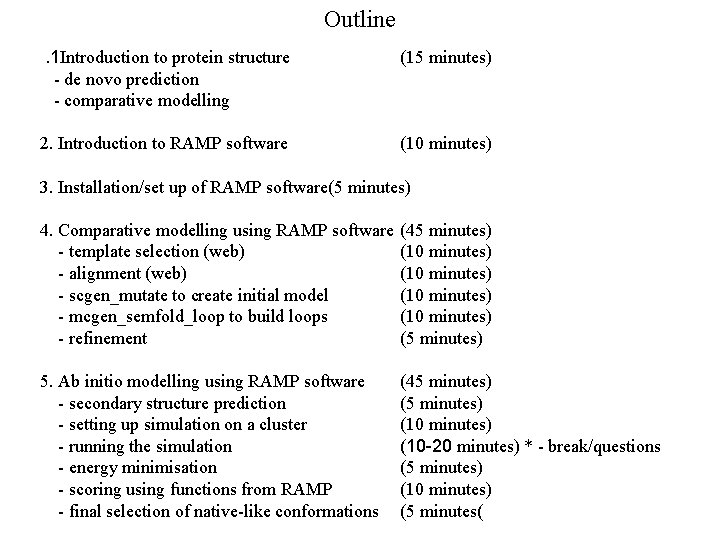
Outline. 1 Introduction to protein structure - de novo prediction - comparative modelling (15 minutes) 2. Introduction to RAMP software (10 minutes) 3. Installation/set up of RAMP software(5 minutes) 4. Comparative modelling using RAMP software - template selection (web) - alignment (web) - scgen_mutate to create initial model - mcgen_semfold_loop to build loops - refinement (45 minutes) (10 minutes) (5 minutes) 5. Ab initio modelling using RAMP software - secondary structure prediction - setting up simulation on a cluster - running the simulation - energy minimisation - scoring using functions from RAMP - final selection of native-like conformations (45 minutes) (10 -20 minutes) * - break/questions (5 minutes) (10 minutes) (5 minutes(

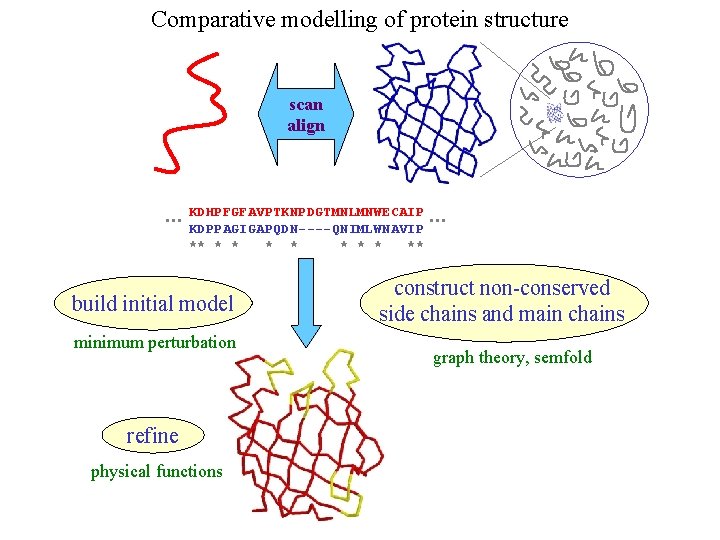
Comparative modelling of protein structure scan align … KDHPFGFAVPTKNPDGTMNLMNWECAIP KDPPAGIGAPQDN----QNIMLWNAVIP ** * * * ** build initial model minimum perturbation refine physical functions … construct non-conserved side chains and main chains graph theory, semfold

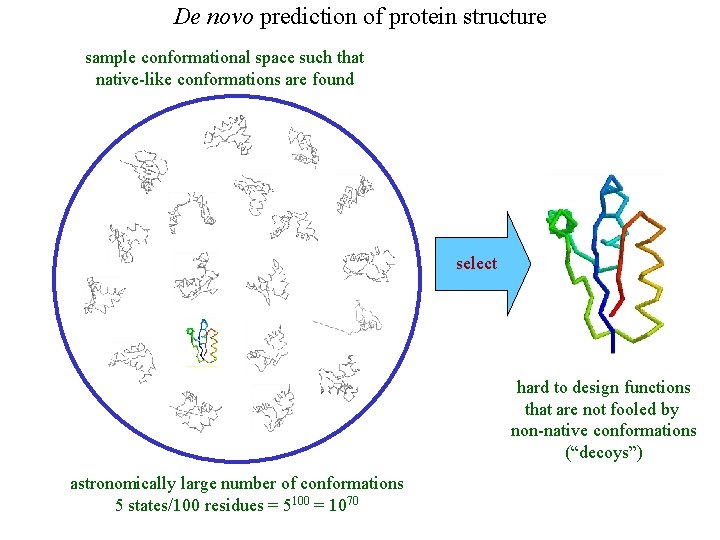
De novo prediction of protein structure sample conformational space such that native-like conformations are found select hard to design functions that are not fooled by non-native conformations (“decoys”) astronomically large number of conformations 5 states/100 residues = 5100 = 1070

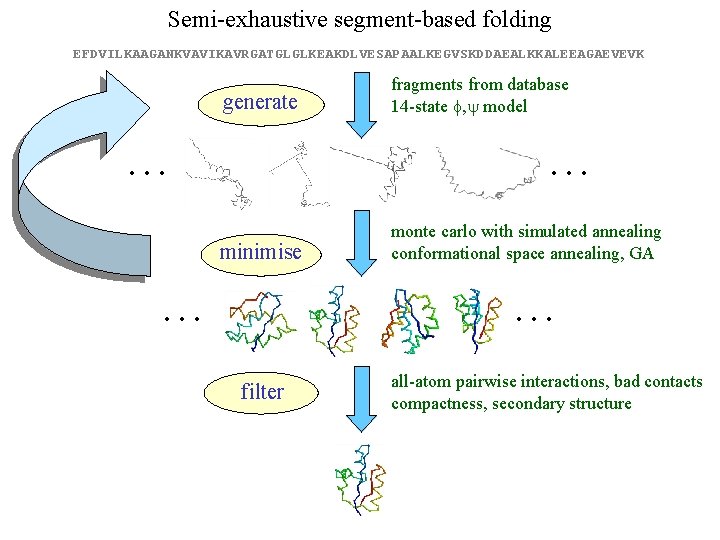
Semi-exhaustive segment-based folding EFDVILKAAGANKVAVIKAVRGATGLGLKEAKDLVESAPAALKEGVSKDDAEALKKALEEAGAEVEVK generate … fragments from database 14 -state f, y model … minimise … monte carlo with simulated annealing conformational space annealing, GA … filter all-atom pairwise interactions, bad contacts compactness, secondary structure

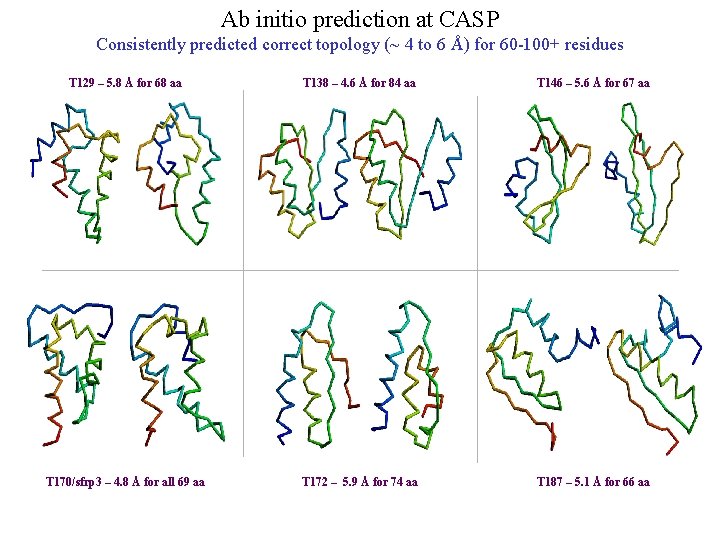
Ab initio prediction at CASP Consistently predicted correct topology (~ 4 to 6 Å) for 60 -100+ residues T 129 – 5. 8 Å for 68 aa T 138 – 4. 6 Å for 84 aa T 146 – 5. 6 Å for 67 aa T 170/sfrp 3 – 4. 8 Å for all 69 aa T 172 – 5. 9 Å for 74 aa T 187 – 5. 1 Å for 66 aa

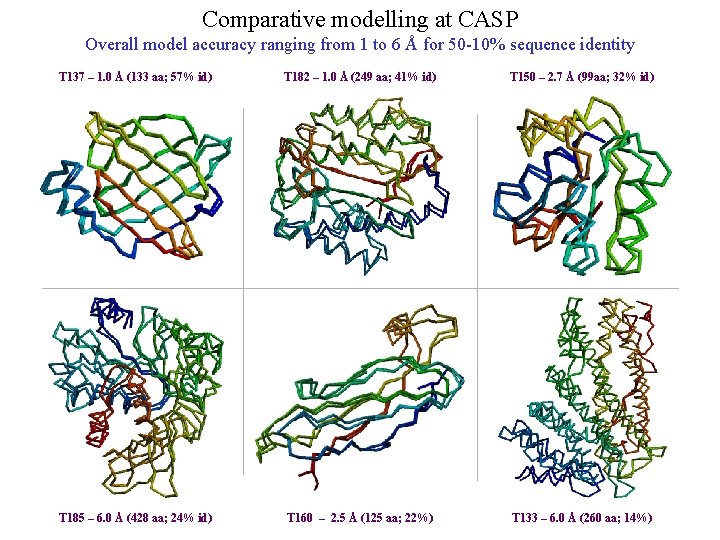
Comparative modelling at CASP Overall model accuracy ranging from 1 to 6 Å for 50 -10% sequence identity T 137 – 1. 0 Å (133 aa; 57% id) T 182 – 1. 0 Å (249 aa; 41% id) T 150 – 2. 7 Å (99 aa; 32% id) T 185 – 6. 0 Å (428 aa; 24% id) T 160 – 2. 5 Å (125 aa; 22%) T 133 – 6. 0 Å (260 aa; 14%)