Proteomics What is a proteome Proteome Characterization Proteomic










![Value of Proteome Data • Contains info not in m. RNA! – [m. RNA] Value of Proteome Data • Contains info not in m. RNA! – [m. RNA]](https://slidetodoc.com/presentation_image/5799b0e2e89393fdf53a55e8927e0e61/image-32.jpg)
- Slides: 32

Proteomics What is a proteome? Proteome Characterization Proteomic Projects Use of the Proteome Reading: Ch 15. 2 BIO 520 Bioinformatics Jim Lund

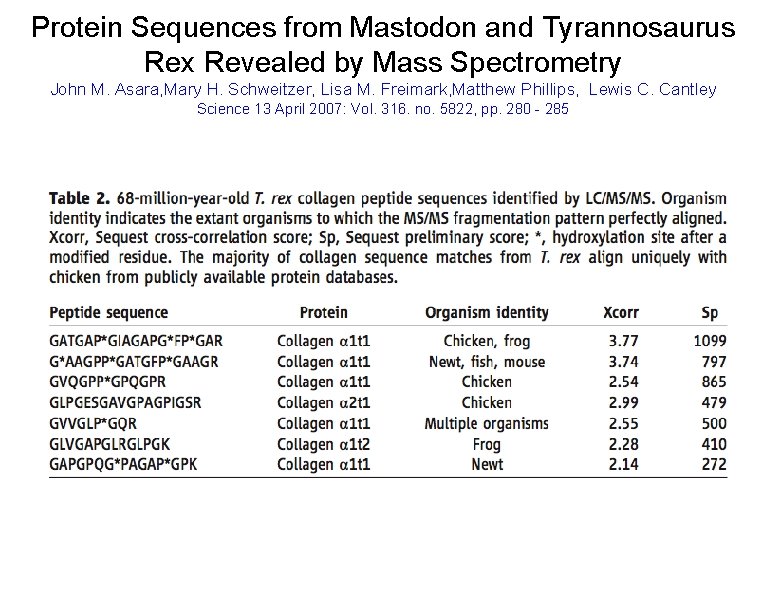
Protein Sequences from Mastodon and Tyrannosaurus Rex Revealed by Mass Spectrometry John M. Asara, Mary H. Schweitzer, Lisa M. Freimark, Matthew Phillips, Lewis C. Cantley Science 13 April 2007: Vol. 316. no. 5822, pp. 280 - 285

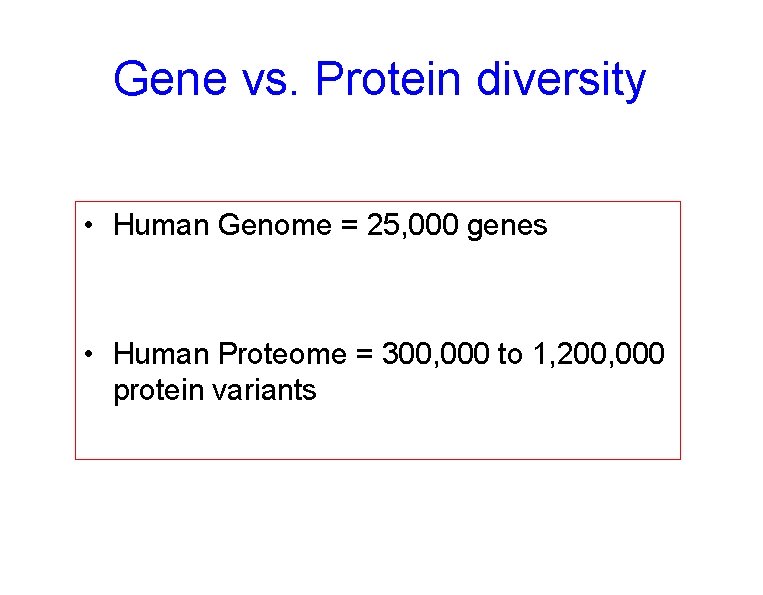
Gene vs. Protein diversity • Human Genome = 25, 000 genes • Human Proteome = 300, 000 to 1, 200, 000 protein variants

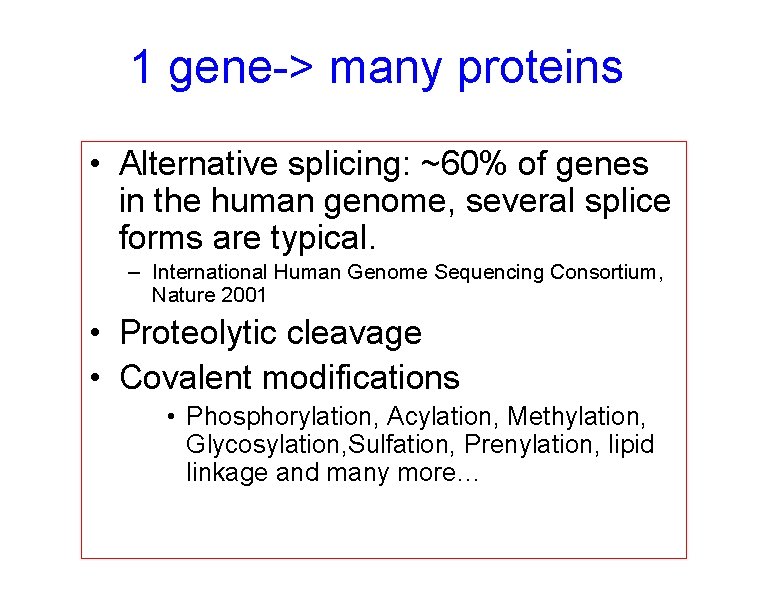
1 gene-> many proteins • Alternative splicing: ~60% of genes in the human genome, several splice forms are typical. – International Human Genome Sequencing Consortium, Nature 2001 • Proteolytic cleavage • Covalent modifications • Phosphorylation, Acylation, Methylation, Glycosylation, Sulfation, Prenylation, lipid linkage and many more…

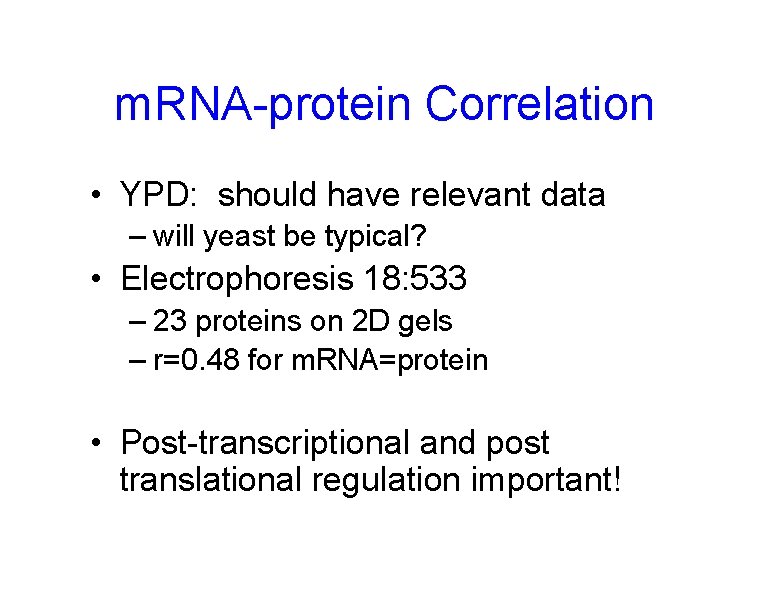
m. RNA-protein Correlation • YPD: should have relevant data – will yeast be typical? • Electrophoresis 18: 533 – 23 proteins on 2 D gels – r=0. 48 for m. RNA=protein • Post-transcriptional and post translational regulation important!

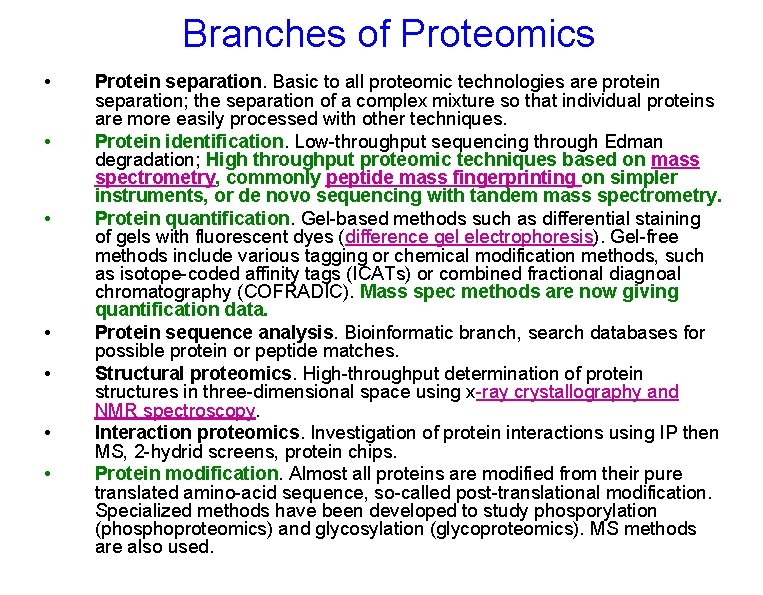
Branches of Proteomics • • Protein separation. Basic to all proteomic technologies are protein separation; the separation of a complex mixture so that individual proteins are more easily processed with other techniques. Protein identification. Low-throughput sequencing through Edman degradation; High throughput proteomic techniques based on mass spectrometry, commonly peptide mass fingerprinting on simpler instruments, or de novo sequencing with tandem mass spectrometry. Protein quantification. Gel-based methods such as differential staining of gels with fluorescent dyes (difference gel electrophoresis). Gel-free methods include various tagging or chemical modification methods, such as isotope-coded affinity tags (ICATs) or combined fractional diagnoal chromatography (COFRADIC). Mass spec methods are now giving quantification data. Protein sequence analysis. Bioinformatic branch, search databases for possible protein or peptide matches. Structural proteomics. High-throughput determination of protein structures in three-dimensional space using x-ray crystallography and NMR spectroscopy. Interaction proteomics. Investigation of protein interactions using IP then MS, 2 -hydrid screens, protein chips. Protein modification. Almost all proteins are modified from their pure translated amino-acid sequence, so-called post-translational modification. Specialized methods have been developed to study phosporylation (phosphoproteomics) and glycosylation (glycoproteomics). MS methods are also used.

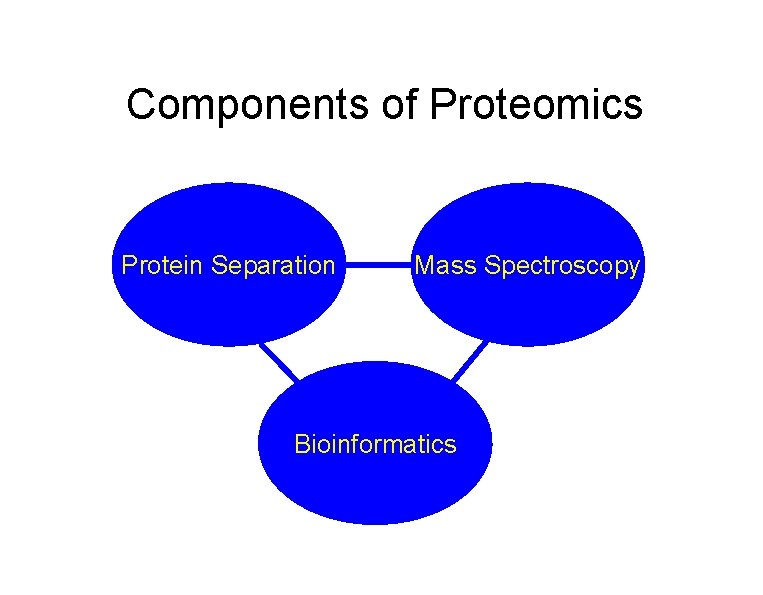
Components of Proteomics Protein Separation Mass Spectroscopy Bioinformatics

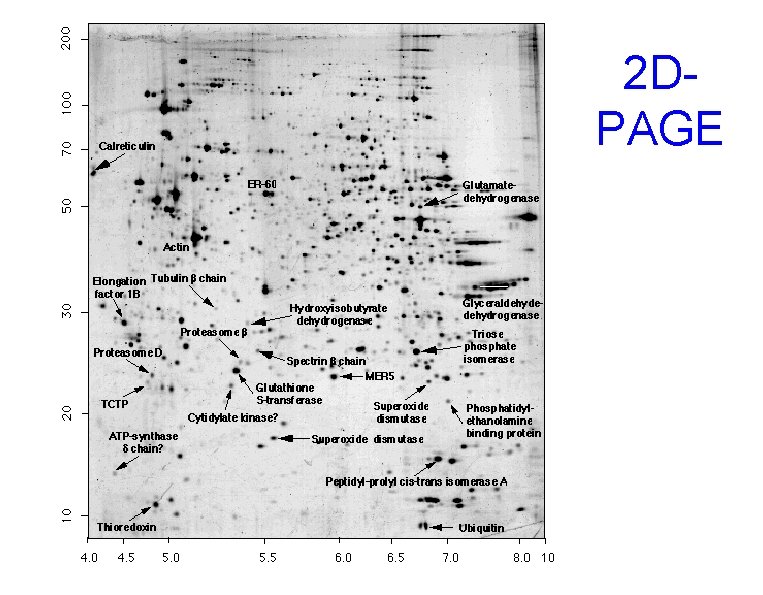
Protein separation • 2 D-PAGE – Separate proteins based on size and charge – Types of 2 D-PAGE gels: • IEF/SDS • NEPHGE/SDS • HPLC

2 DPAGE

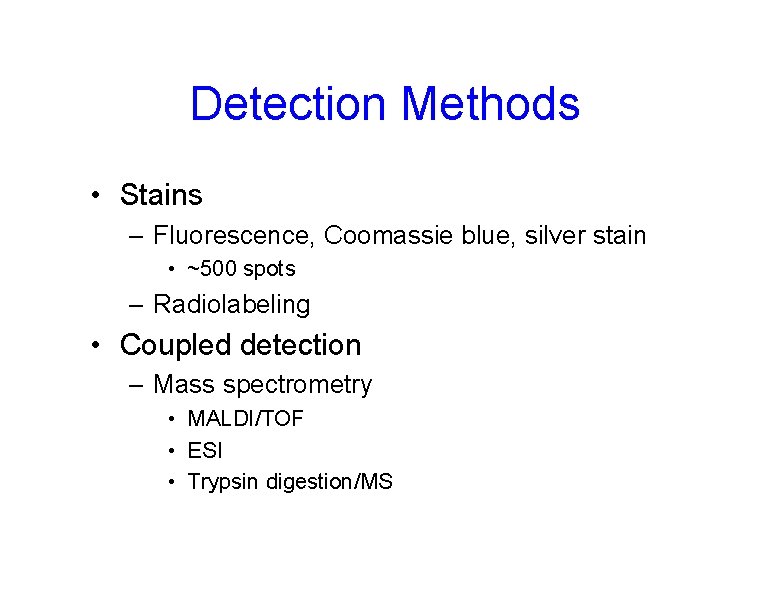
Detection Methods • Stains – Fluorescence, Coomassie blue, silver stain • ~500 spots – Radiolabeling • Coupled detection – Mass spectrometry • MALDI/TOF • ESI • Trypsin digestion/MS

Instrumentation • 2 D gels – Simple • MALDI-TOF and variants – $200, 000+, benchtop

2 D Gel Results • Swiss. Prot – www. expasy. ch/ch 2 d/ • 2 DWG Meta-database of 2 D-gels – http: //www-lecb. ncifcrf. gov/2 dwg. DB/

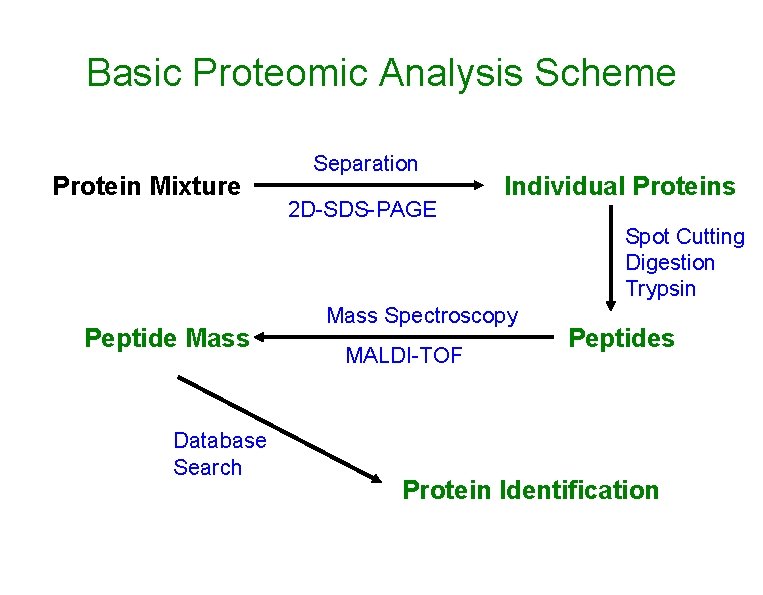
Basic Proteomic Analysis Scheme Protein Mixture Separation 2 D-SDS-PAGE Individual Proteins Spot Cutting Digestion Trypsin Peptide Mass Database Search Mass Spectroscopy MALDI-TOF Peptides Protein Identification

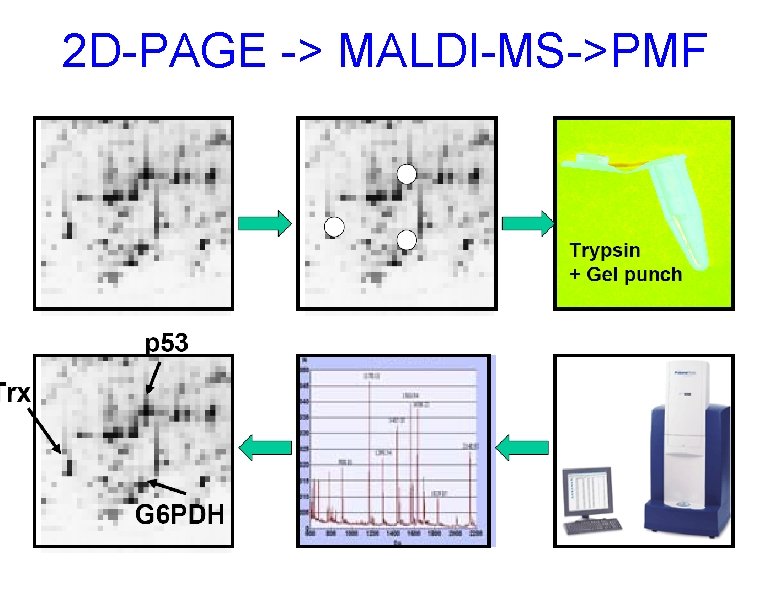
2 D-PAGE -> MALDI-MS->PMF

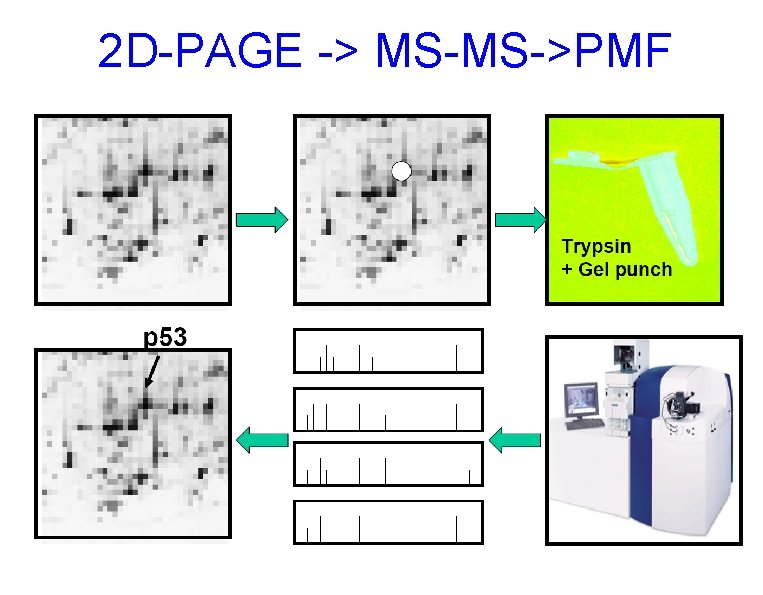
2 D-PAGE -> MS-MS->PMF

MS-MS

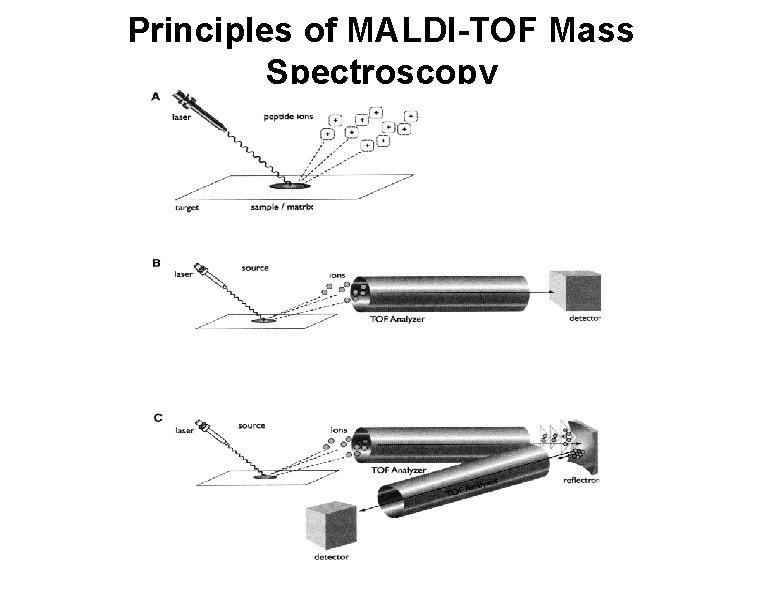
Principles of MALDI-TOF Mass Spectroscopy

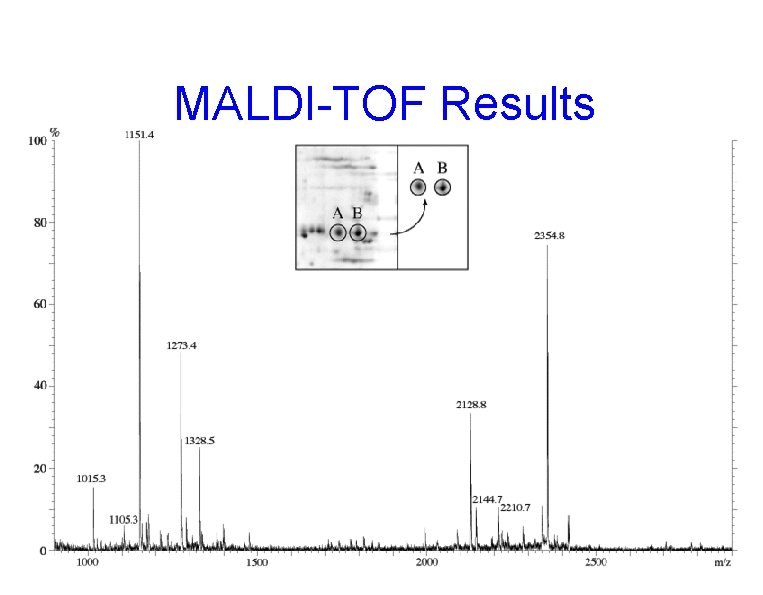
MALDI-TOF Results

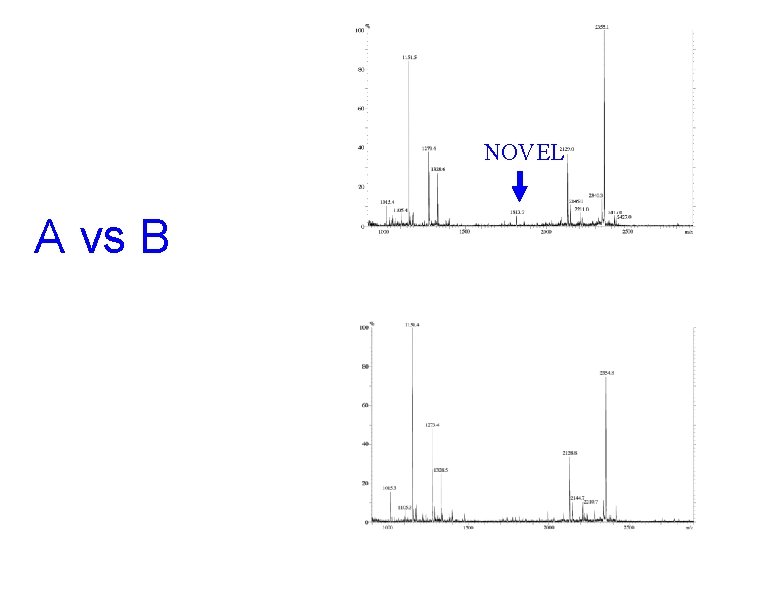
NOVEL A vs B

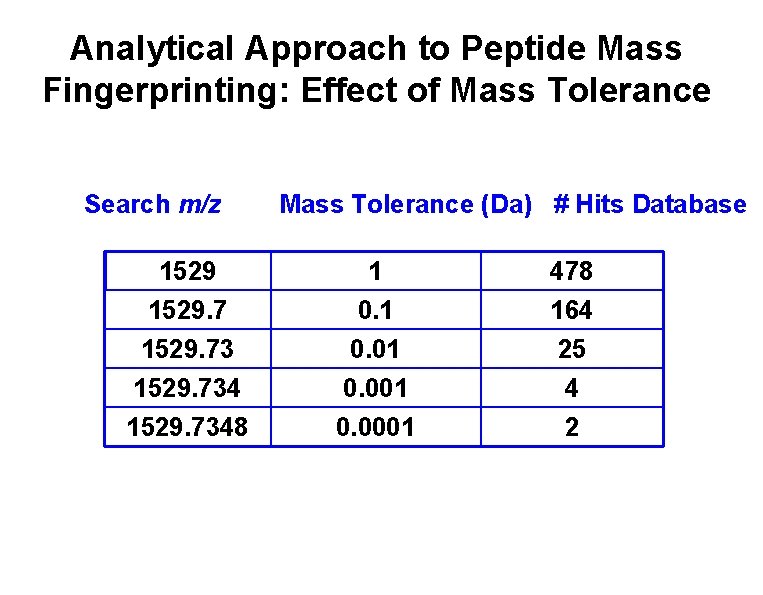
Analytical Approach to Peptide Mass Fingerprinting: Effect of Mass Tolerance Search m/z 1529. 734 1529. 7348 Mass Tolerance (Da) # Hits Database 1 0. 01 0. 0001 478 164 25 4 2

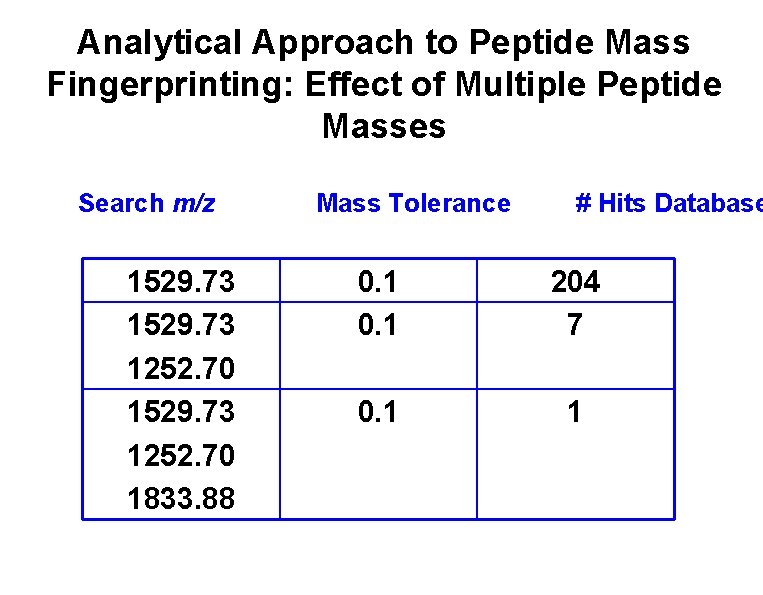
Analytical Approach to Peptide Mass Fingerprinting: Effect of Multiple Peptide Masses Search m/z 1529. 73 1252. 70 1833. 88 Mass Tolerance # Hits Database 0. 1 204 7 0. 1 1

Peptide Mass Fingerprinting • Find peptide fragments from MS spectra. – Charge state deconvolution. • Make peptide fragment list. • Check versus list of all possible polypeptides. • Need to have Protein database!

Methanol m/z spectra

Peptide Mass Fingerprinting

Protein Sequences from Mastodon and Tyrannosaurus Rex Revealed by Mass Spectrometry

Peptide Search Programs • Mascot – http: //www. matrixscience. com/ • SEQUEST – http: //fields. scripps. edu/sequest/ (Thermo Scientific) • X!Tandem – http: //thegpm. org/ – Free and Open Source • Pept. Ident – Uses Swiss-Prot protein database – http: //au. expasy. org/tools/peptident. html – Free and Open Source • Protein. Prospector – Searches user supplied protein database. – http: //falcon. ludwig. ucl. ac. uk/mshome 3. 2. htm – Free and Open Source • GFS – Uses raw genome sequence. – http: //gfs. unc. edu/cgi-bin/Web. Objects/GFSWeb – Free and Open Source

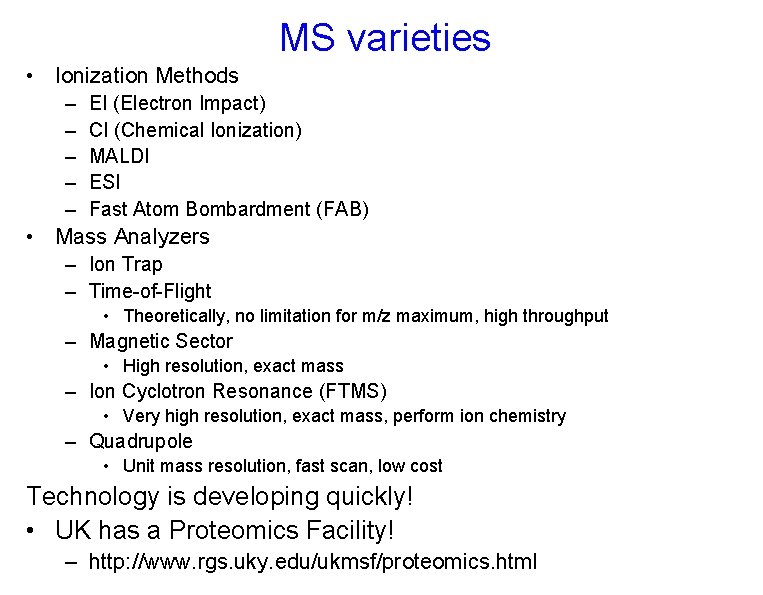
MS varieties • Ionization Methods – – – EI (Electron Impact) CI (Chemical Ionization) MALDI ESI Fast Atom Bombardment (FAB) • Mass Analyzers – Ion Trap – Time-of-Flight • Theoretically, no limitation for m/z maximum, high throughput – Magnetic Sector • High resolution, exact mass – Ion Cyclotron Resonance (FTMS) • Very high resolution, exact mass, perform ion chemistry – Quadrupole • Unit mass resolution, fast scan, low cost Technology is developing quickly! • UK has a Proteomics Facility! – http: //www. rgs. uky. edu/ukmsf/proteomics. html

Searching parameters • Modifications (e. g. cysteine residues, etc. ) • Number of allowable missed cleavages • Data properties: monoisotopic/average, charge state, amino acid composition • MS/MS data

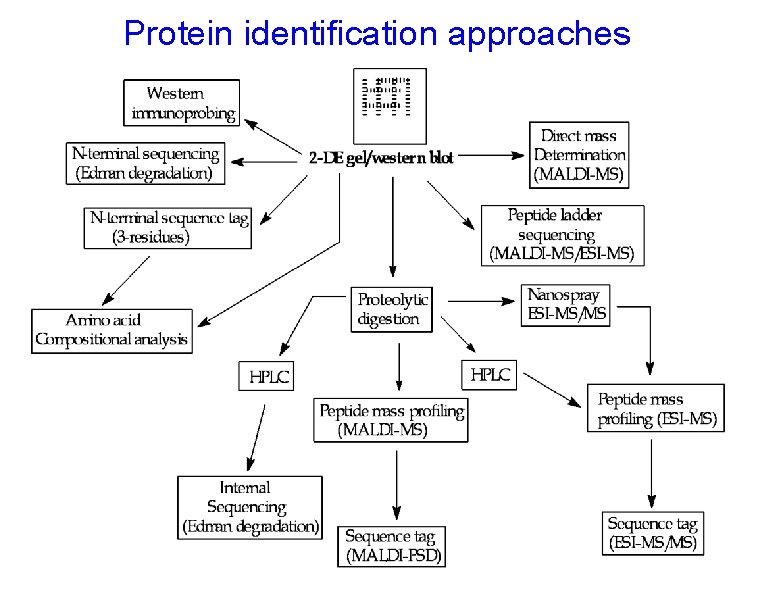
Protein identification approaches

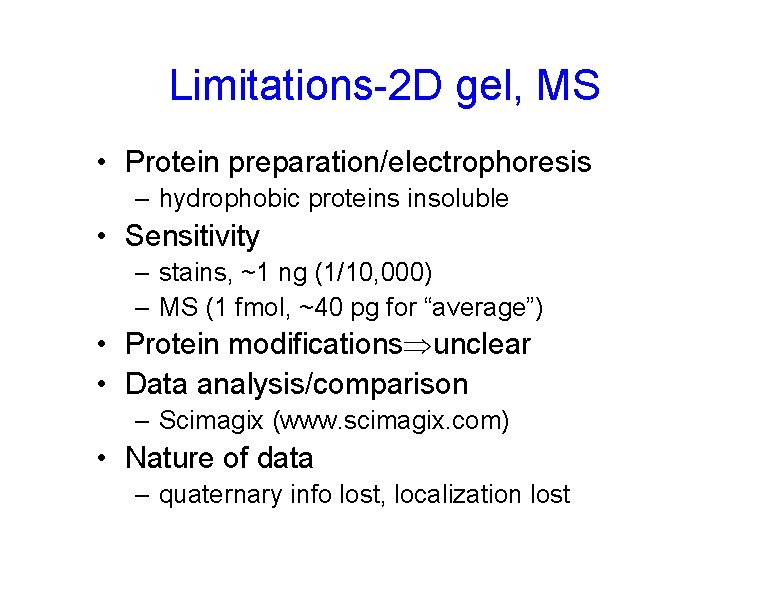
Limitations-2 D gel, MS • Protein preparation/electrophoresis – hydrophobic proteins insoluble • Sensitivity – stains, ~1 ng (1/10, 000) – MS (1 fmol, ~40 pg for “average”) • Protein modifications unclear • Data analysis/comparison – Scimagix (www. scimagix. com) • Nature of data – quaternary info lost, localization lost

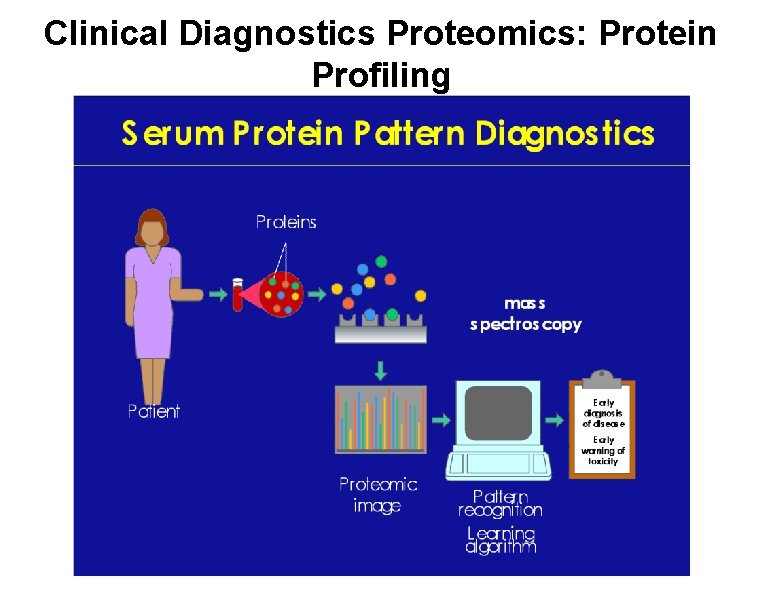
Clinical Diagnostics Proteomics: Protein Profiling
![Value of Proteome Data Contains info not in m RNA m RNA Value of Proteome Data • Contains info not in m. RNA! – [m. RNA]](https://slidetodoc.com/presentation_image/5799b0e2e89393fdf53a55e8927e0e61/image-32.jpg)
Value of Proteome Data • Contains info not in m. RNA! – [m. RNA] != [protein] – Covalent modification of proteins critical to regulation, often with constant expression – Association state of proteins critical • How can we use this information?