1 INTRODUCTION Proteomics Tech Overview Yonsei Proteome Research
























- Slides: 45

프로테옴 연구동향과 정부의 정책방향 1. INTRODUCTION: Proteomics Tech Overview 백융기 Yonsei Proteome Research Center, Yonsei University, Seoul, Korea 120 -749 http: //kr. expasy. org, www. proteomix. org, paikyk@yonsei. ac. kr

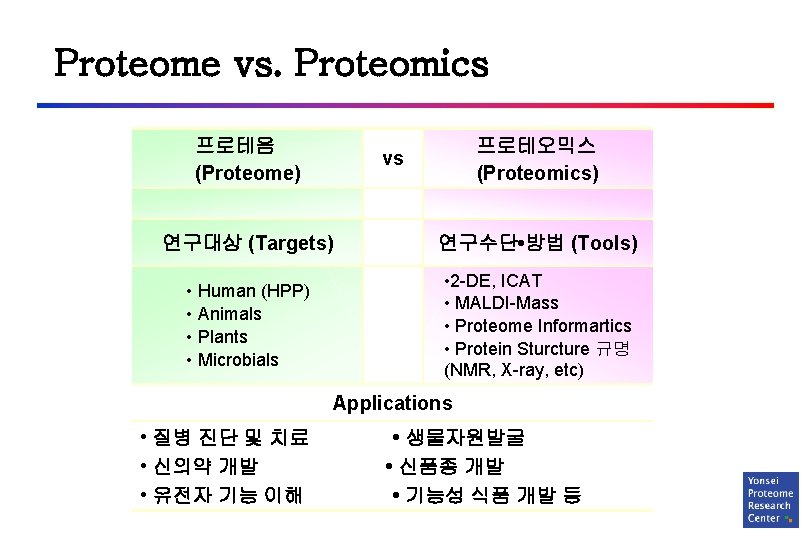
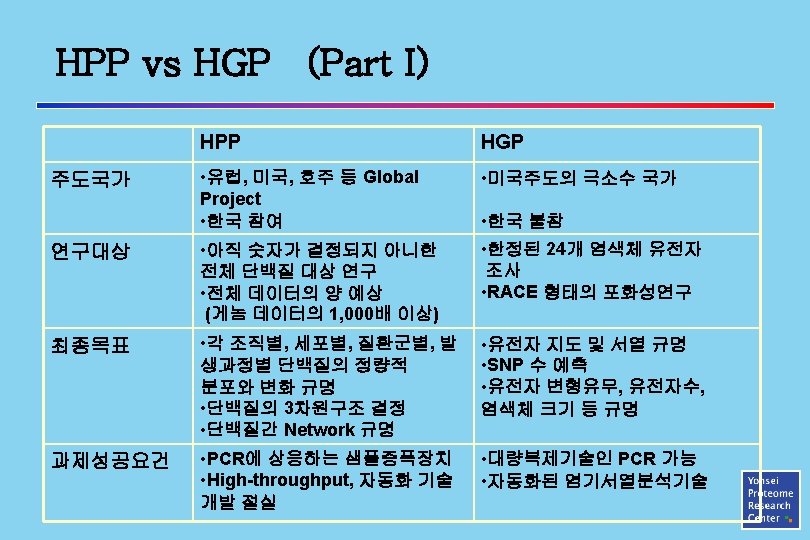
Proteome vs. Proteomics 프로테옴 (Proteome) 프로테오믹스 (Proteomics) vs 연구대상 (Targets) 연구수단 • 방법 (Tools) • Human (HPP) • Animals • Plants • Microbials • 2 -DE, ICAT • MALDI-Mass • Proteome Informartics • Protein Sturcture 규명 (NMR, X-ray, etc) Applications • 질병 진단 및 치료 • 신의약 개발 • 유전자 기능 이해 • 생물자원발굴 • 신품종 개발 • 기능성 식품 개발 등

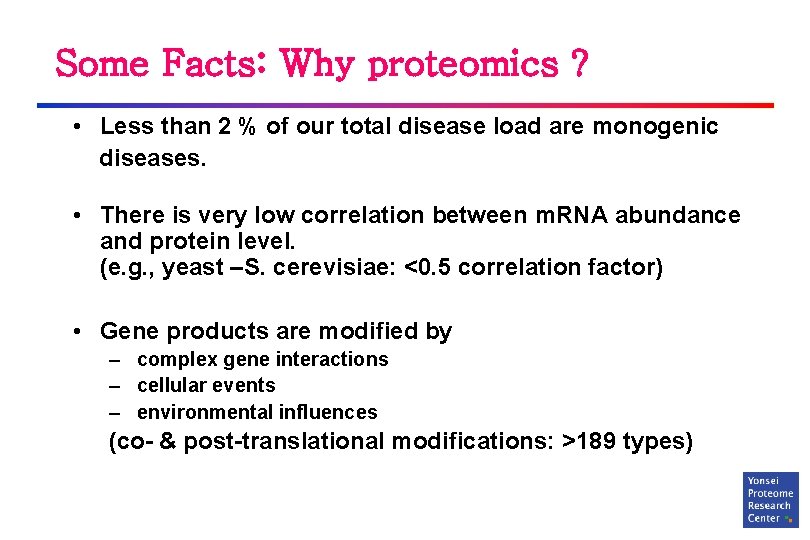
Some Facts: Why proteomics ? • Less than 2 % of our total disease load are monogenic diseases. • There is very low correlation between m. RNA abundance and protein level. (e. g. , yeast –S. cerevisiae: <0. 5 correlation factor) • Gene products are modified by – complex gene interactions – cellular events – environmental influences (co- & post-translational modifications: >189 types)

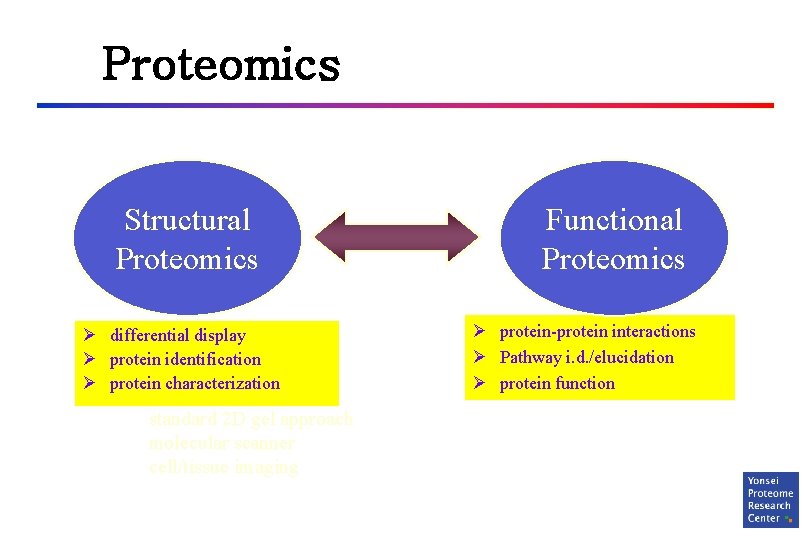
Proteomics Structural Proteomics Ø differential display Ø protein identification Ø protein characterization standard 2 D gel approach molecular scanner cell/tissue imaging Functional Proteomics Ø protein-protein interactions Ø Pathway i. d. /elucidation Ø protein function

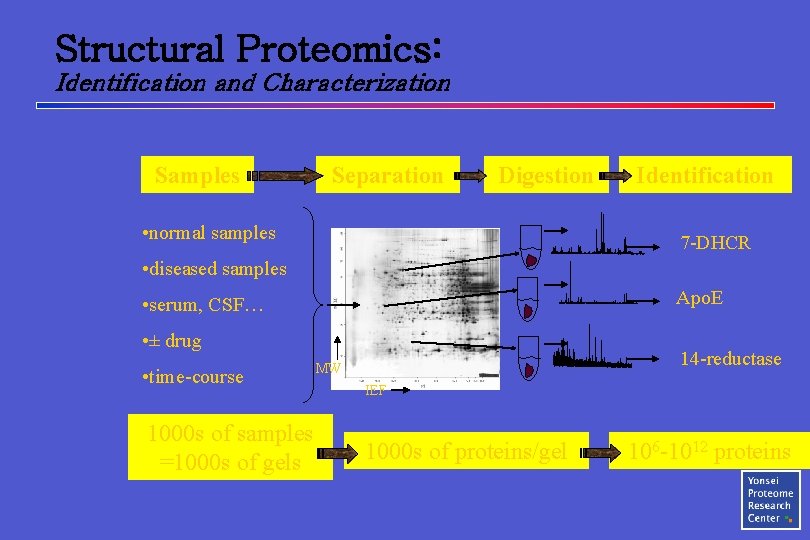
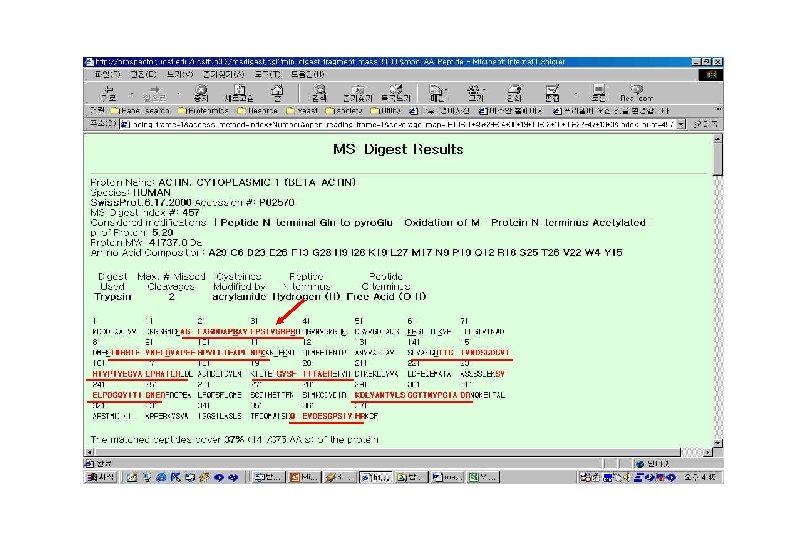
Structural Proteomics: Identification and Characterization Samples Separation Digestion • normal samples Identification 7 -DHCR • diseased samples Apo. E • serum, CSF… • ± drug • time-course 1000 s of samples =1000 s of gels 14 -reductase MW IEF 1000 s of proteins/gel 106 -1012 proteins

Principle of 2 -D Electrophoresis 1. First dimension: denaturing isoelectric focusing separation according to the p. I 2. Second dimension: SDS electrophoresis separation according to the MW The 2 -D electrophoresis gel resolves thousands of protein spots each time.

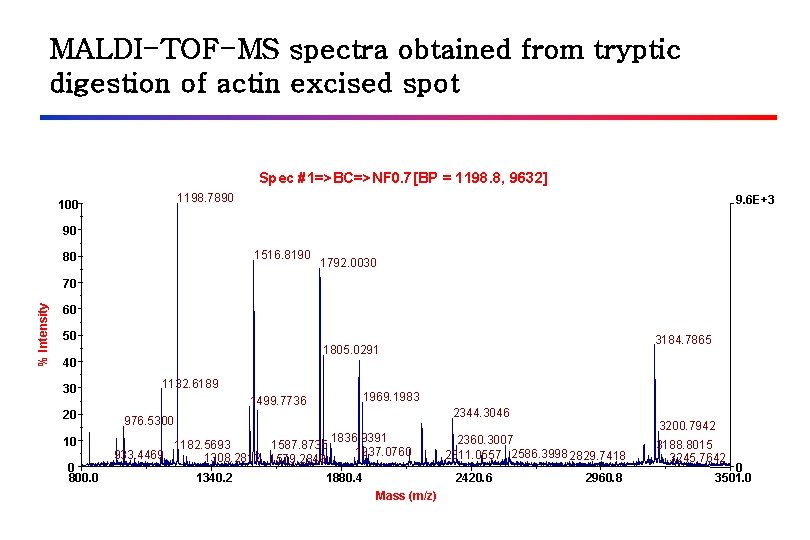
MALDI-TOF-MS spectra obtained from tryptic digestion of actin excised spot Spec #1=>BC=>NF 0. 7[BP = 1198. 8, 9632] 1198. 7890 100 9. 6 E+3 90 1516. 8190 80 1792. 0030 % Intensity 70 60 50 40 30 20 10 0 800. 0 3184. 7865 1805. 0291 1132. 6189 1969. 1983 1499. 7736 2344. 3046 976. 5300 1836. 9391 1182. 5693 1587. 8735 1937. 0760 933. 4469 1308. 2812 1579. 2849 1340. 2 1880. 4 2360. 3007 2311. 0557 2586. 3998 2829. 7418 2420. 6 Mass (m/z) 3200. 7942 2960. 8 3188. 8015 3245. 7642 0 3501. 0


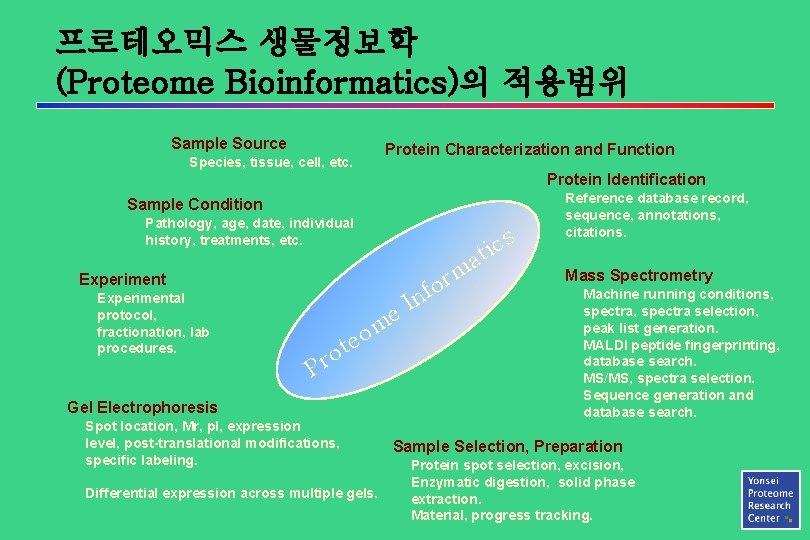
프로테오믹스 생물정보학 (Proteome Bioinformatics)의 적용범위 Sample Source Protein Characterization and Function Species, tissue, cell, etc. Protein Identification Sample Condition Pathology, age, date, individual history, treatments, etc. m s c i at Experimental protocol, fractionation, lab procedures. e m o e t o r P Gel Electrophoresis Spot location, Mr, p. I, expression level, post-translational modifications, specific labeling. Differential expression across multiple gels. In r fo Reference database record, sequence, annotations, citations. Mass Spectrometry Machine running conditions, spectra selection, peak list generation. MALDI peptide fingerprinting, database search. MS/MS, spectra selection. Sequence generation and database search. Sample Selection, Preparation Protein spot selection, excision, Enzymatic digestion, solid phase extraction. Material, progress tracking.





II. Proteomics applications, examples Studies of basic cell function and molecular organisation The discovery of molecular markers for diagnosis and monitoring disease The discovery of novel biologically active molecules and drugs The study of drug modes of actions and mechanisms of toxicity Investigations of pathophysiology Subtype individuals to predict response to therapy The discovery of novel drug targets The molecular dissection of genetic or pharmacological perturbations

프로테오믹스 관련 미국 특허사례 • USP 6, 084, 069, Autotaxin: motility stimulating protein useful in cacner diagnosis and therapy • USP 6, 027, 905, Methods for the detection of cervical cancer • USP 5, 914, 238, Materials and methods for detection of breast cancer (2 DE etc) • USP 5, 792, 744, Proteins from mammalian liver • USP 6, 127, 134, Different gel electrophoresis using matched multiple dyes • USP 6, 165, 709, Methods for drug target screening • USP 5, 976, 543, 12 -k. Da protein derived from M. Tuberculosis useful for treatment of autoimmunie disease • USP 6, 229, 911, Methods and apparatus for providing a bioinformatics database

III. Future Directions • Automations: Robotics • Highthroughput : PCR-version of proteins? -2 DE 대체 기술(ICAT, Flu-dye tech etc) • Integrated Data Base System -LIMS (SQL 4, Worksbase, YPRC-PDB etc) • Regional Cooperations (e. g. , AOHUPO) • Public Sector vs Private Companies -[NIH, MIT HGP Gr. ] vs [Celera Genomics, LSB, OGS] • Funds 1 W. P. Balckstock, M. P. Weir, TIBTECH, p 121 -127, March 1999. 2 J. E. Celis, Electrophoresis, 12, 2177 -2240, 1995.

New Trends in Proteomics Automation • Automation - High-throughput processing of spot cutting Enzyme Digestion, Molecular Scanner Sample Cleanup Robotics • Data Management - LIMS, Works. Base

Label & Quantify Isotope–Coded Affinity Tags ICAT™ • Breakthrough approach to quantitative protein expression analysis - 2 D gel alternative • Currently in Development • Test Kits available December with Pro. ICAT SW for Qstar Pulsar • Kit launch in early 2001 heavy reagent: d 8 -ICAT (X=deuterium) light reagent: d 0 -ICAT (X=hydrogen) O NH HN S biotin tag O XX N H X XO O XX O N XX H I linker (heavy or reactive group light) Emerging Technology S H cys

Protein Quantification & Identification w/ICAT Determine differences in protein expression by measuring relative intensities of light vs. heavy 100 Mixture 1 MS Light Heavy ICAT Reagentlabeled cysteines Optional fractionation Quantitation 0 550 100 570 m/z Identification MDLC or MDLC with 1 D gel Slide courtesy of Ruedi Aebersold 580 NH 2 -EACDPLR-COOH Affinity separation Digest Mixture 2 560 MS/MS 0 200 400 600 m/z 800 Quantitation and protein identification

New Trends in Proteome Informatics • • • High-throughput Data Quality Data Management-Consistency Data Exchange, DB, Standard Annotation Data Mining

IV. 인간 프로테옴 프로젝트 -Human Proteome Project. Is Human Protoeme Project Next? (The Scientists, April 2, 2001) The Protoeme isn’t genome II (Nature, 410, 725 -, 2001) In Hot Pursuit of the Proteome (Wall Street J. , April 5, 2001) “Goodbye race to decode the human genome; Hello race to decode human proteome” (Time, Feb. 18, 2001)




Human Proteins • 38, 000 – 43, 000 proteins predicted (Based on ORF) • Actual Proteins : 43, 000 x 10 ~ 20 = 1 ~ x 106 proteins (due to Post-Translational Modifications) (Source : Swiss Prot, Ex. PASy etc. ) • Currently only <10, 000 proteins identified (< 1. 0%) • More than 99 % of total human proteins unknown.

Where is this market opportunity coming from ? • Completion of Human Genome Project, funding is going to Proteomics. • Enable development of drugs for previously untreatable diseases. • Big Pharma and biotechs must accelerate drug development to maintain current valuations therefore using high throughput systems.

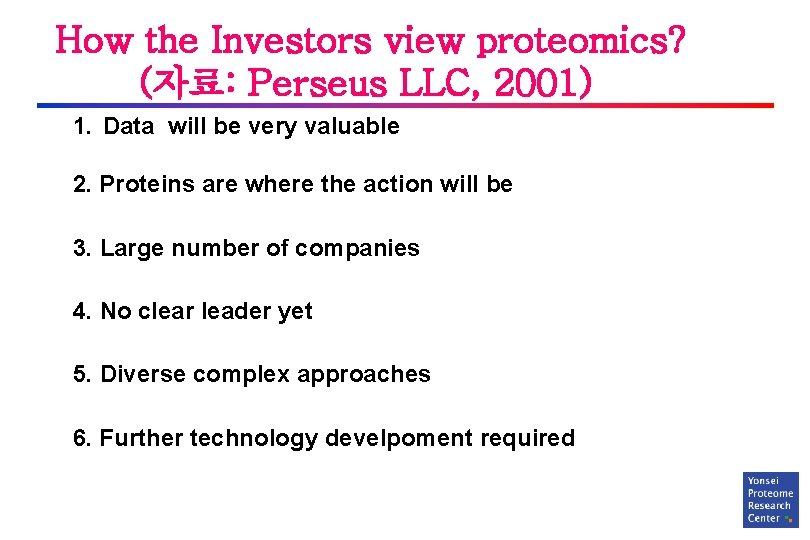
How the Investors view proteomics? (자료: Perseus LLC, 2001) 1. Data will be very valuable 2. Proteins are where the action will be 3. Large number of companies 4. No clear leader yet 5. Diverse complex approaches 6. Further technology develpoment required

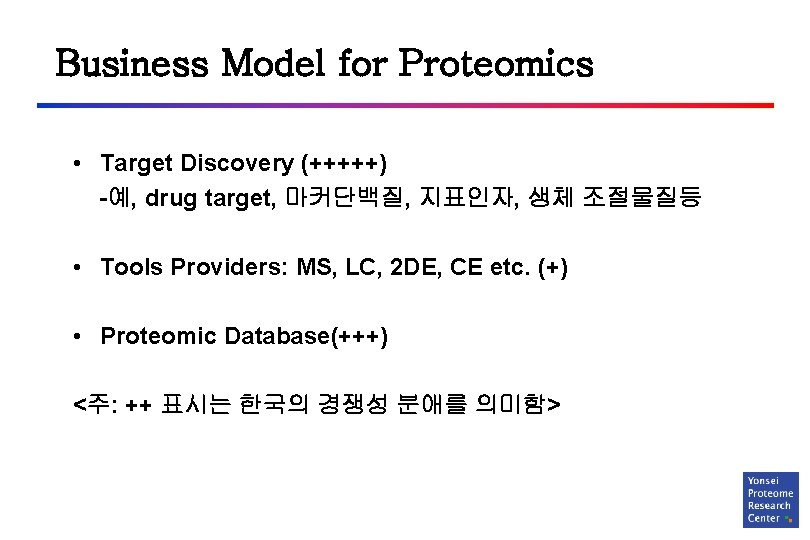
Business Model for Proteomics • Target Discovery (+++++) -예, drug target, 마커단백질, 지표인자, 생체 조절물질등 • Tools Providers: MS, LC, 2 DE, CE etc. (+) • Proteomic Database(+++) <주: ++ 표시는 한국의 경쟁성 분애를 의미함>

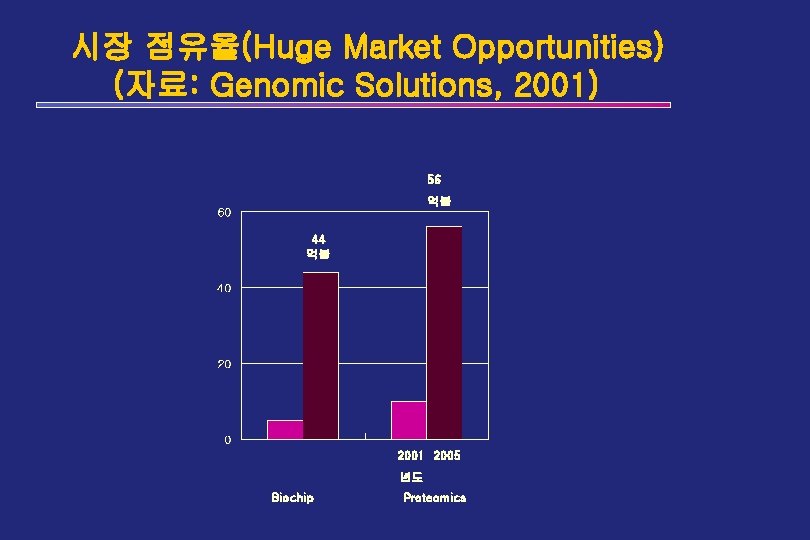
시장 점유율(Huge Market Opportunities) (자료: Genomic Solutions, 2001) 56 억불 44 억불 2001 2005 년도 Biochip Proteomics

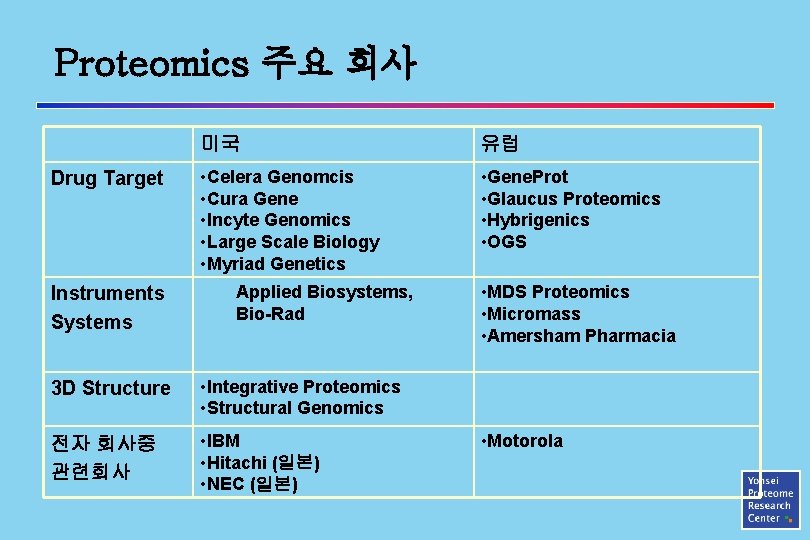
Proteomics 주요 회사 Drug Target Instruments Systems 미국 유럽 • Celera Genomcis • Cura Gene • Incyte Genomics • Large Scale Biology • Myriad Genetics • Gene. Prot • Glaucus Proteomics • Hybrigenics • OGS Applied Biosystems, Bio-Rad 3 D Structure • Integrative Proteomics • Structural Genomics 전자 회사중 관련회사 • IBM • Hitachi (일본) • NEC (일본) • MDS Proteomics • Micromass • Amersham Pharmacia • Motorola




Global Project-HPP를 수행하는 중요한 이유 1. One gene = > 10 proteins • PTM (Post-Translational Modification) • Isoforms • Cell or Tissue-specific expressions 2. > 1 x 106 proteins in Human (Predicted) 3. Annotation 뿐 만 아니라, 구조-기능 규명작업의 병행이 필수 4. 인간 Genome 처럼 시작과 끝이 미리 정해져 있지 않고, 작업의 saturation point 예측 불가 5. 신규성 단백질 (Novel protein)의 다양한 patent화 가능성 • Druggable / Diagnostic / Therapeutic / Regulatory Proteins 6. 게놈 프로젝트의 공헌기회 상실을 훌륭하게 대신해 줄 수 있는 기회 부여 ►► 첨단기술 축적 및 공유 7. 집약적이고, High-throughput 개념의 목표지향적 과제수행의 필요성 대두










