COMPUTATIONAL VACCINE DESIGN RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY






- Slides: 22

COMPUTATIONAL VACCINE DESIGN RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF WASHINGTON How can we design vaccines based on conformational epitopes and protein structure prediction simulations?

GENOME SEQUENCE TO PROTEIN AND PROTEOME… STRUCTURE FUNCTION INTERACTION DNA/RNA PROTEIN COMPOUND SYSTEMS INFRASTRUCTURE APPLICATIONS RICE THERAPEUTICS NANOTECHNOLOGY DESIGN EVOLUTION

INTRODUCTION • Conformational epitopes are two or more nonlocal regions of an antigen that interact structurally at the atomic level, together with each other and with the antibody. • Majority of B-cell epitopes are conformational. • Protective antibodies recognize structural elements in the context of complete antigen structure.

PROBLEMS • Linear peptides corresponding to the epitopes are devoid of structural context of native antigen. • Immune evasion mechanisms: • Conformational flexibility. • Steric masking. • Antigen variation. • Presence of immunodominant decoy elements.

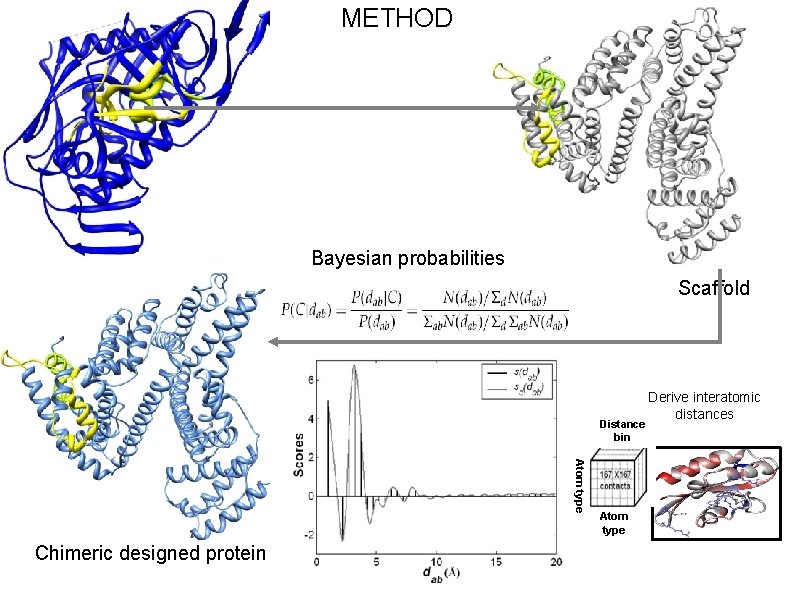
RESEARCH DESIGN Aim: • Transform an immunological region (i. e. , region which can induce antibody) to an antigenic region (i. e. , region which can bind with antibody). Objectives: • Retaining the native structure of epitopes. • Presenting the epitopes exposed to aqueous environment. Method: • Computational design of chimeric constructs by grafting epitopes in soluble/stable scaffolds.

METHOD Bayesian probabilities Scaffold Distance bin Atom type Chimeric designed protein Atom type Derive interatomic distances

APPLICATIONS • HIV • Influenza • Syphilis • Anthrax

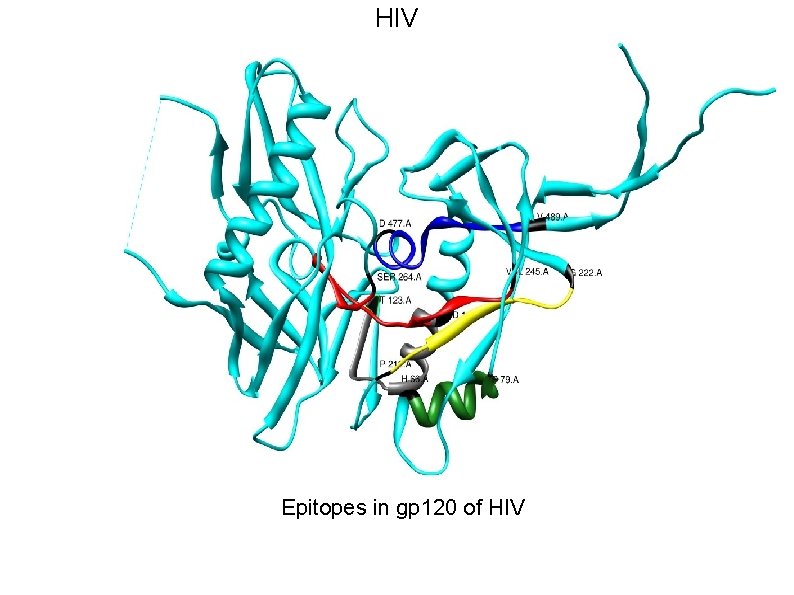
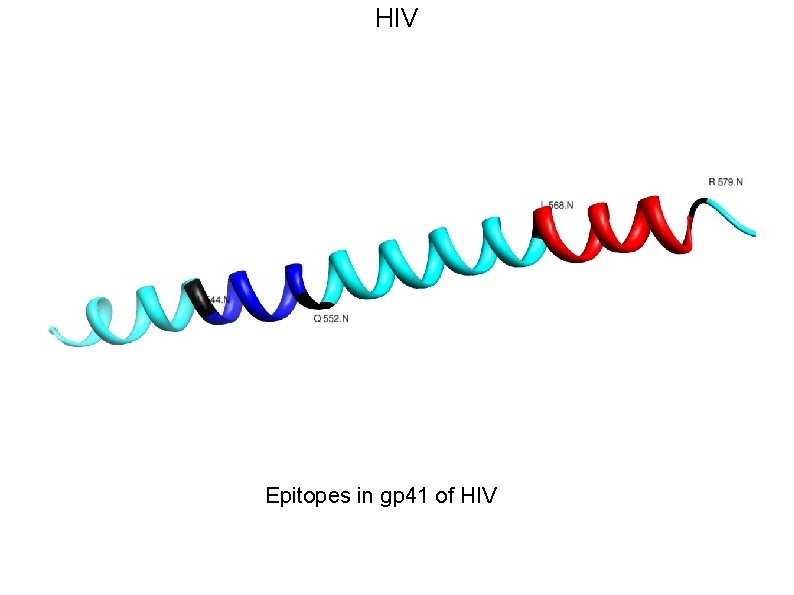
HIV • HIV-1’s extensive diversity is a major challenge for vaccine design strategies. • The presence of segments that are nearly invariant in all HIV-1 M group strongly suggests that these conserved elements are both obligatory for viral viability and are therefore potential Achilles’ Heel of the virus. • Our scaffold based vaccine design is based on conserved elements of viral spike protein gp 120 and gp 41.

HIV Epitopes in gp 120 of HIV

HIV Epitopes in gp 41 of HIV

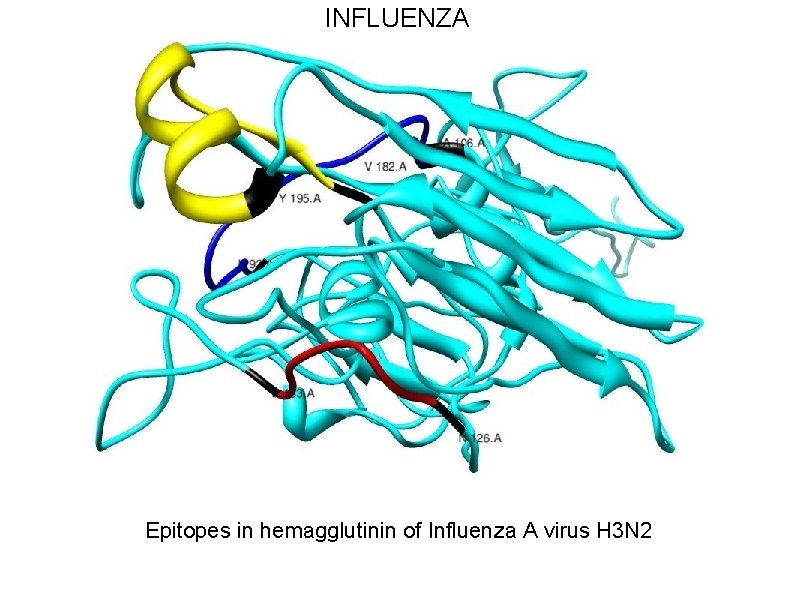
INFLUENZA • For Influenza, we are utilizing the epitopes in viral surface protein hemagglutinin(HA). • Hemagglutin is responsible for receptor binding and membrane fusion of viral particles. • We have selected 3 protective epitopes from HA 1 of Influenza A virus H 3 N 2 A/Wuhan/359/95 strain.

INFLUENZA Epitopes in hemagglutinin of Influenza A virus H 3 N 2

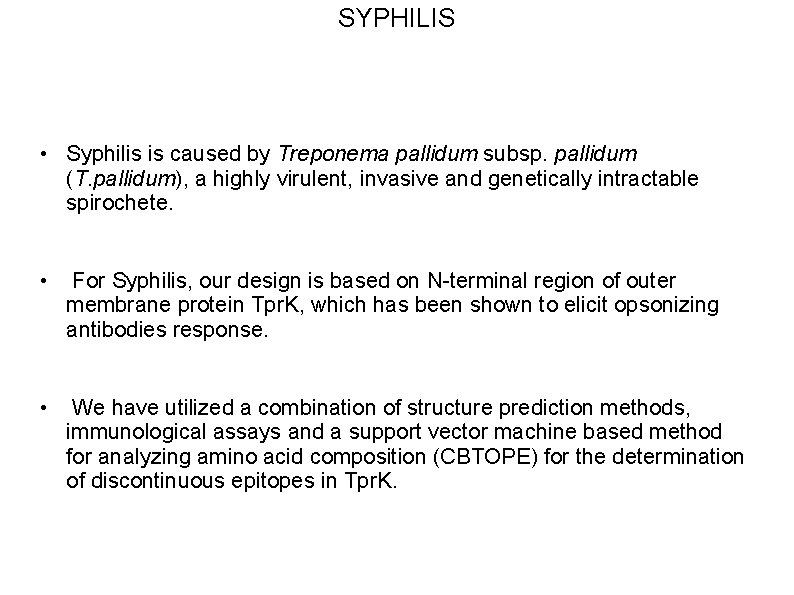
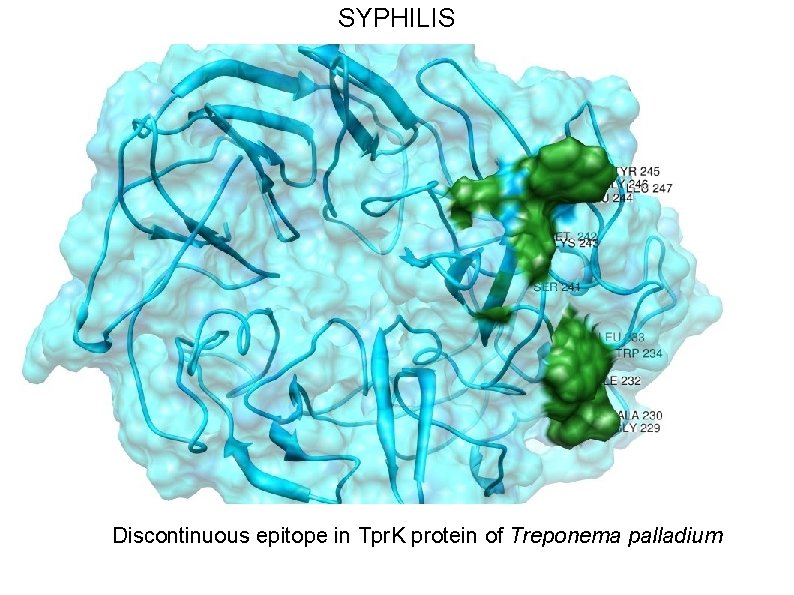
SYPHILIS • Syphilis is caused by Treponema pallidum subsp. pallidum (T. pallidum), a highly virulent, invasive and genetically intractable spirochete. • For Syphilis, our design is based on N-terminal region of outer membrane protein Tpr. K, which has been shown to elicit opsonizing antibodies response. • We have utilized a combination of structure prediction methods, immunological assays and a support vector machine based method for analyzing amino acid composition (CBTOPE) for the determination of discontinuous epitopes in Tpr. K.

SYPHILIS Tpr. K of Treponema palladium

SYPHILIS Discontinuous epitope in Tpr. K protein of Treponema palladium

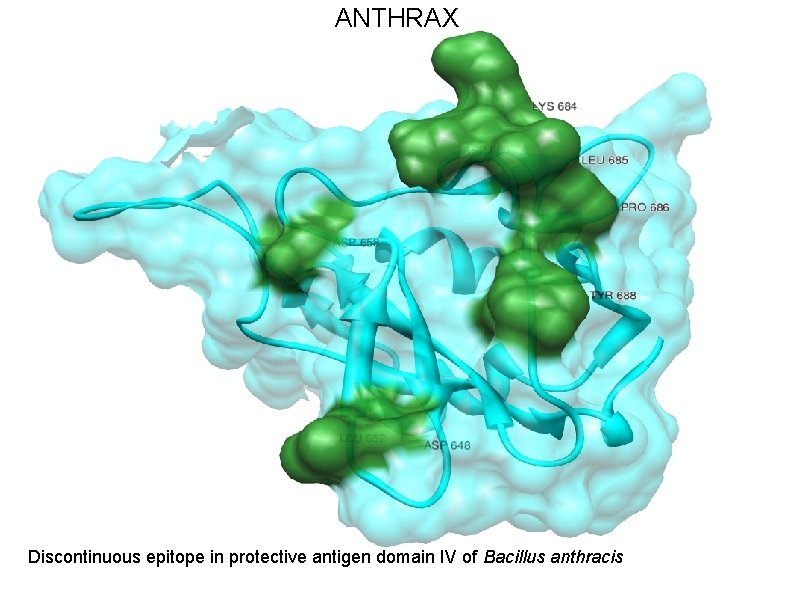
ANTHRAX • Anthrax is caused by Bacillus anthracis, a gram-positive, spore forming, and rod-shaped bacterium. • Anthrax toxin belongs to the family of bacterial AB toxins, composed of a single B subunit, protective antigen and two alternative A subunits: edema factor and lethal factor. • Protective antigen (PA) is the dominant antigen in both natural and vaccine-induced immunity to anthrax infection. • We are exploiting the epitopes from receptor binding domain (Domain IV) of protective antigen.

ANTHRAX Protective antigen of Bacillus anthracis

ANTHRAX Discontinuous epitope in protective antigen domain IV of Bacillus anthracis

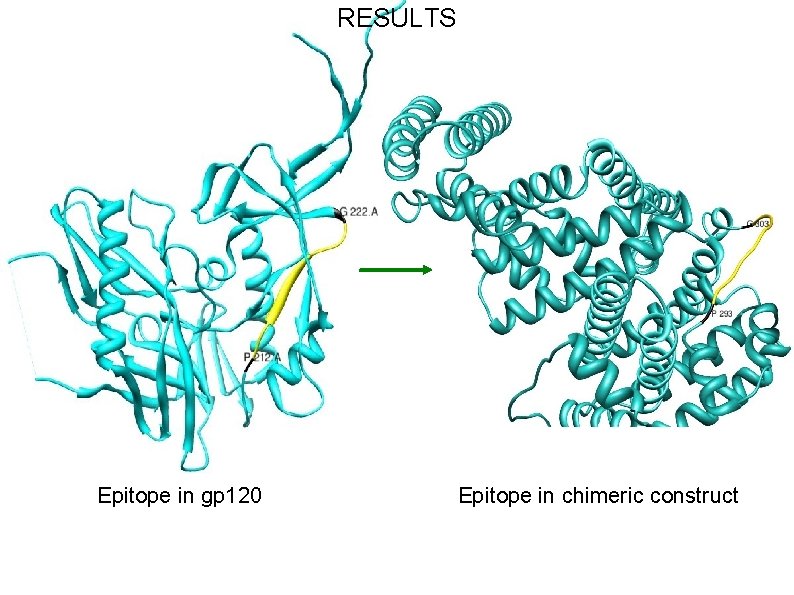
RESULTS Epitope in gp 120 Epitope in chimeric construct

RESULTS Epitope in hemagglutinin Epitope in chimeric construct

ACKNOWLEDGEMENTS Current group members: Past group members: • Adrian Laurenzi • Brian Buttrick • Chuck Mader • Dominic Fisher • Emilia Gan • Ersin Emre Oren • Gaurav Chopra • George White • Hernan Zamalloa • Jason North • Jeremy Horst • Ling-Hong Hung • Matthew Clark • Manish • Michael Shannon • Michael Zhou • Omid Zarei • Raymond Zhang • Stewart Moughon • Thomas Wood • Weerayuth Kittichotirat • Aaron Chang • Aaron Goldman • Brady Bernard • Cyrus Hui • David Nickle • Duangdao Wichadukul • Duncan Milburn • Ekachai Jenwitheesuk • Gong Cheng • Imran Rashid • Jason Mc. Dermott • Juni Lee • Kai Wang • Marissa La. Madrid • Michael Inouye • Michal Guerquin • Nipa Jongkon • Rob Braiser • Renee Ireton • Shu Feng • Sarunya Suebtragoon • Shing-Chung Ngan • Shyamala Iyer • Siriphan Manocheewa • Somsak Phattarasukol • Tianyun Liu • Vanessa Steinhilb • Vania Wang • Yi-Ling Cheng • Zach Frazier

ACKNOWLEDGEMENTS Collaborators: Funding agencies: • BGI - Gane Wong - Jun Yu - Jun Wang - et al. • BIOTEC/KMUTT • MSE - Mehmet Sarikaya - Candan Tamerler - et al. • UW Microbiology - James Staley - John Mittler - Michael Lagunoff - Roger Bumgarner - Wesley Van Voorhis - et al. • National Institutes of Health • National Science Foundation -DBI -IIS • Searle Scholars Program • Puget Sound Partners in Global Health • Washington Research Foundation • UW -Advanced Technology Initiative -TGIF Budget: • ~US$1 million/year total costs