SHOTGUN STRUCTURAL PROTEOMICS RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY






- Slides: 18

SHOTGUN STRUCTURAL PROTEOMICS RAM SAMUDRALA ASSOCIATE PROFESSOR UNIVERSITY OF WASHINGTON Given a heterogeneous mixture of proteins, how can we determine all their structures in a high-throughput and high-resolution manner?

MOTIVATION FOR DETERMINING PROTEIN STRUCTURE The functions necessary for life are undertaken by proteins. Protein function is mediated by protein three-dimensional structure. Knowing protein structure at high resolution will enable us to: Determine and understand molecular function. Understand substrate and ligand binding. Devise intelligent mutagenesis and biochemical experiments to understand biological function. Design therapeutics rationally. Design novel proteins. Knowing the structures of all proteins encoded by an organism’s genome will enable us to understand complex pathways and systems, and ultimately organismal behaviour and evolution. Applications in the area of medicine, nanotechnology, and biological computing.

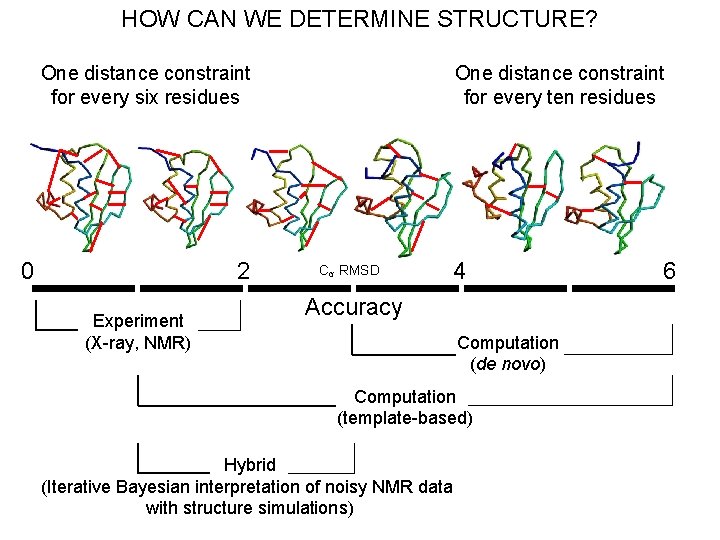
HOW CAN WE DETERMINE STRUCTURE? One distance constraint for every six residues 0 2 Experiment (X-ray, NMR) One distance constraint for every ten residues Cα RMSD 4 Accuracy Computation (de novo) Computation (template-based) Hybrid (Iterative Bayesian interpretation of noisy NMR data with structure simulations) 6

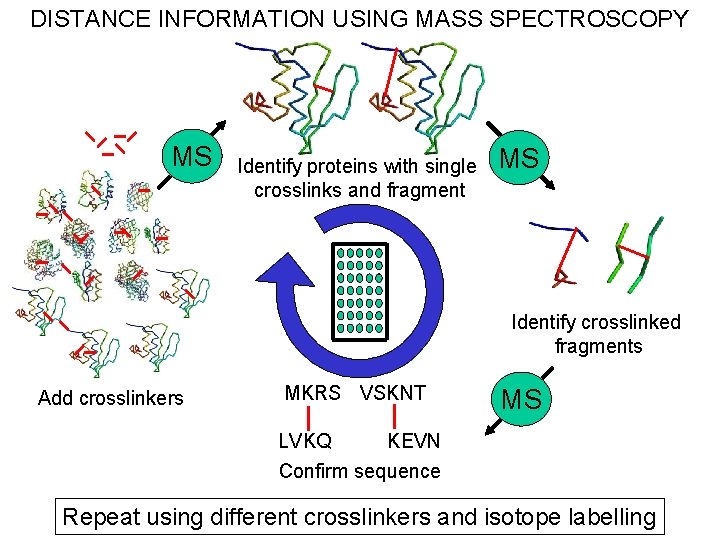
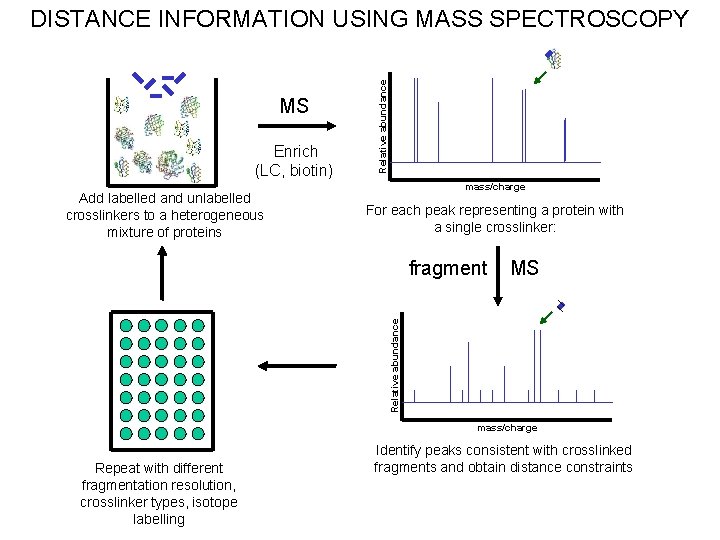
DISTANCE INFORMATION USING MASS SPECTROSCOPY MS Identify proteins with single crosslinks and fragment MS Identify crosslinked fragments Add crosslinkers MKRS VSKNT MS KEVN LVKQ Confirm sequence Repeat using different crosslinkers and isotope labelling

HOW AND WHY WILL THIS WORK? Perform experiments to obtain a number of distance constraints (one for every six residues for medium to high-resolution structures). Perform simulations based on high confidence constraints and use distance distributions from resulting structures to iteratively reinterpret the spectra (without repeating experiment) until we obtain a high-resolution structure. Computational aspects largely complete. Components of approach have been implemented by others in a limited way but are assembled here in a robust and unique manner. Method can handle: Impure protein purification (ex: structural genomics failures). Environment-dependent structures (ex: chaperones + effectors). Partially disordered proteins. Several proteins simultaneously (large scale). No need for proteolytic digestion (complicates things). Focus on structures from noisy data, unlike X-ray diffraction and NMR.

PLAN OF ACTION Begin computational studies using simulated data (with noise) and develop software to prioritise experiments (ex: crosslinker choices). Initial studies using UW Mass Spectrometry Center: Start with fairly pure mixtures >> not-so-pure mixtures >> 2 -3 proteins >> handful of proteins >> Difficult proteins >> heterogenous mixtures >> whole proteomes. Advice from Aebersold, Kelleher. Team of 10 -20 personnel working on crosslinking technology, protein enrichment, mass spectroscopy, structure calculation, parameterisation. Dedicated instrumentation through Pioneer Award, startup, MRI. Bayesian framework will be utilised to estimate accuracy/error: Avoid repeating past oversight with NMR. Obtain an R-factor like estimate as in X-ray diffraction. Comparison of generated spectra from models to actual spectra. Iterative reinterpretation of experimental data.

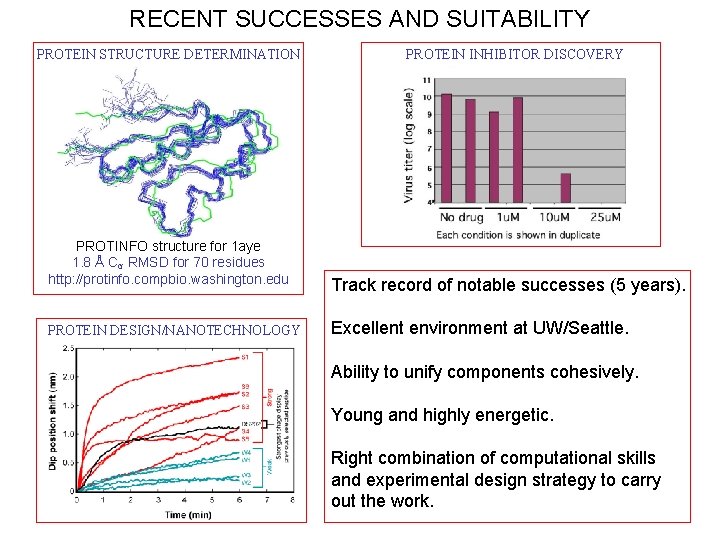
RECENT SUCCESSES AND SUITABILITY PROTEIN STRUCTURE DETERMINATION PROTINFO structure for 1 aye 1. 8 Å Cα RMSD for 70 residues http: //protinfo. compbio. washington. edu PROTEIN DESIGN/NANOTECHNOLOGY PROTEIN INHIBITOR DISCOVERY Track record of notable successes (5 years). Excellent environment at UW/Seattle. Ability to unify components cohesively. Young and highly energetic. Right combination of computational skills and experimental design strategy to carry out the work.

OUTCOME AND EXPECTATIONS Structural genomics projects aim to obtain a representative structure of every protein family using X-ray diffraction and NMR methods and employ computational methods to fill in the gaps. However, several families of proteins will not be accessible by these structure determination methodologies, and computational methods alone are far from capable of consistently producing high resolution structures. Even in successful cases, the effect of the biological environment on protein structure is not accounted for. Our hybrid approach, which complements existing structural genomics efforts, will be used to rapidly obtain structures for entire proteomes in biologically relevant environments.


WHY ARE CURRENT METHODS NOT ADEQUATE? The major bottlenecks for both X-ray diffraction and NMR studies is producing sufficient quantities of the protein in a pure form to perform the experiments. Deviations from ideal behaviour in a protein sample result in slow and labourintensive structure determination, if at all possible. These major structure determination techniques were developed at a time when our worldview of proteins was simple and did not account for environment-dependent structure formation, protein dynamics and conformational changes, and post-translational modifications. The vast majority of proteins will therefore be inaccessible to X-ray diffraction and NMR studies. Computational approaches do not have the resolution of experimental approaches and lack consistency.

CROSSLINKING POSSIBILITIES Seven chemical groups that can be crosslinked: amines (2), carboxyls (3), and thiols (2). Numerous distances for the ~42 (7 x 6) possible pairs of groups. For every 100 residues, there may be up to ten members of each group, but typically one crosslink is possible at a particular distance out of the ~100 possible pairs. For every 100 residues, the total number of groups is ~20 -40, resulting in a potential yield of 400 -1600 distance constraints if all crosslink possibilities can occur.

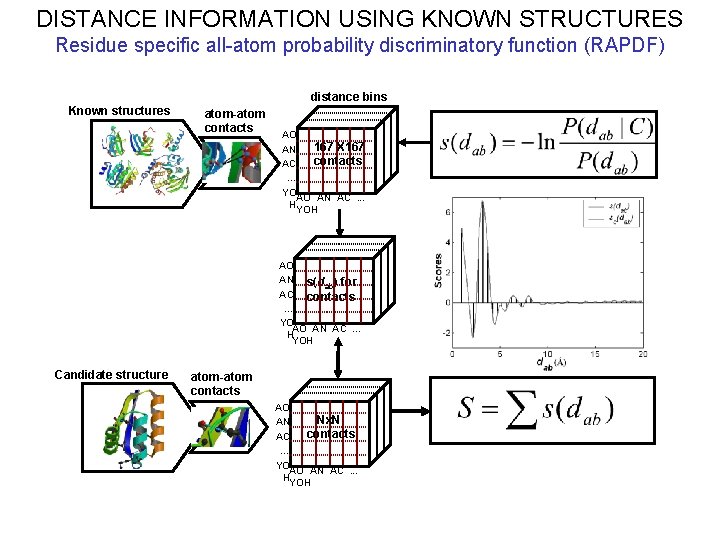
DISTANCE INFORMATION USING KNOWN STRUCTURES Residue specific all-atom probability discriminatory function (RAPDF) distance bins Known structures atom-atom contacts AO 167 X 167 AN contacts AC … YO AO AN AC. . . H YOH AO AN s(d ) for ab AC contacts … YO AO AN AC. . . H YOH Candidate structure atom-atom contacts AO Nx. N AN contacts AC … YO AO AN AC. . . H YOH

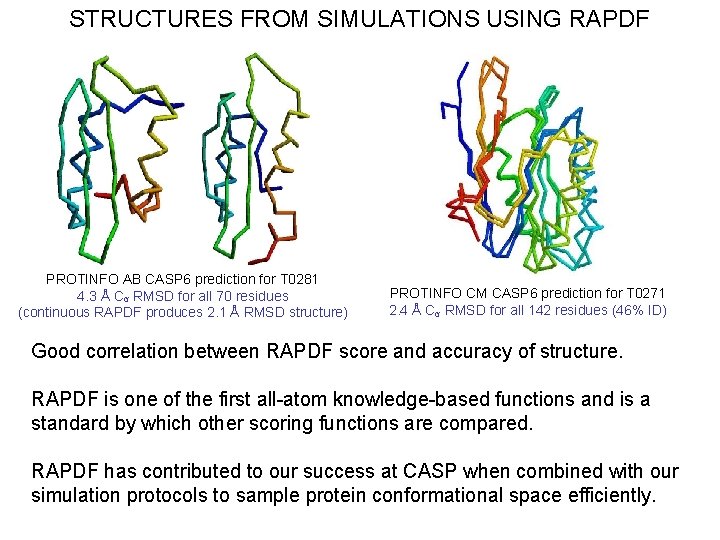
STRUCTURES FROM SIMULATIONS USING RAPDF PROTINFO AB CASP 6 prediction for T 0281 4. 3 Å Cα RMSD for all 70 residues (continuous RAPDF produces 2. 1 Å RMSD structure) PROTINFO CM CASP 6 prediction for T 0271 2. 4 Å Cα RMSD for all 142 residues (46% ID) Good correlation between RAPDF score and accuracy of structure. RAPDF is one of the first all-atom knowledge-based functions and is a standard by which other scoring functions are compared. RAPDF has contributed to our success at CASP when combined with our simulation protocols to sample protein conformational space efficiently.

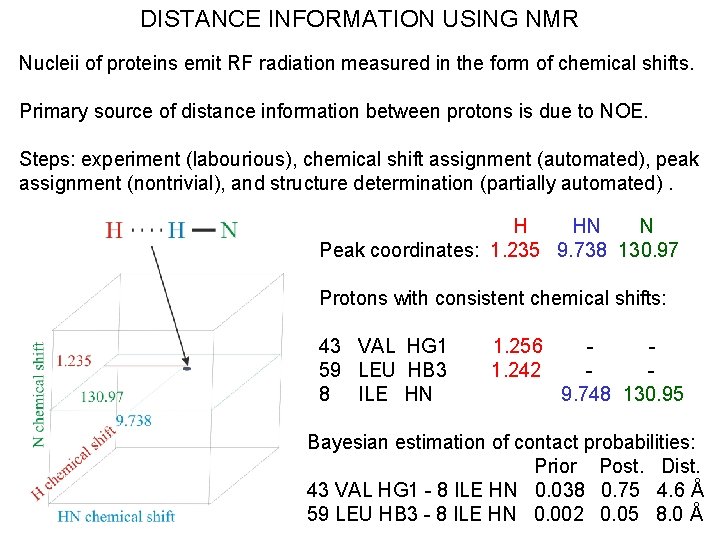
DISTANCE INFORMATION USING NMR Nucleii of proteins emit RF radiation measured in the form of chemical shifts. Primary source of distance information between protons is due to NOE. Steps: experiment (labourious), chemical shift assignment (automated), peak assignment (nontrivial), and structure determination (partially automated). H HN N Peak coordinates: 1. 235 9. 738 130. 97 Protons with consistent chemical shifts: 43 VAL HG 1 59 LEU HB 3 8 ILE HN 1. 256 1. 242 9. 748 130. 95 Bayesian estimation of contact probabilities: Prior Post. Dist. 43 VAL HG 1 - 8 ILE HN 0. 038 0. 75 4. 6 Å 59 LEU HB 3 - 8 ILE HN 0. 002 0. 05 8. 0 Å

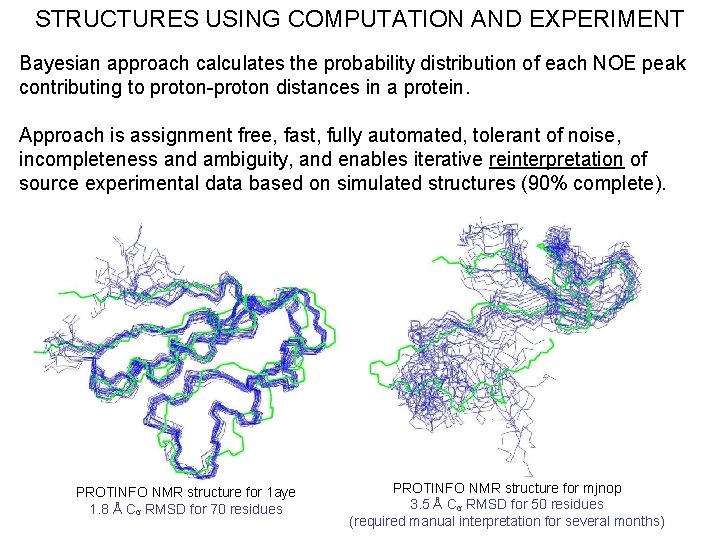
STRUCTURES USING COMPUTATION AND EXPERIMENT Bayesian approach calculates the probability distribution of each NOE peak contributing to proton-proton distances in a protein. Approach is assignment free, fast, fully automated, tolerant of noise, incompleteness and ambiguity, and enables iterative reinterpretation of source experimental data based on simulated structures (90% complete). PROTINFO NMR structure for 1 aye 1. 8 Å Cα RMSD for 70 residues PROTINFO NMR structure for mjnop 3. 5 Å Cα RMSD for 50 residues (required manual interpretation for several months)

MS Enrich (LC, biotin) Add labelled and unlabelled crosslinkers to a heterogeneous mixture of proteins Relative abundance DISTANCE INFORMATION USING MASS SPECTROSCOPY mass/charge For each peak representing a protein with a single crosslinker: MS Relative abundance fragment mass/charge Repeat with different fragmentation resolution, crosslinker types, isotope labelling Identify peaks consistent with crosslinked fragments and obtain distance constraints

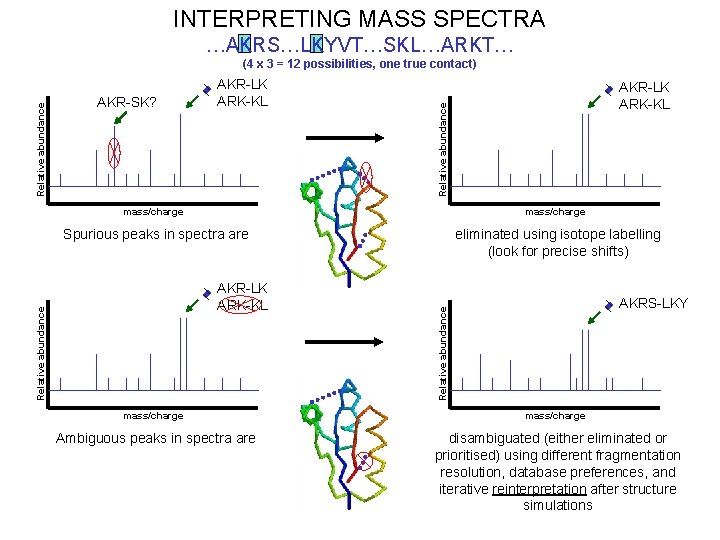
INTERPRETING MASS SPECTRA …AKRS…LKYVT…SKL…ARKT… AKR-LK ARK-KL Relative abundance AKR-SK? mass/charge Spurious peaks in spectra are eliminated using isotope labelling (look for precise shifts) AKRS-LKY Relative abundance AKR-LK ARK-KL Relative abundance (4 x 3 = 12 possibilities, one true contact) mass/charge Ambiguous peaks in spectra are disambiguated (either eliminated or prioritised) using different fragmentation resolution, database preferences, and iterative reinterpretation after structure simulations

DISTANCE INFORMATION USING FRET Analogous to MS approach, but instead of peaks representing mass/charge ratios that identify two crosslinked residues (indirect distance information), we can obtain direct distance information. Express protein in an in vitro system to ensure single flurophore donor/acceptor pair for two residues in a protein. Use confocal microscopy setup to measure energy transfer for many donor/acceptor pairs. Distance is based on donor/acceptor type can be obtained for any pair of residues that do not cause loss of structure (determined by consistency across many pairs); tangential benefit of identifying structurally important residues. Ideal for measurement of long range distances and for large proteins.