Kmeans properties Pasi Frnti 11 10 2017 Kmeans











![Better initialization technique Simple initializations: • Random centroids (Random) [Forgy][Mac. Queen] • Further point Better initialization technique Simple initializations: • Random centroids (Random) [Forgy][Mac. Queen] • Further point](https://slidetodoc.com/presentation_image_h2/97d3f61e3ce03055831a667ef70fecc4/image-53.jpg)


- Slides: 61

K-means properties Pasi Fränti 11. 10. 2017 K-means properties on six clustering benchmark datasets Pasi Fränti and Sami Sieranoja Algorithms, 2017.

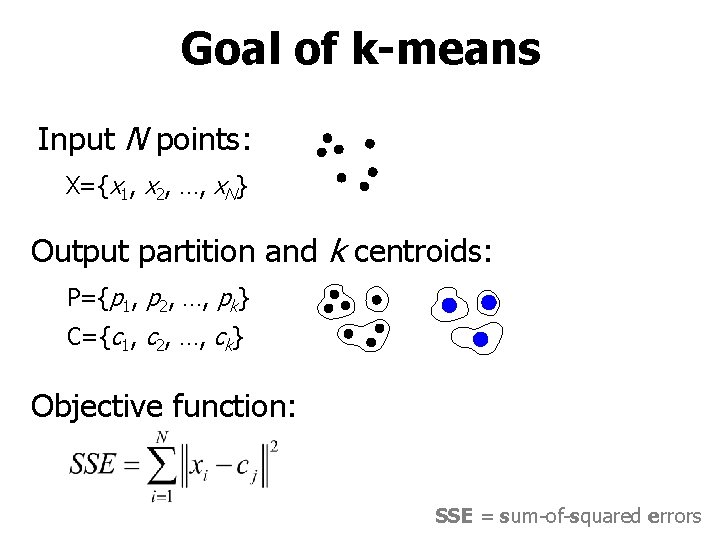
Goal of k-means Input N points: X={x 1, x 2, …, x. N} Output partition and k centroids: P={p 1, p 2, …, pk} C={c 1, c 2, …, ck} Objective function: SSE = sum-of-squared errors

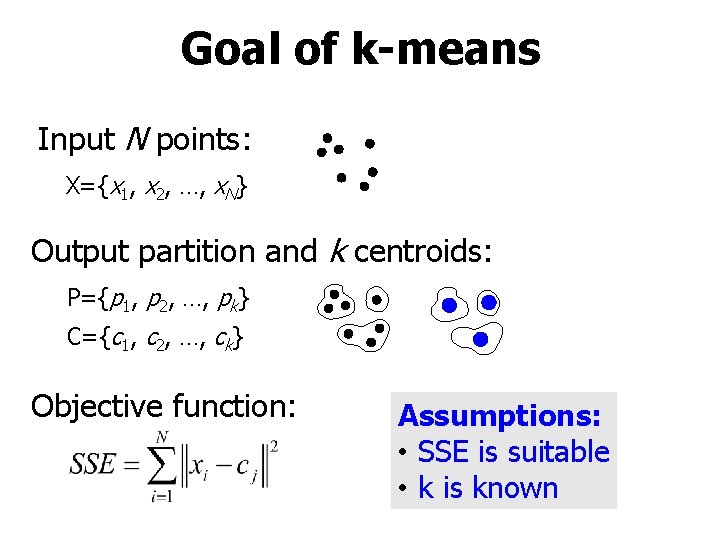
Goal of k-means Input N points: X={x 1, x 2, …, x. N} Output partition and k centroids: P={p 1, p 2, …, pk} C={c 1, c 2, …, ck} Objective function: Assumptions: • SSE is suitable • k is known

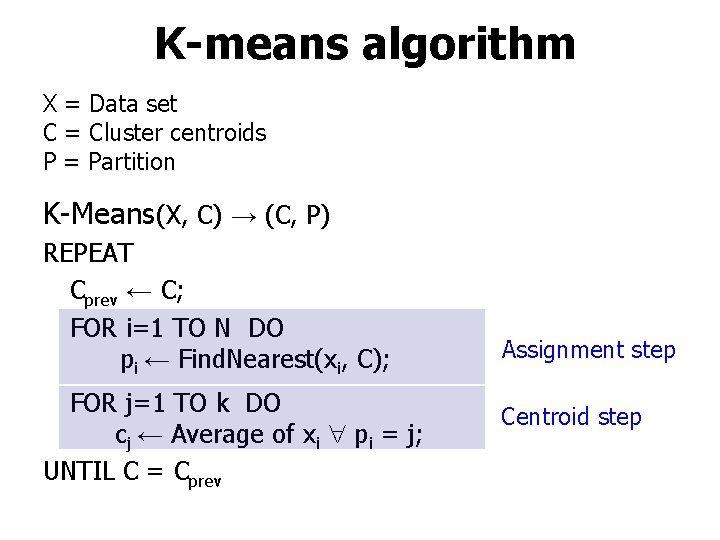
K-means algorithm X = Data set C = Cluster centroids P = Partition K-Means(X, C) → (C, P) REPEAT Cprev ← C; FOR i=1 TO N DO pi ← Find. Nearest(xi, C); FOR j=1 TO k DO cj ← Average of xi pi = j; UNTIL C = Cprev Assignment step Centroid step

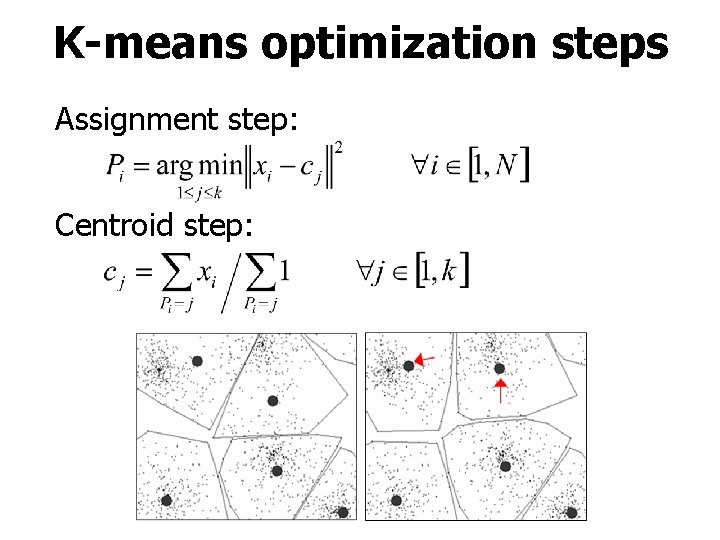
K-means optimization steps Assignment step: Centroid step:

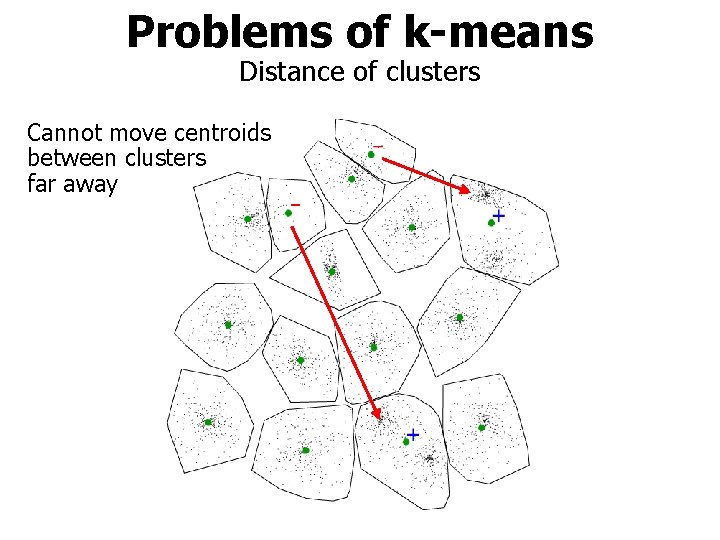
Problems of k-means Distance of clusters Cannot move centroids between clusters far away

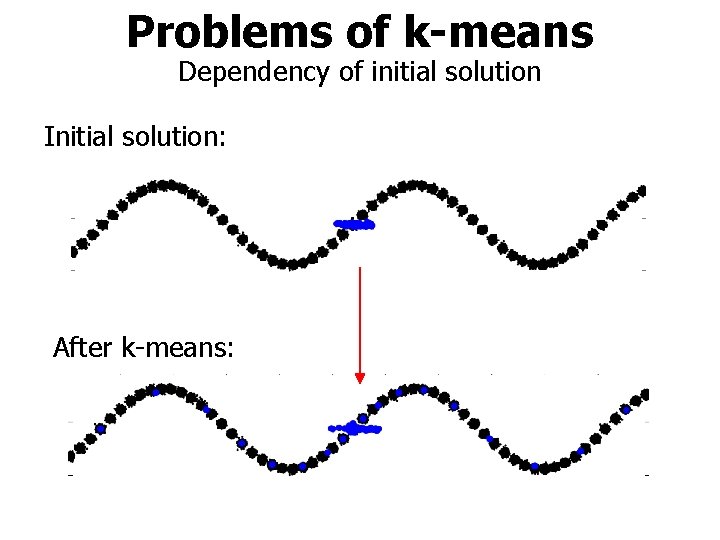
Problems of k-means Dependency of initial solution Initial solution: After k-means:

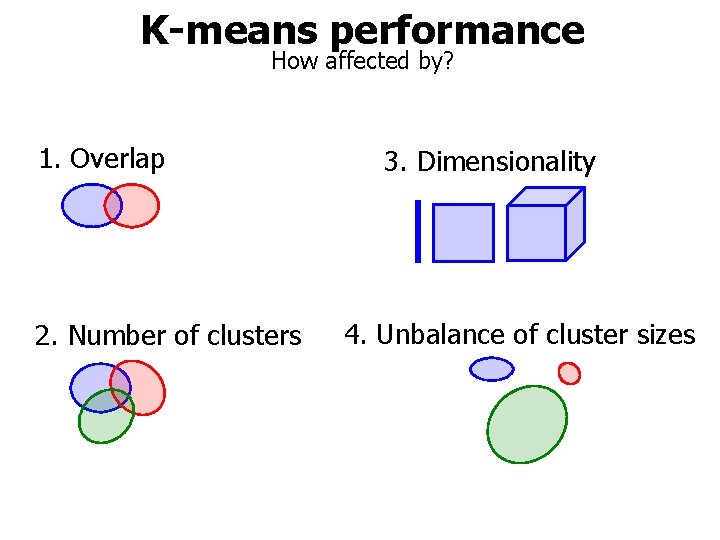
K-means performance How affected by? 1. Overlap 2. Number of clusters 3. Dimensionality 4. Unbalance of cluster sizes

Basic Benchmark

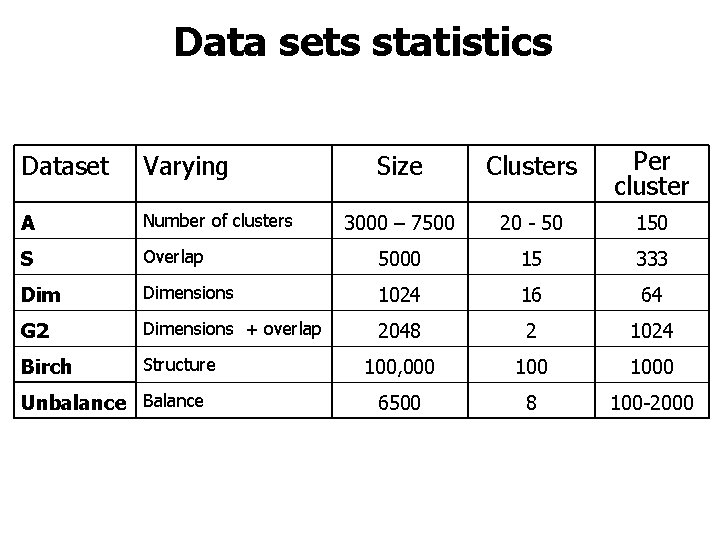
Data sets statistics Size Clusters Per cluster 3000 – 7500 20 - 50 150 Overlap 5000 15 333 Dimensions 1024 16 64 G 2 Dimensions + overlap 2048 2 1024 Birch Structure 100, 000 1000 6500 8 100 -2000 Dataset Varying A Number of clusters S Unbalance Balance

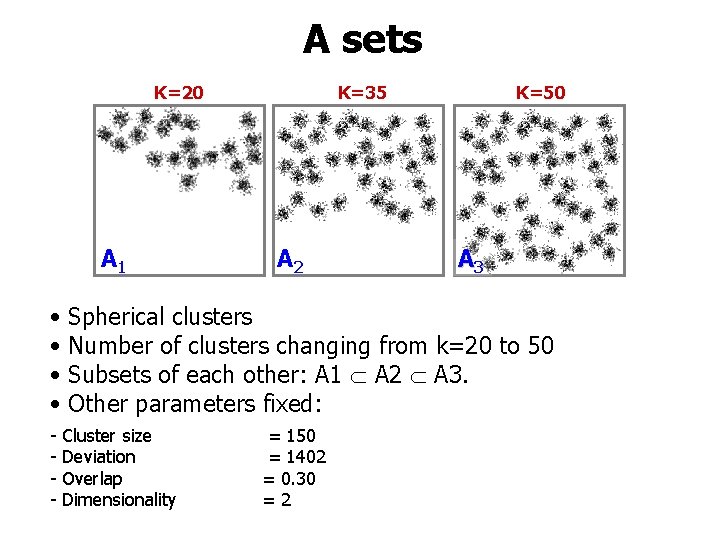
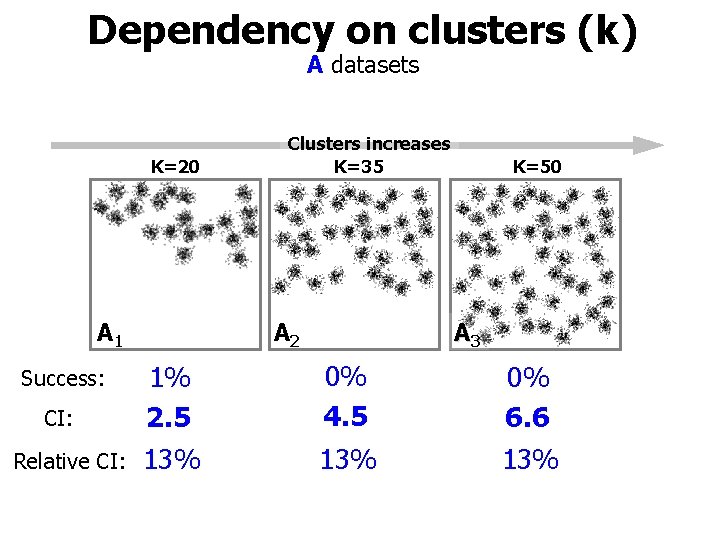
A sets K=20 A 1 • • K=35 A 2 K=50 A 3 Spherical clusters Number of clusters changing from k=20 to 50 Subsets of each other: A 1 A 2 A 3. Other parameters fixed: - Cluster size - Deviation - Overlap - Dimensionality = 150 = 1402 = 0. 30 =2

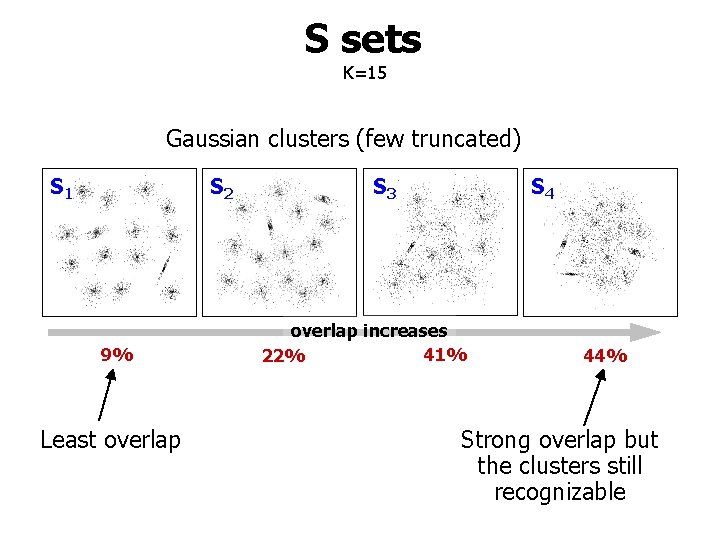
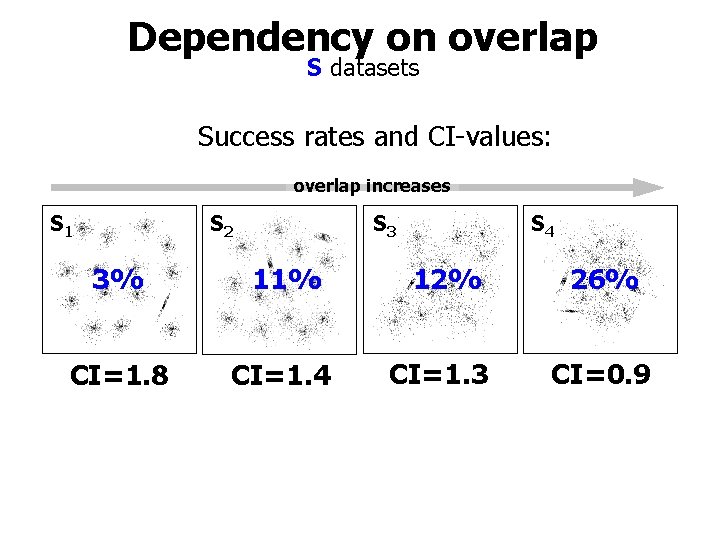
S sets K=15 Gaussian clusters (few truncated) S 1 S 2 9% Least overlap S 3 S 4 overlap increases 41% 22% 44% Strong overlap but the clusters still recognizable

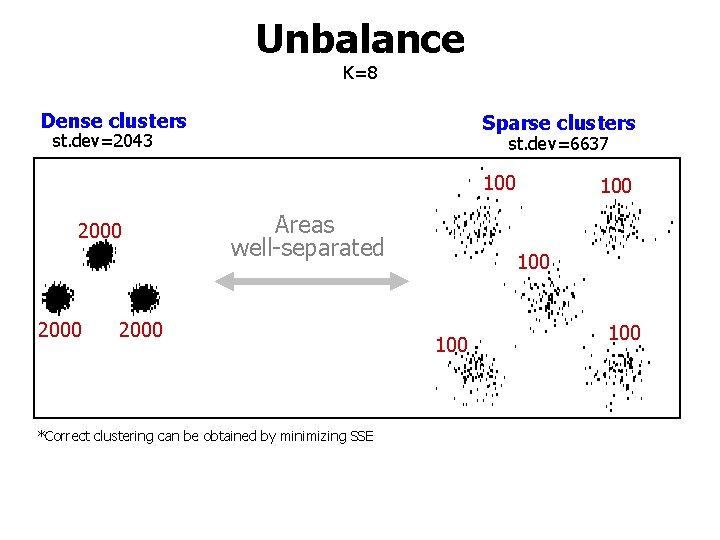
Unbalance K=8 Dense clusters Sparse clusters st. dev=2043 st. dev=6637 100 2000 Areas well-separated 2000 *Correct clustering can be obtained by minimizing SSE 100 100

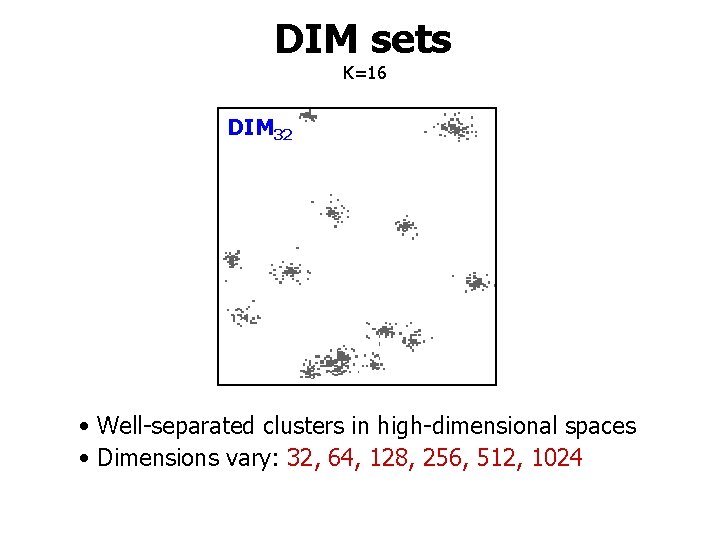
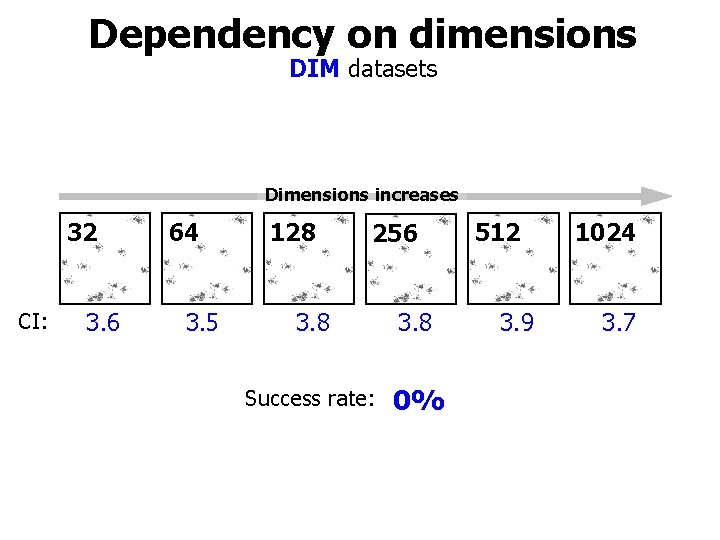
DIM sets K=16 DIM 32 • Well-separated clusters in high-dimensional spaces • Dimensions vary: 32, 64, 128, 256, 512, 1024

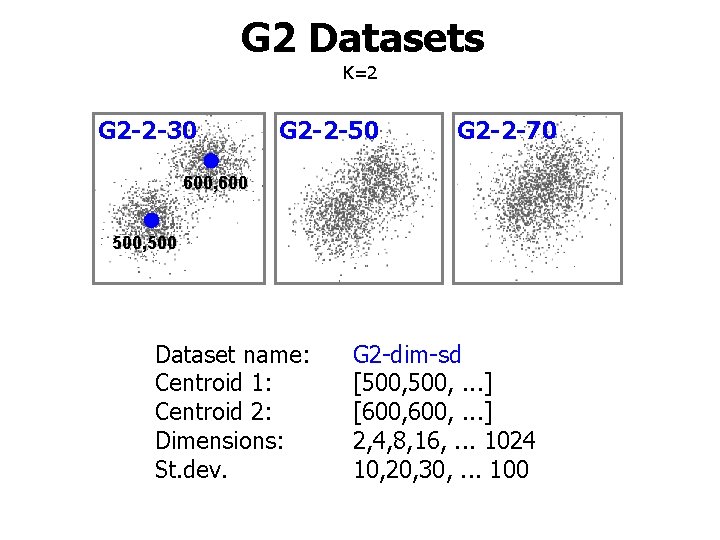
G 2 Datasets K=2 G 2 -2 -30 G 2 -2 -50 G 2 -2 -70 600, 600 500, 500 Dataset name: Centroid 1: Centroid 2: Dimensions: St. dev. G 2 -dim-sd [500, . . . ] [600, . . . ] 2, 4, 8, 16, . . . 1024 10, 20, 30, . . . 100

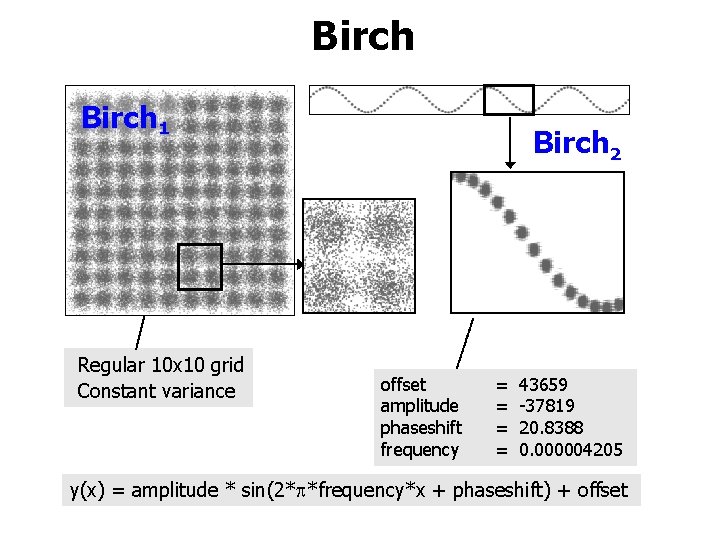
Birch 1 Regular 10 x 10 grid Constant variance Birch 2 offset amplitude phaseshift frequency = = 43659 -37819 20. 8388 0. 000004205 y(x) = amplitude * sin(2* *frequency*x + phaseshift) + offset

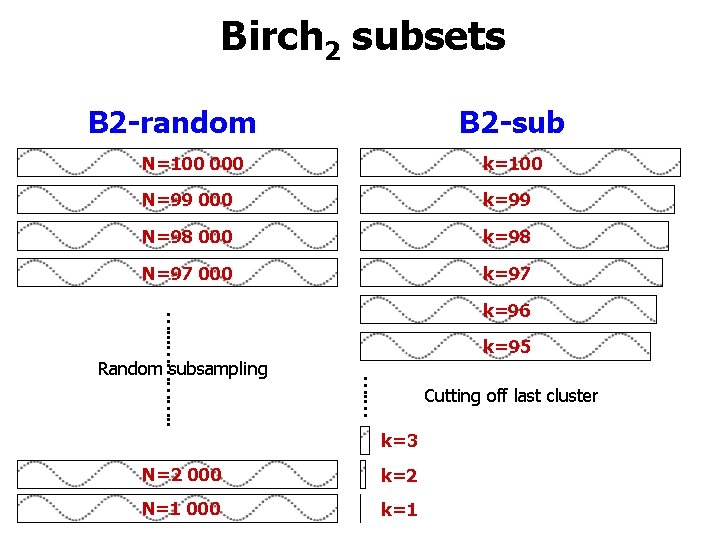
Birch 2 subsets B 2 -random B 2 -sub N=100 000 k=100 N=99 000 k=99 N=98 000 k=98 N=97 000 k=97 k=95 …. … …. . . …………. . Random subsampling k=96 Cutting off last cluster k=3 N=2 000 k=2 N=1 000 k=1

Properties

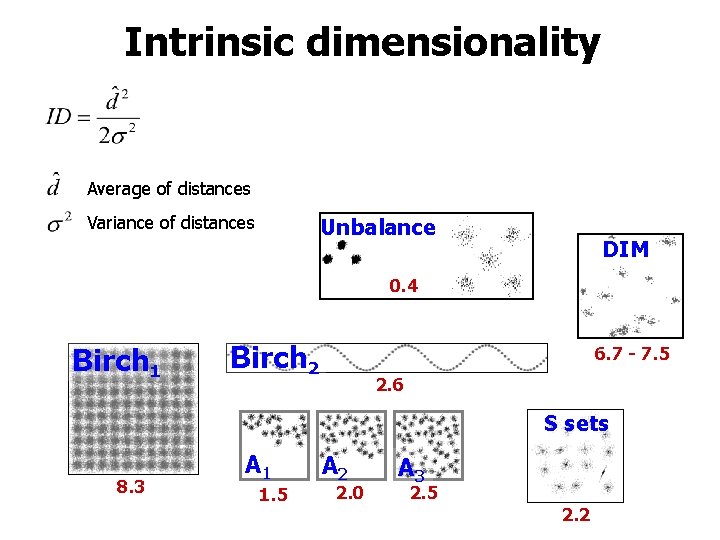
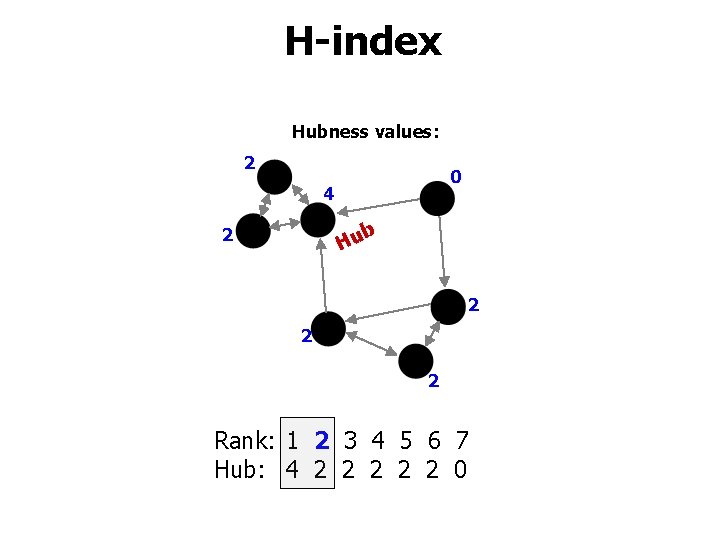
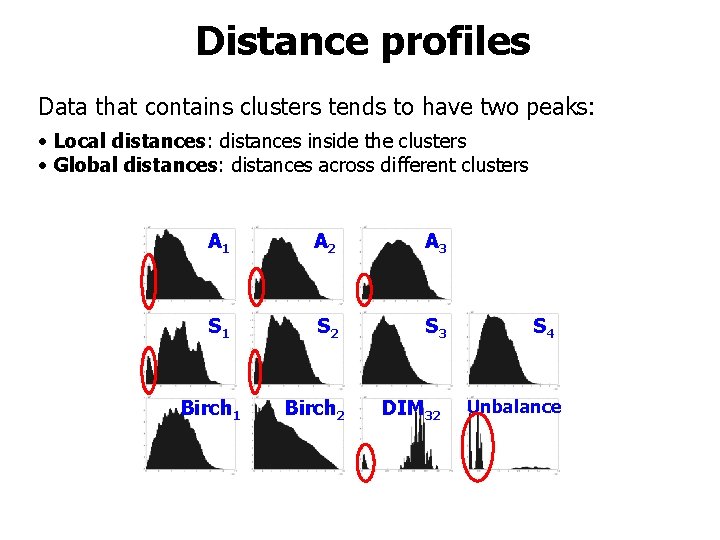
Measured properties • • • Overlap Contrast Intrinsic dimensionality H-index Distance profiles

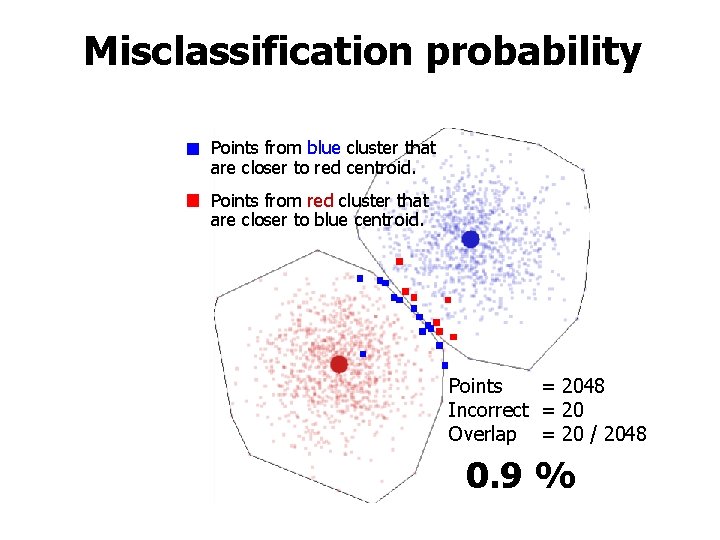
Misclassification probability Points from blue cluster that are closer to red centroid. Points from red cluster that are closer to blue centroid. Points = 2048 Incorrect = 20 Overlap = 20 / 2048 0. 9 %

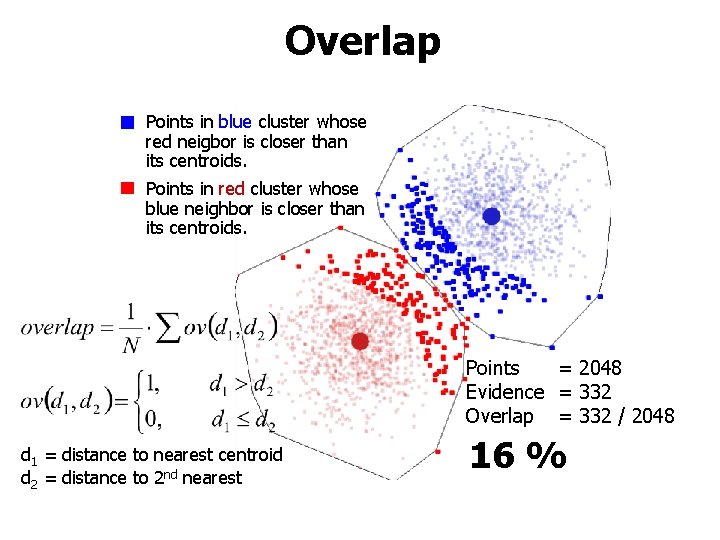
Overlap Points in blue cluster whose red neigbor is closer than its centroids. Points in red cluster whose blue neighbor is closer than its centroids. Points = 2048 Evidence = 332 Overlap = 332 / 2048 d 1 = distance to nearest centroid d 2 = distance to 2 nd nearest 16 %

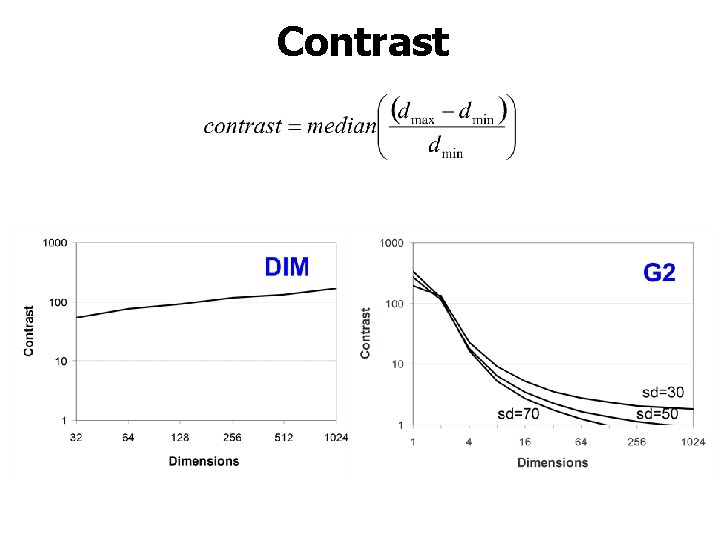
Contrast

Intrinsic dimensionality Average of distances Variance of distances Unbalance DIM 0. 4 Birch 1 Birch 2 6. 7 - 7. 5 2. 6 S sets 8. 3 A 1 1. 5 A 2 2. 0 A 3 2. 5 2. 2

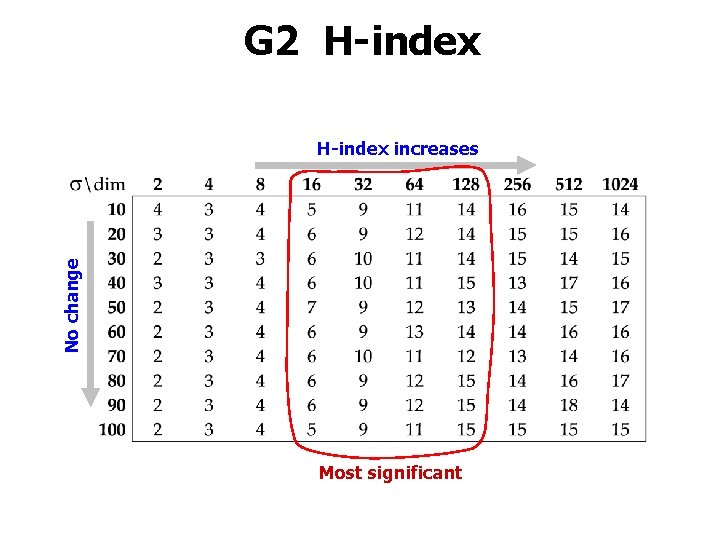
H-index Hubness values: 2 0 4 b 2 Hu 2 2 2 Rank: 1 2 3 4 5 6 7 Hub: 4 2 2 2 0

Distance profiles Data that contains clusters tends to have two peaks: • Local distances: distances inside the clusters • Global distances: distances across different clusters A 1 A 2 A 3 S 1 S 2 S 3 Birch 2 DIM 32 Birch 1 S 4 Unbalance

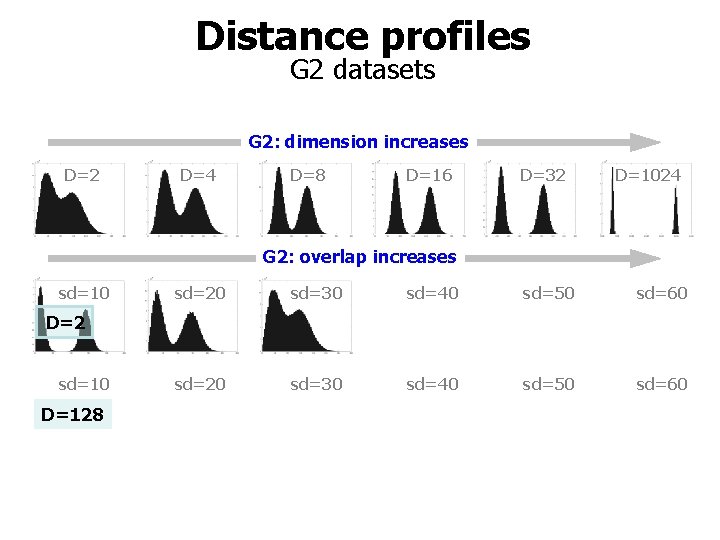
Distance profiles G 2 datasets G 2: dimension increases D=2 D=4 D=8 D=16 D=32 D=1024 G 2: overlap increases sd=10 sd=20 sd=30 sd=40 sd=50 sd=60 D=2 sd=10 D=128

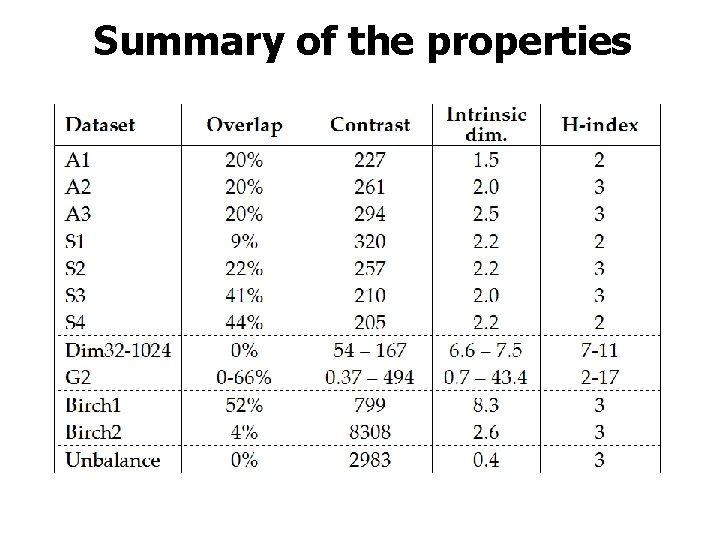
Summary of the properties

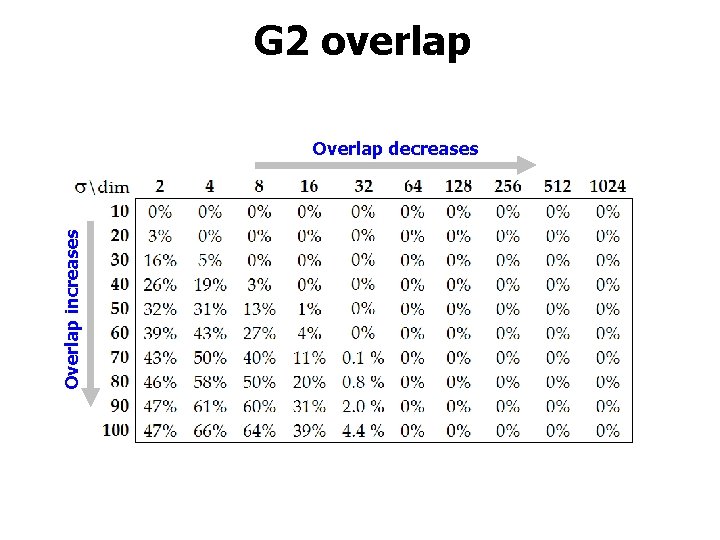
G 2 overlap Overlap increases Overlap decreases

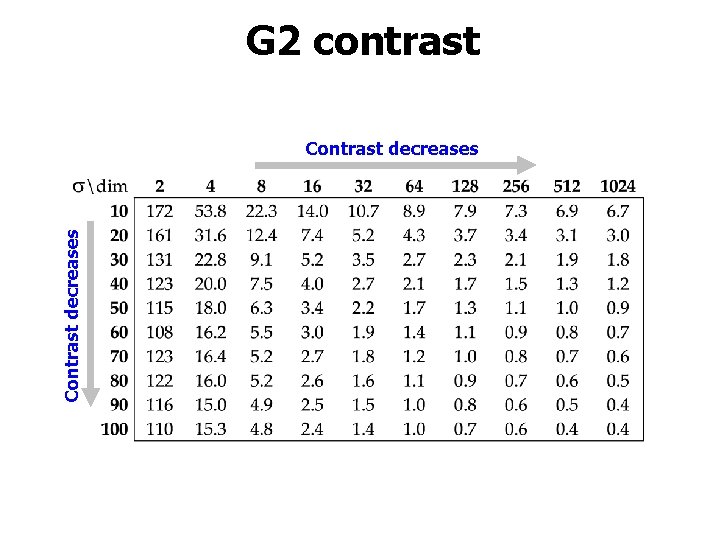
G 2 contrast Contrast decreases

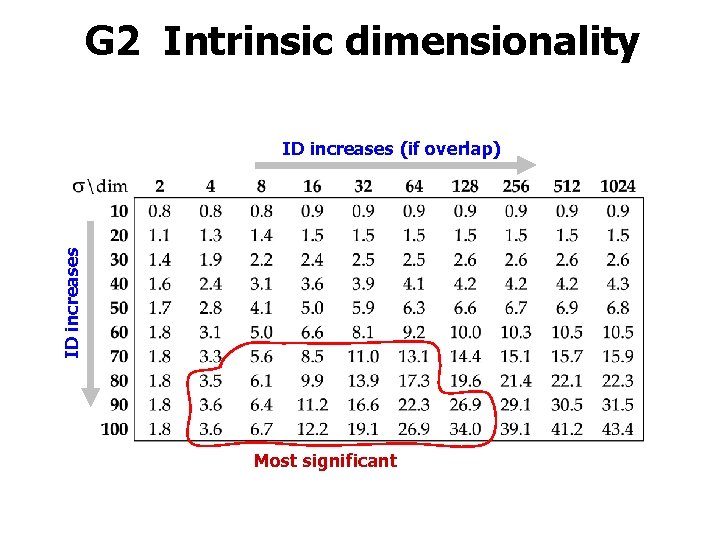
G 2 Intrinsic dimensionality ID increases (if overlap) Most significant

G 2 H-index No change H-index increases Most significant

Evaluation

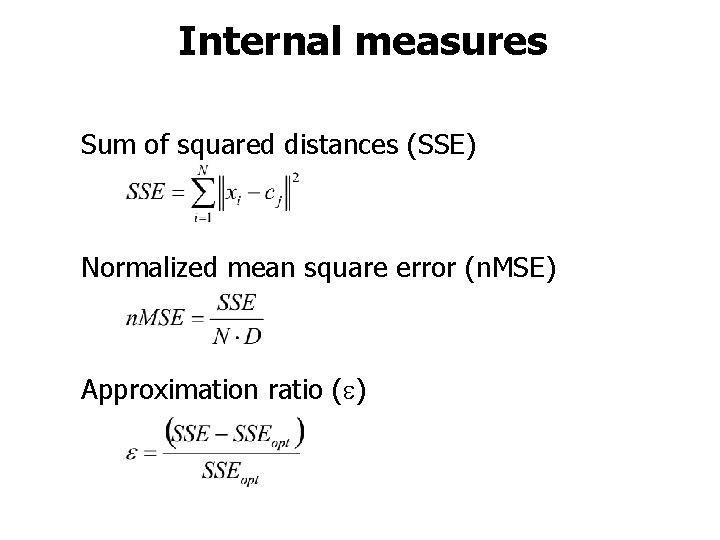
Internal measures Sum of squared distances (SSE) Normalized mean square error (n. MSE) Approximation ratio ( )

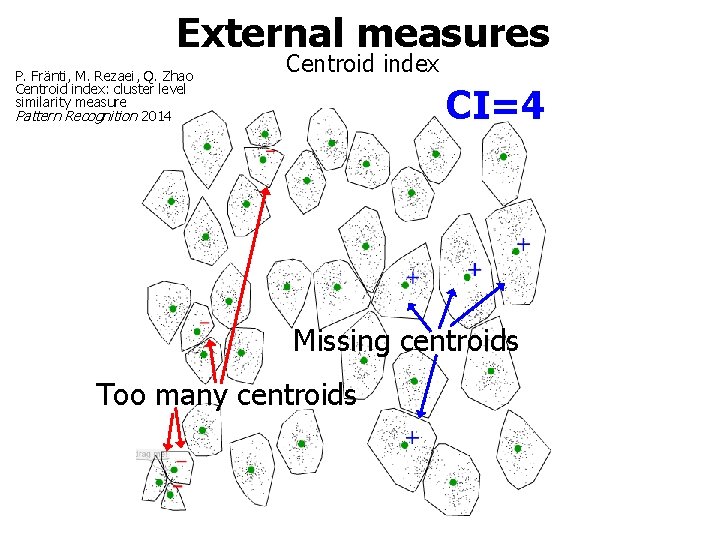
External measures P. Fränti, M. Rezaei, Q. Zhao Centroid index: cluster level similarity measure Pattern Recognition 2014 Centroid index CI=4 Missing centroids Too many centroids

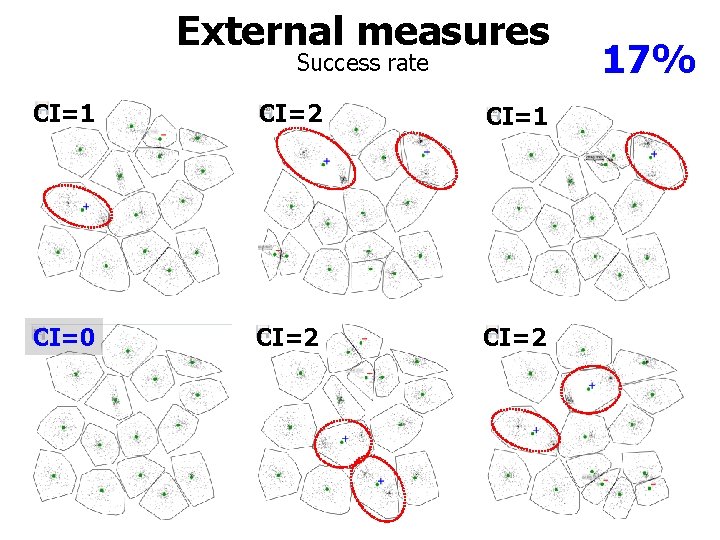
External measures Success rate CI=1 CI=2 CI=1 CI=0 CI=2 17%

Results

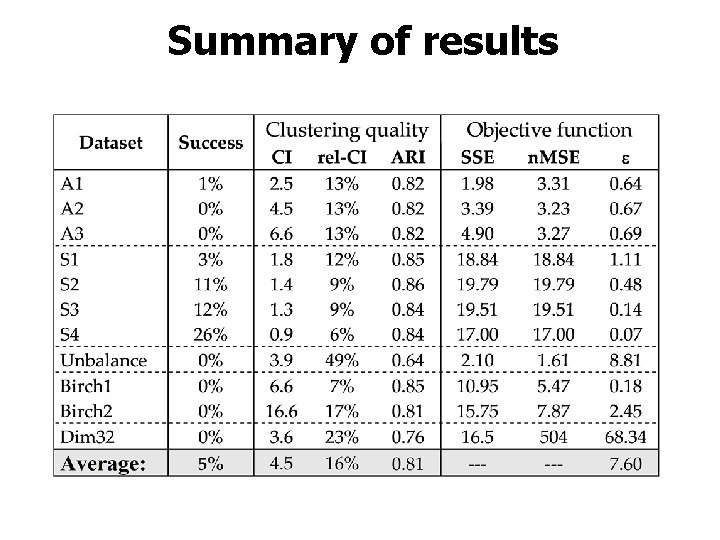
Summary of results

Dependency on overlap S datasets Success rates and CI-values: overlap increases S 1 S 2 S 3 S 4 3% 11% 12% 26% CI=1. 8 CI=1. 4 CI=1. 3 CI=0. 9

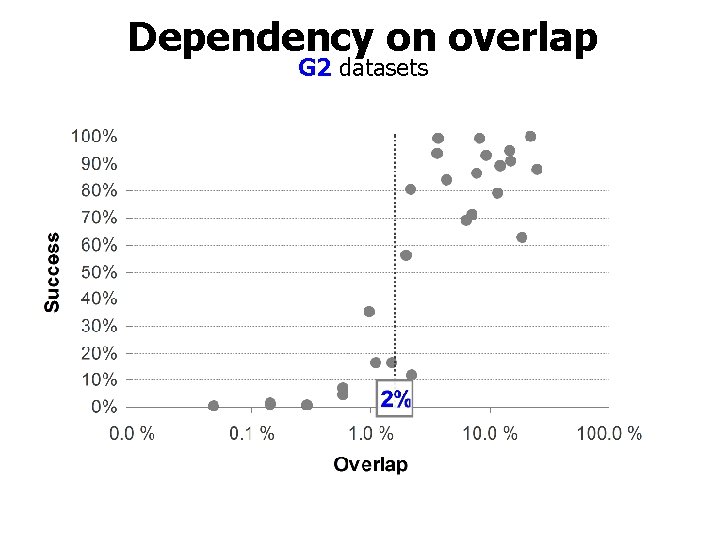
Dependency on overlap G 2 datasets

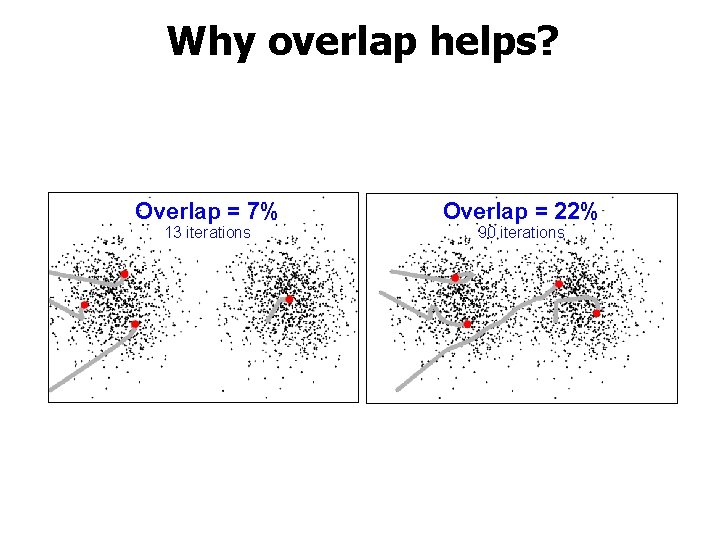
Why overlap helps? Overlap = 7% 13 iterations Overlap = 22% 90 iterations

Main observation 1. Overlap is good!

Dependency on clusters (k) A datasets K=20 A 1 Success: CI: Relative CI: Clusters increases K=35 A 2 K=50 A 3 1% 2. 5 0% 4. 5 0% 6. 6 13% 13%

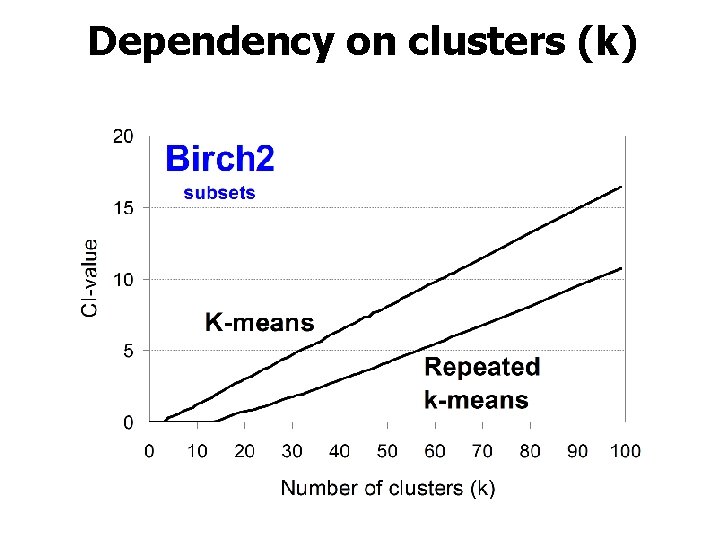
Dependency on clusters (k)

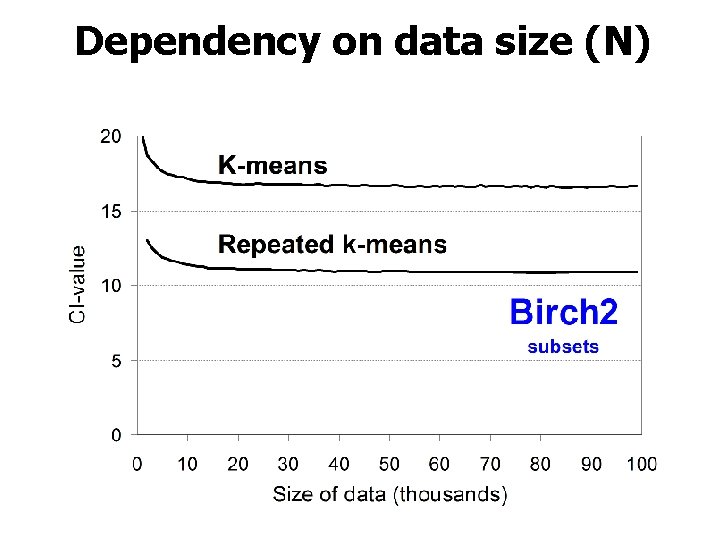
Dependency on data size (N)

Main observation 2. Number of clusters Linear increase with k!

Dependency on dimensions DIM datasets Dimensions increases 32 CI: 3. 6 64 3. 5 128 256 3. 8 Success rate: 0% 512 3. 9 1024 3. 7

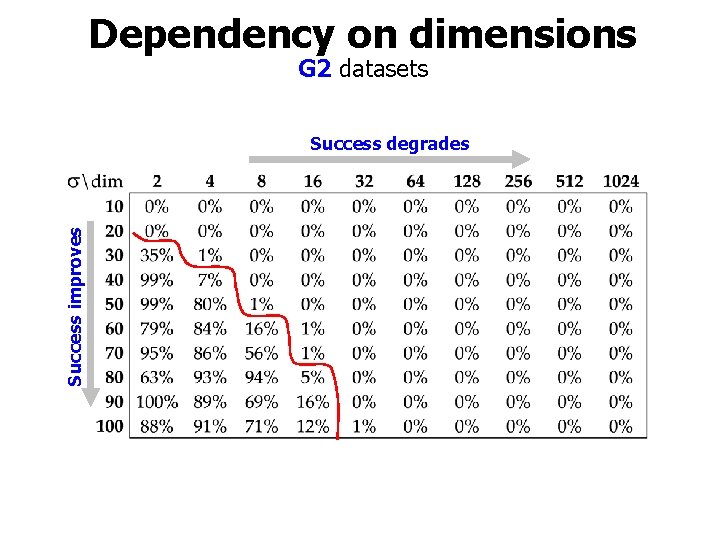
Dependency on dimensions G 2 datasets Success improves Success degrades

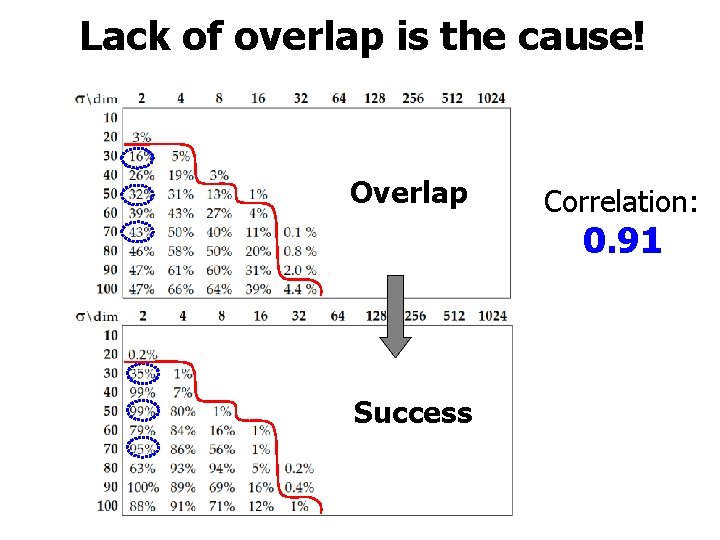
Lack of overlap is the cause! Overlap Correlation: 0. 91 Success

Main observation 3. Dimensionality No direct effect!

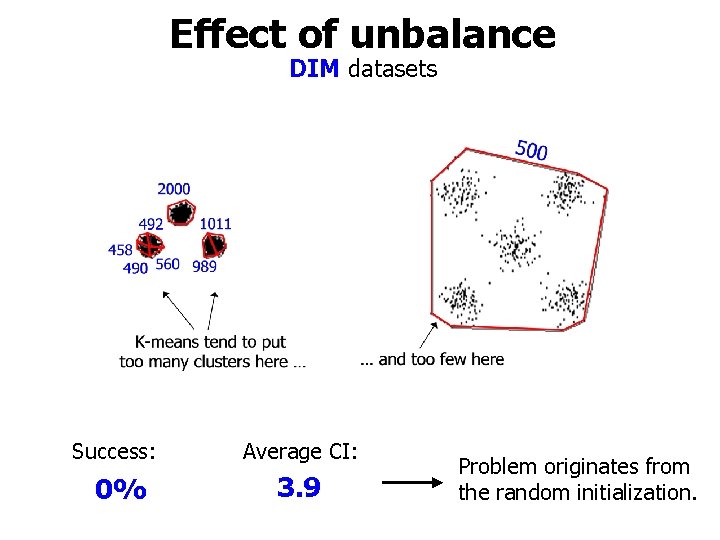
Effect of unbalance DIM datasets Success: Average CI: 0% 3. 9 Problem originates from the random initialization.

Main observation 4. Unbalance of cluster sizes Unbalance is bad!

Improving k-means
![Better initialization technique Simple initializations Random centroids Random ForgyMac Queen Further point Better initialization technique Simple initializations: • Random centroids (Random) [Forgy][Mac. Queen] • Further point](https://slidetodoc.com/presentation_image_h2/97d3f61e3ce03055831a667ef70fecc4/image-53.jpg)
Better initialization technique Simple initializations: • Random centroids (Random) [Forgy][Mac. Queen] • Further point heuristic (max) [Gonzalez] More complex: • K-means++ [Vasilievski] • Luxburg [Luxburg]

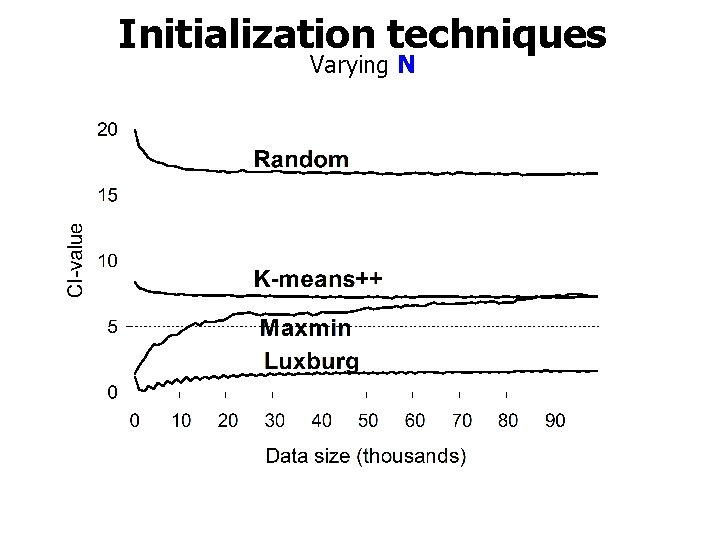
Initialization techniques Varying N

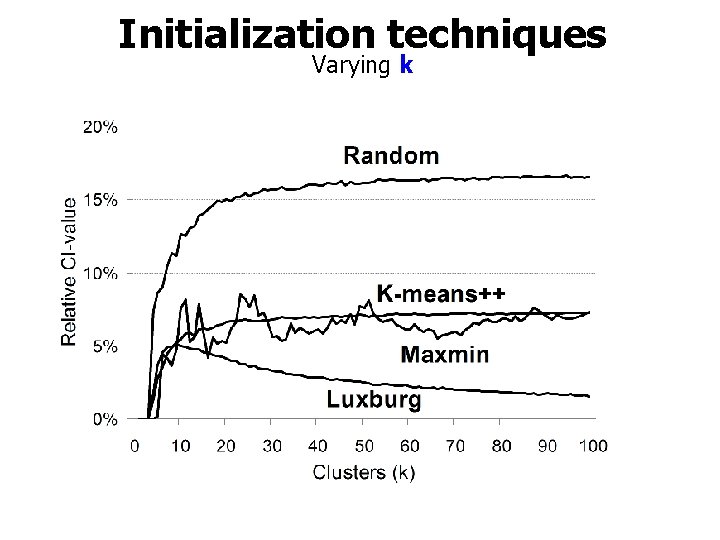
Initialization techniques Varying k

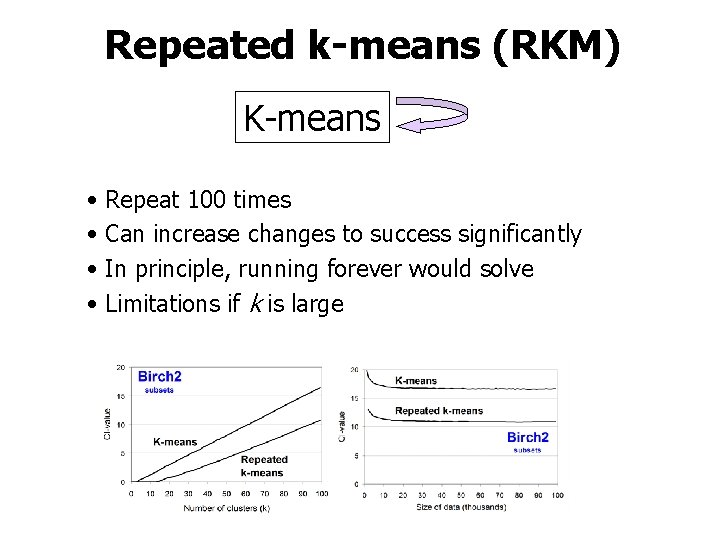
Repeated k-means (RKM) K-means • • Repeat 100 times Can increase changes to success significantly In principle, running forever would solve Limitations if k is large

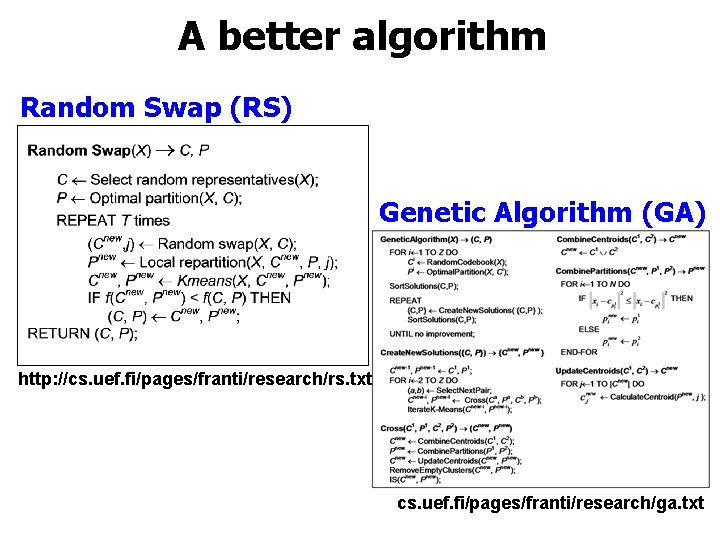
A better algorithm Random Swap (RS) Genetic Algorithm (GA) http: //cs. uef. fi/pages/franti/research/rs. txt cs. uef. fi/pages/franti/research/ga. txt

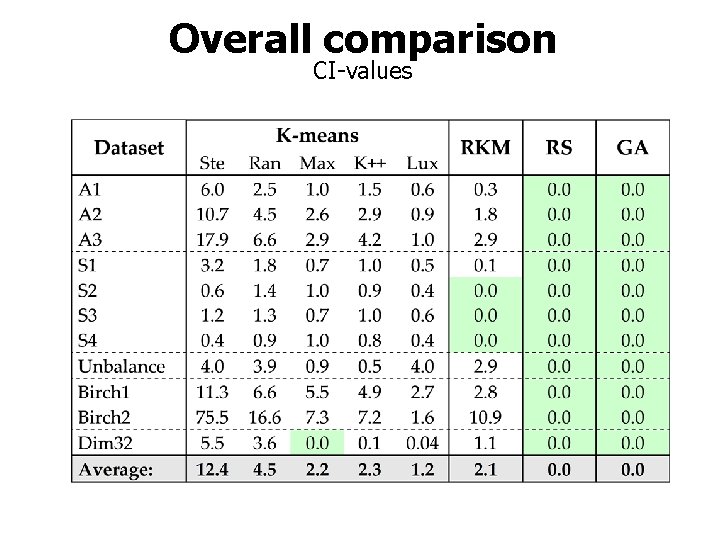
Overall comparison CI-values

Conclusions

Conclusions How did K-means perform? 1. Overlap 3. Dimensionality Good! No change 2. Number of clusters Bad! 4. Unbalance of cluster sizes Bad!

References • • • • • J. Mac. Queen, Some methods for classification and analysis of multivariate observations, Berkeley Symposium on Mathematical Statistics and Probability, Volume 1: Statistics, pp. 281 -297, University of California Press, Berkeley, Calif. , 1967. S. P. Lloyd, Least squares quantization in PCM, IEEE Trans. on Information Theory, 28 (2), 129– 137, 1982. Forgy, E. (1965). Cluster analysis of multivariate data: Efficiency vs. interpretability of classification. Biometrics, 21, 768. M. Steinbach, L. Ertöz, V. Kumar, The challenges of clustering high dimensional data, New Vistas in Statistical Physics -Applications in Econophysics, Bioinformatics, and Pattern Recognition, Springer-Verlag, 2003. U. Luxburg, R. C. Williamson, I. Guyon, "Clustering: Science or Art? ", J. Machine Learning Research, 27: 65– 79, 2012. P. Fränti, "Genetic algorithm with deterministic crossover for vector quantization", Pattern Recognition Letters, 21 (1), 61 -68, 2000 P. Fränti and J. Kivijärvi, "Randomized local search algorithm for the clustering problem", Pattern Analysis and Applications, 3 (4), 358 -369, 2000. P. Fränti, O. Virmajoki and V. Hautamäki, Fast agglomerative clustering using a k-nearest neighbor graph, IEEE Trans. on Pattern Analysis and Machine Intelligence, 28 (11), 1875 -1881, November 2006. Zhang R. Ramakrishnan and M. Livny, BIRCH: A new data clustering algorithm and its applications, Data Mining and Knowledge Discovery, 1 (2), 141 -182, 1997. I. Kärkkäinen and P. Fränti, Dynamic local search algorithm for the clustering problem, Research Report A-2002 -6. P. Fränti and O. Virmajoki, Iterative shrinking method for clustering problems, Pattern Recognition, 39 (5), 761 -765, May 2006. P. Fränti R. Mariescu-Istodor and C. Zhong, XNN graph IAPR Joint Int. Workshop on Structural, Syntactic, and Statistical Pattern Recognition Merida, Mexico, LNCS 10029, 207 -217, November 2016. M. Rezaei and P. Fränti, "Set-matching methods for external cluster validity", IEEE Trans. on Knowledge and Data Engineering, 28 (8), 2173 -2186, August 2016. E. Chavez and G. Navarro, A probabilistic spell for the curse of dimensionality. Workshop on Algorithm Engineering and Experimentation, LNCS 2153, 147 -160, 2001. N. Tomasev, M. Radovanovi, D. Mladeni and M. Ivanovi, “The role of hubness in clustering high-dimensional data”, IEEE Trans. on Knowledge and Data Engineering, 26 (3), 739 -751, March 2014. D. Steinley, Local optima in k-means clustering: what you don’t know may hurt you”, Psychological Methods, 8, 294– 304, 2003. P. Fränti, M. Rezaei and Q. Zhao, "Centroid index: Cluster level similarity measure", Pattern Recognition, 47 (9), 3034 -3045, 2014. T. Gonzalez, Clustering to minimize the maximum intercluster distance. Theoretical Computer Science, 38 (2– 3), 293– 306, 1985.
 Thrust kmeans
Thrust kmeans Sota clustering
Sota clustering Javascript kmeans
Javascript kmeans Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Pasi hongisto
Pasi hongisto Cook’s theorem
Cook’s theorem Frnti
Frnti Frnti
Frnti Tsp
Tsp Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Semaine 332021
Semaine 332021 Regulamentul baschetului
Regulamentul baschetului Pasi fränti
Pasi fränti Plan sagittal
Plan sagittal Turvavalaistus
Turvavalaistus Pasi puranen
Pasi puranen Pasi kaipainen
Pasi kaipainen Pasi näkki
Pasi näkki Pasi kaipainen
Pasi kaipainen Structura repetitiva cu numar cunoscut de pasi
Structura repetitiva cu numar cunoscut de pasi Fjali nenrenditese vendore
Fjali nenrenditese vendore Typasi
Typasi Pasi rantahalvari
Pasi rantahalvari Pasi
Pasi Pasi fränti
Pasi fränti Extensive and intensive properties
Extensive and intensive properties Physical properties of ice cube
Physical properties of ice cube B13l
B13l Test regnskapsprogram 2018
Test regnskapsprogram 2018 Duracion contrato de obras ley 9/2017
Duracion contrato de obras ley 9/2017 Ncaa clearinghouse 2017
Ncaa clearinghouse 2017 2. daļas 8. uzd. procenti tekstā (2017)
2. daļas 8. uzd. procenti tekstā (2017) 2017 english standards of learning curriculum framework
2017 english standards of learning curriculum framework Ilae clasificacion
Ilae clasificacion Vcaa 2017 english exam
Vcaa 2017 english exam Job meeting bari 2017
Job meeting bari 2017 Arbeit.swiss jobroom
Arbeit.swiss jobroom Morten meen gallefos
Morten meen gallefos 2017 pearson education inc
2017 pearson education inc Lebah jantan 2017
Lebah jantan 2017 2000 rzymskie cyfry
2000 rzymskie cyfry Fyss 2020
Fyss 2020 Visual studio code oracle
Visual studio code oracle Vtc 2017
Vtc 2017 Pearson education
Pearson education Emp 2017
Emp 2017 648 de 2017
648 de 2017 Acr tirads 2017
Acr tirads 2017 2017 pearson education inc
2017 pearson education inc Forrester wave predictive analytics 2016
Forrester wave predictive analytics 2016 Dm 910 del 2017
Dm 910 del 2017 Reizināšanas likums
Reizināšanas likums Cost of downtime 2017
Cost of downtime 2017 Smx advanced 2017 slides
Smx advanced 2017 slides Informe técnico 1005-2018-servir-gpgsc
Informe técnico 1005-2018-servir-gpgsc