Centroid index Cluster level quality measure Pasi Frnti


![External index Selection of existed methods Pair-counting measures • Rand index (RI) [Rand, 1971] External index Selection of existed methods Pair-counting measures • Rand index (RI) [Rand, 1971]](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-9.jpg)


![Centroid index (CI) [Fränti, Rezaei, Zhao, Pattern Recognition 2014] CI = 4 empty 15 Centroid index (CI) [Fränti, Rezaei, Zhao, Pattern Recognition 2014] CI = 4 empty 15](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-16.jpg)

![Adjusted Rand Index [Hubert & Arabie, 1985] Data set Adjusted Rand Index (ARI) KM Adjusted Rand Index [Hubert & Arabie, 1985] Data set Adjusted Rand Index (ARI) KM](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-26.jpg)
![Normalized Mutual information [Kvalseth, 1987] Data set KM Normalized Mutual Information (NMI) RKM KM++ Normalized Mutual information [Kvalseth, 1987] Data set KM Normalized Mutual Information (NMI) RKM KM++](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-27.jpg)
![Normalized Van Dongen [Kvalseth, 1987] Data set Bridge House Miss America House Birch 1 Normalized Van Dongen [Kvalseth, 1987] Data set Bridge House Miss America House Birch 1](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-28.jpg)
![Centroid Similarity Index [Fränti, Rezaei, Zhao, 2014] Centroid Similarity Index (CSI) Data set Bridge Centroid Similarity Index [Fränti, Rezaei, Zhao, 2014] Centroid Similarity Index (CSI) Data set Bridge](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-29.jpg)
![Centroid Index [Fränti, Rezaei, Zhao, 2014] C-Index (CI 2) Data set Bridge House Miss Centroid Index [Fränti, Rezaei, Zhao, 2014] C-Index (CI 2) Data set Bridge House Miss](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-30.jpg)



- Slides: 51

Centroid index Cluster level quality measure Pasi Fränti 3. 9. 2018

Clustering accuracy

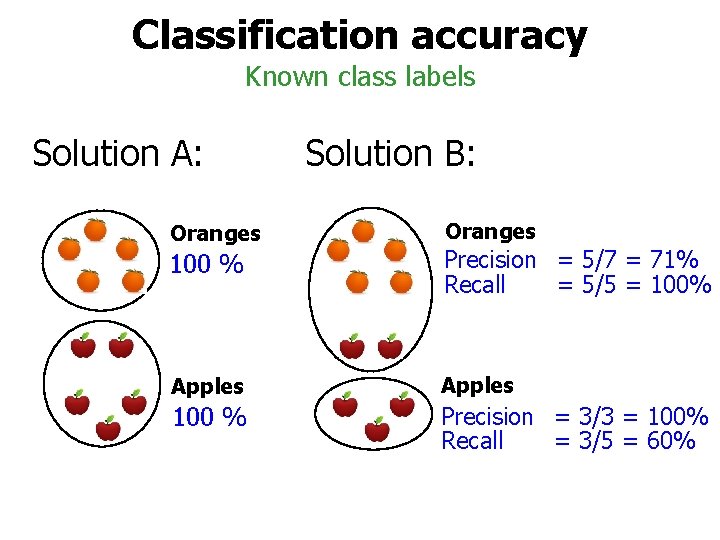
Classification accuracy Known class labels Solution A: Solution B: Oranges 100 % Precision = 5/7 = 71% Recall = 5/5 = 100% Apples 100 % P Precision = 3/3 = 100% Recall = 3/5 = 60%

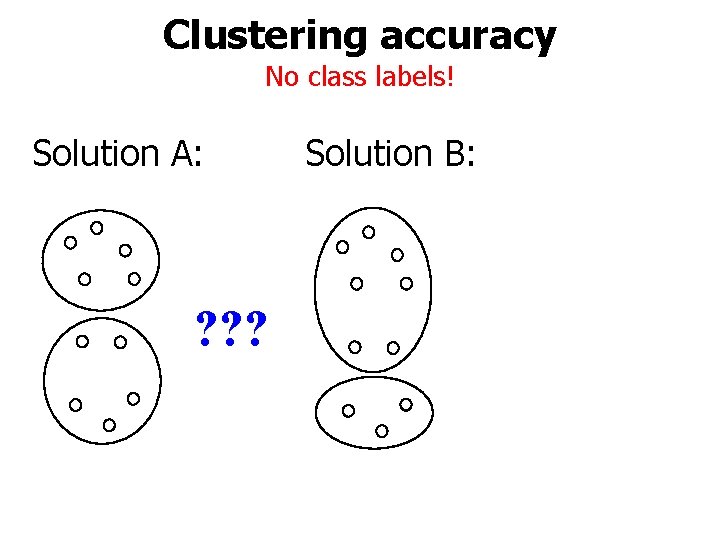
Clustering accuracy No class labels! Solution A: ? ? ? Solution B:

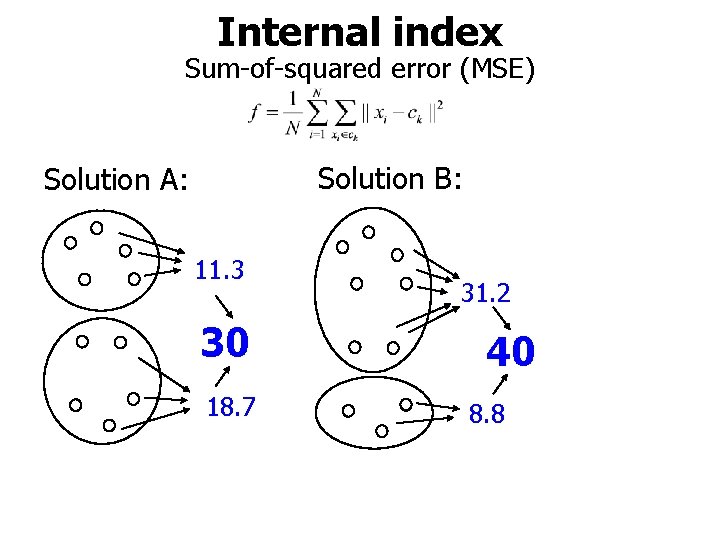
Internal index Sum-of-squared error (MSE) Solution B: Solution A: 11. 3 30 18. 7 31. 2 40 8. 8

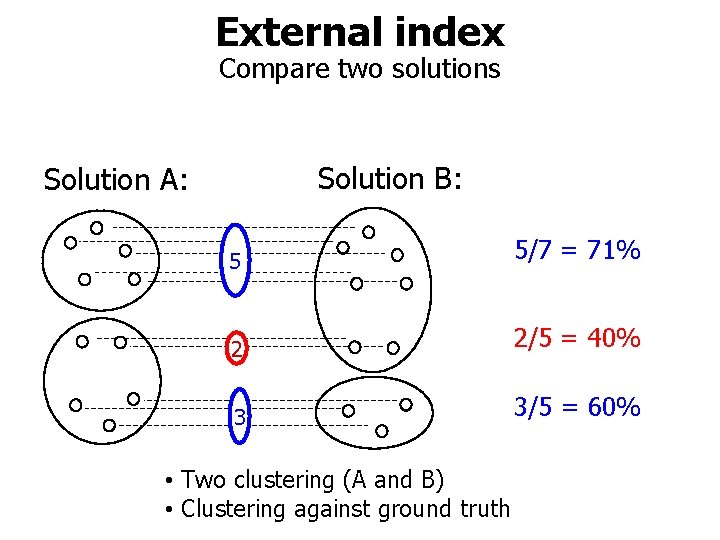
External index Compare two solutions Solution B: Solution A: 5 5/7 = 71% 2 2/5 = 40% 3 3/5 = 60% • Two clustering (A and B) • Clustering against ground truth

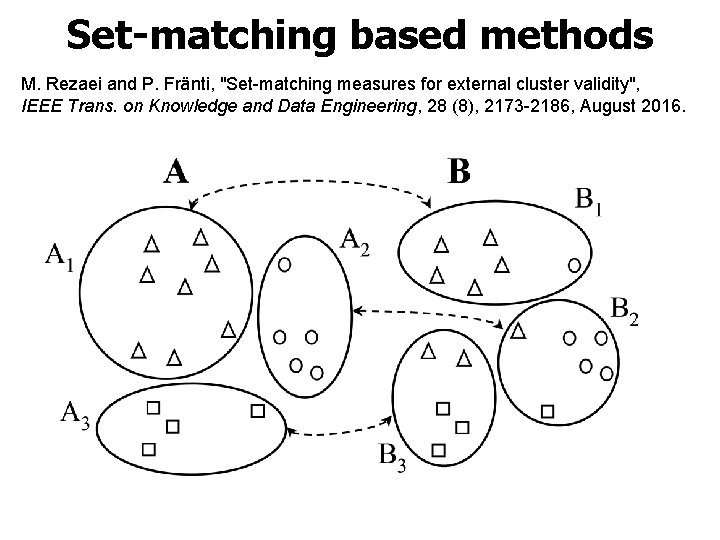
Set-matching based methods M. Rezaei and P. Fränti, "Set-matching measures for external cluster validity", IEEE Trans. on Knowledge and Data Engineering, 28 (8), 2173 -2186, August 2016.

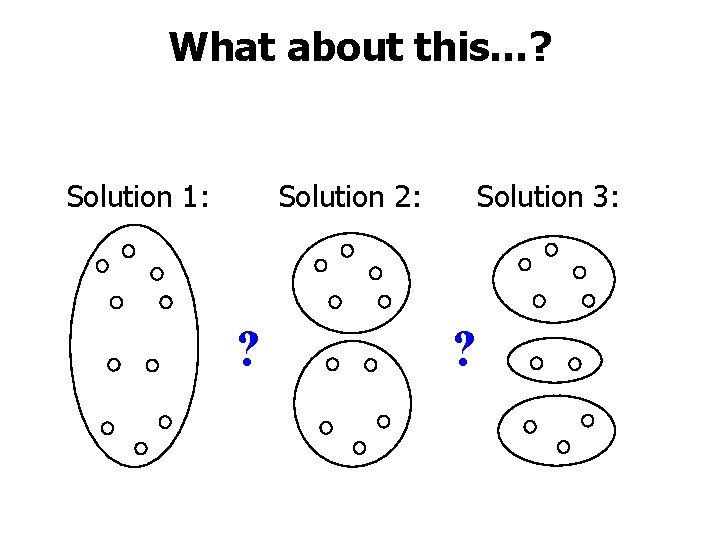
What about this…? Solution 2: Solution 1: ? Solution 3: ?
![External index Selection of existed methods Paircounting measures Rand index RI Rand 1971 External index Selection of existed methods Pair-counting measures • Rand index (RI) [Rand, 1971]](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-9.jpg)
External index Selection of existed methods Pair-counting measures • Rand index (RI) [Rand, 1971] • Adjusted Rand index (ARI) [Hubert & Arabie, 1985] Information-theoretic measures • Mutual information (MI) [Vinh, Epps, Bailey, 2010] • Normalized Mutual information (NMI) [Kvalseth, 1987] Set-matching based measures • • Normalized van Dongen (NVD) [Kvalseth, 1987] Criterion H (CH) [Meila & Heckerman, 2001] Purity [Rendon et al, 2011] Centroid index (CI) [Fränti, Rezaei & Zhao, 2014]

Cluster level measure

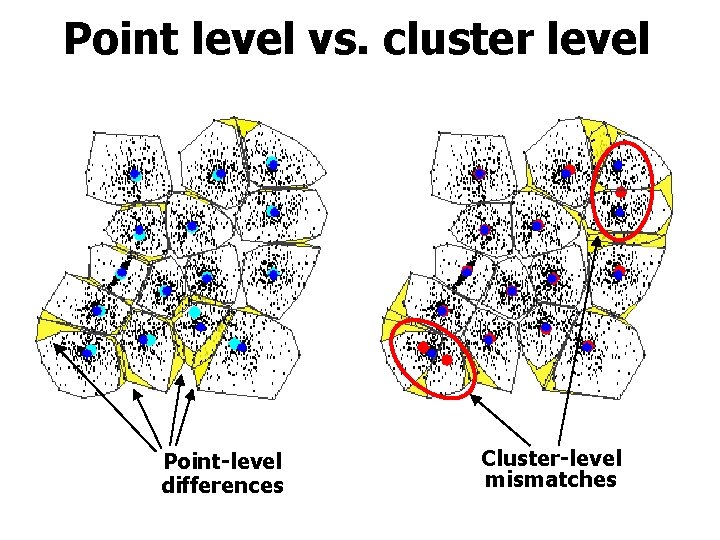
Point level vs. cluster level Point-level differences Cluster-level mismatches

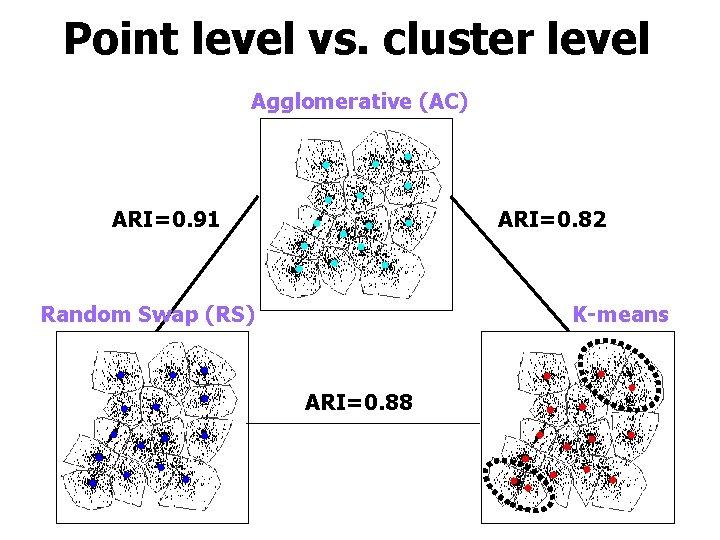
Point level vs. cluster level Agglomerative (AC) ARI=0. 91 ARI=0. 82 Random Swap (RS) K-means ARI=0. 88

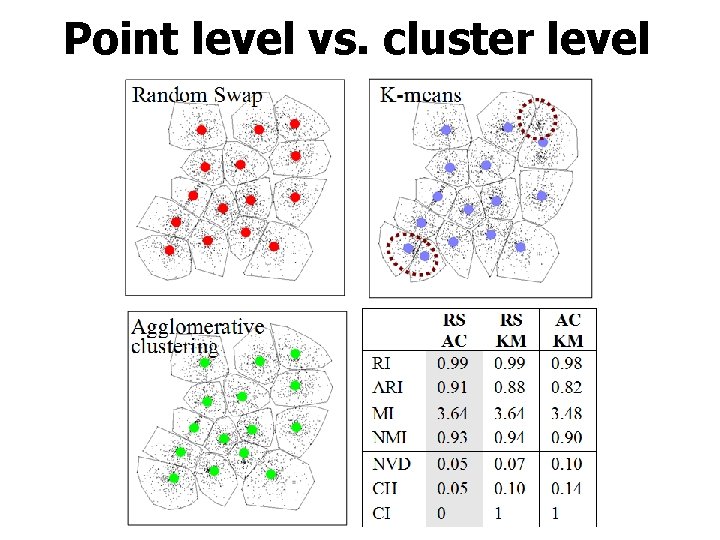
Point level vs. cluster level

Centroid index

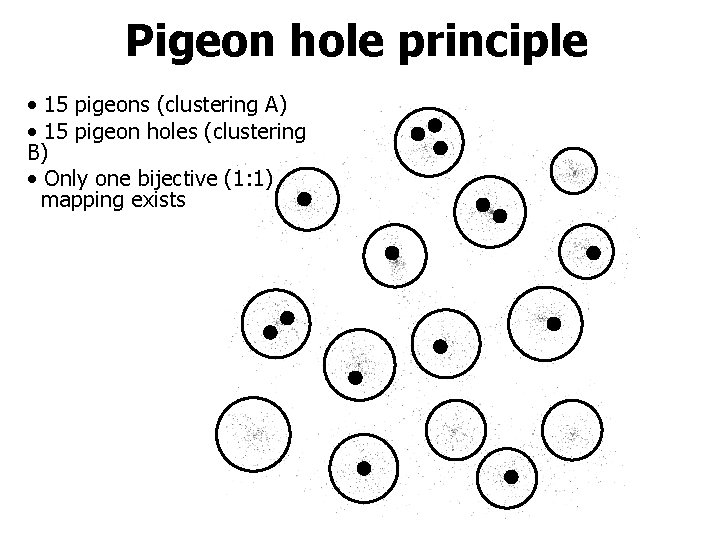
Pigeon hole principle • 15 pigeons (clustering A) • 15 pigeon holes (clustering B) • Only one bijective (1: 1) mapping exists
![Centroid index CI Fränti Rezaei Zhao Pattern Recognition 2014 CI 4 empty 15 Centroid index (CI) [Fränti, Rezaei, Zhao, Pattern Recognition 2014] CI = 4 empty 15](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-16.jpg)
Centroid index (CI) [Fränti, Rezaei, Zhao, Pattern Recognition 2014] CI = 4 empty 15 prototypes (pigeons) 15 real clusters (pigeon holes) empty

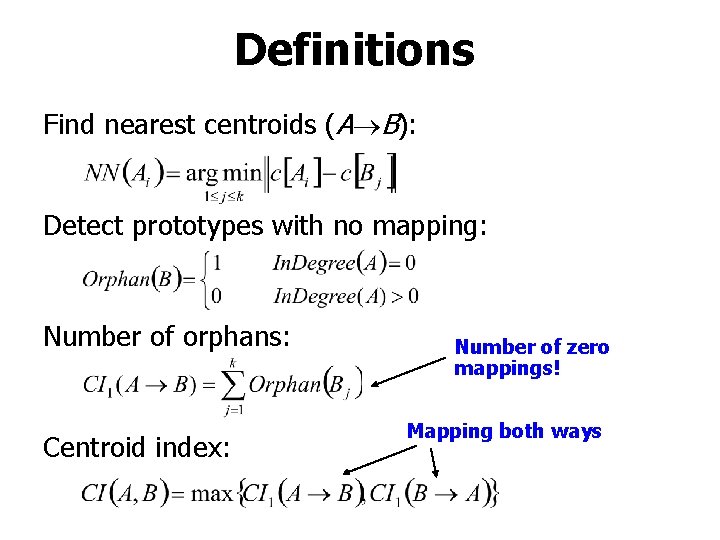
Definitions Find nearest centroids (A B): Detect prototypes with no mapping: Number of orphans: Centroid index: Number of zero mappings! Mapping both ways

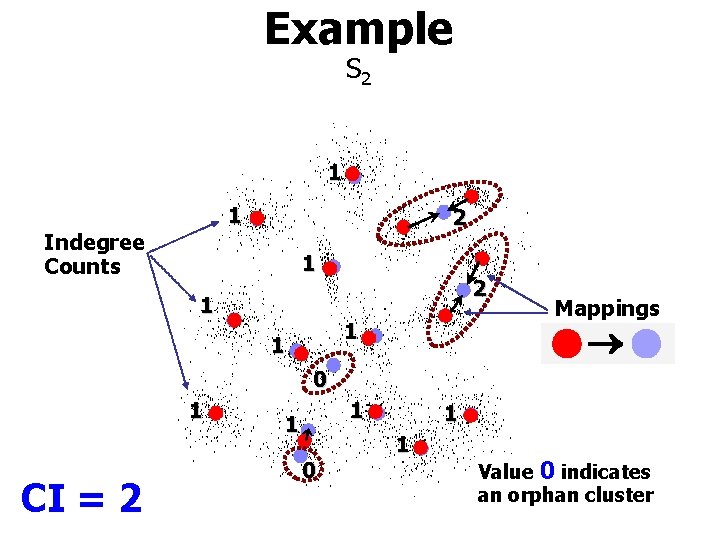
Example S 2 1 1 Indegree Counts 2 1 1 Mappings 0 1 CI = 2 1 1 0 1 1 Value 0 indicates an orphan cluster

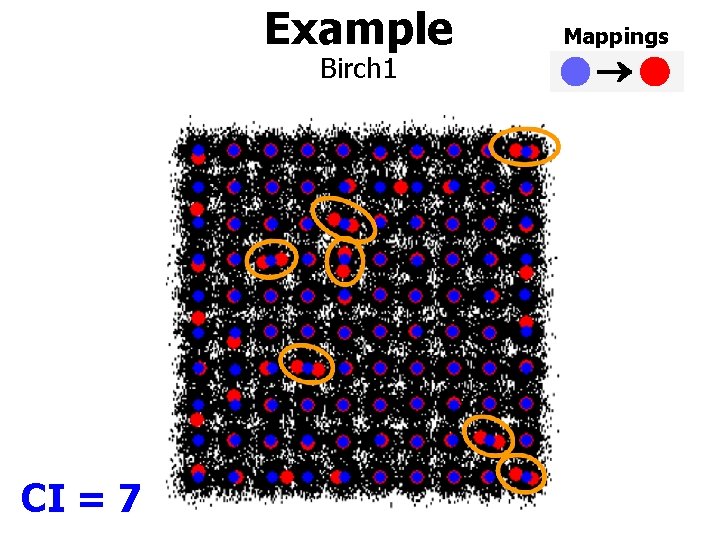
Example Birch 1 CI = 7 Mappings

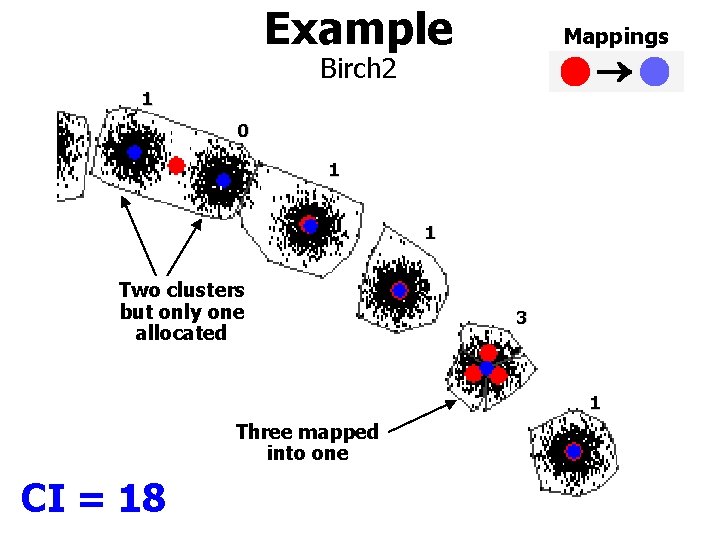
Example Mappings Birch 2 1 0 1 1 Two clusters but only one allocated 3 1 Three mapped into one CI = 18

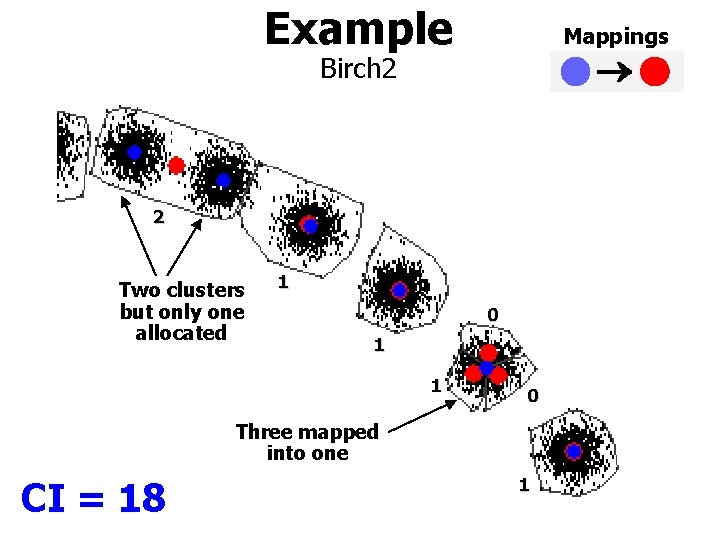
Example Mappings Birch 2 2 Two clusters but only one allocated 1 0 1 1 0 Three mapped into one CI = 18 1

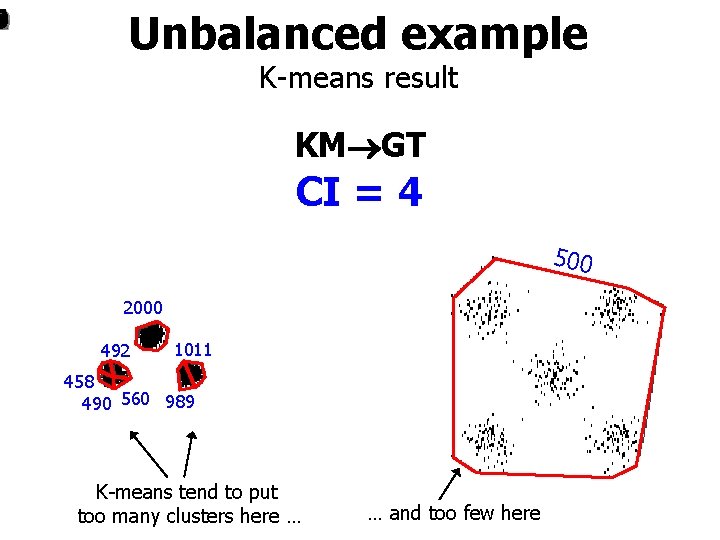
1 4 2 0 Unbalanced example K-means result KM GT CI = 4 500 2000 492 1011 458 490 560 989 K-means tend to put too many clusters here … … and too few here

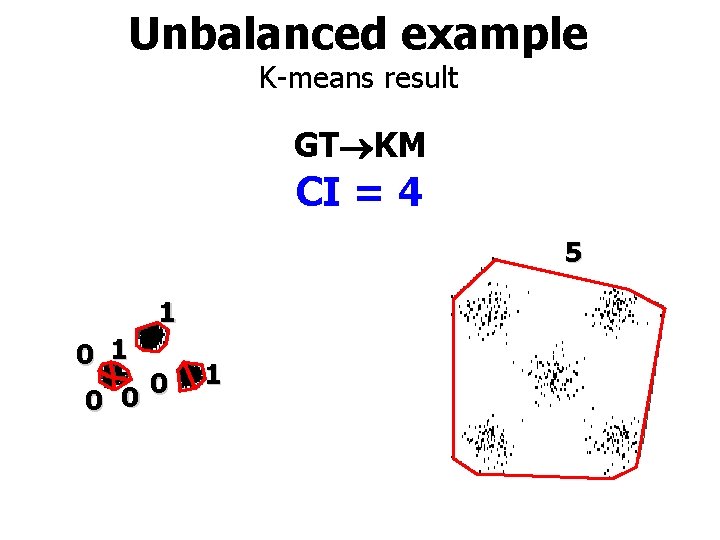
Unbalanced example K-means result GT KM CI = 4 5 1 0 0 0 1

Experiments

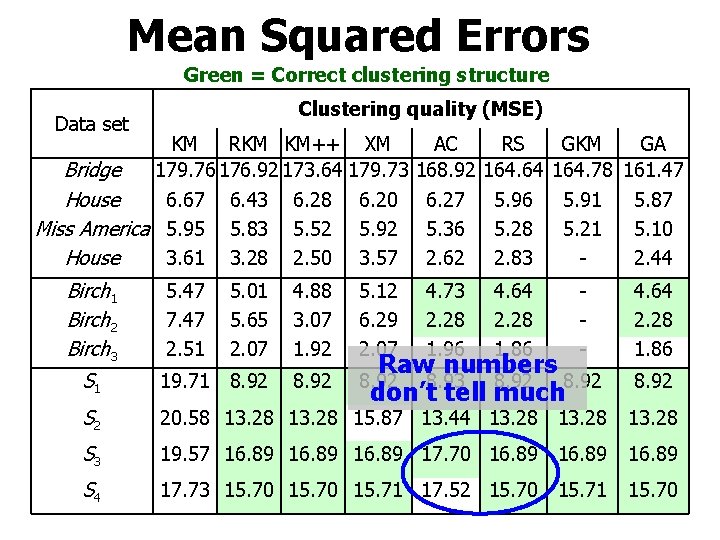
Mean Squared Errors Green = Correct clustering structure Clustering quality (MSE) Data set KM RKM KM++ XM Bridge 179. 76 176. 92 173. 64 179. 73 House 6. 67 6. 43 6. 28 6. 20 Miss America 5. 95 5. 83 5. 52 5. 92 House 3. 61 3. 28 2. 50 3. 57 Birch 1 Birch 2 Birch 3 5. 47 7. 47 2. 51 5. 12 6. 29 2. 07 AC 168. 92 6. 27 5. 36 2. 62 RS 164. 64 5. 96 5. 28 2. 83 4. 73 2. 28 1. 96 4. 64 2. 28 1. 86 GKM GA 164. 78 161. 47 5. 91 5. 87 5. 21 5. 10 2. 44 5. 01 5. 65 2. 07 4. 88 3. 07 1. 92 - S 1 19. 71 8. 92 S 2 20. 58 13. 28 15. 87 13. 44 13. 28 S 3 19. 57 16. 89 17. 70 16. 89 S 4 17. 73 15. 70 15. 71 17. 52 15. 70 15. 71 15. 70 Raw numbers 8. 92 8. 93 8. 92 don’t tell much 4. 64 2. 28 1. 86 8. 92
![Adjusted Rand Index Hubert Arabie 1985 Data set Adjusted Rand Index ARI KM Adjusted Rand Index [Hubert & Arabie, 1985] Data set Adjusted Rand Index (ARI) KM](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-26.jpg)
Adjusted Rand Index [Hubert & Arabie, 1985] Data set Adjusted Rand Index (ARI) KM Bridge 0. 38 House 0. 40 Miss America 0. 19 RKM KM++ XM AC RS GKM GA 0. 40 0. 39 0. 37 0. 43 0. 52 0. 50 1 0. 40 0. 44 0. 47 0. 43 0. 53 1 0. 19 0. 18 0. 20 0. 23 1 House 0. 46 0. 49 0. 52 0. 46 0. 49 - 1 Birch 2 Birch 3 S 1 S 2 S 3 S 4 0. 85 0. 93 0. 98 0. 91 0. 96 1. 00 - 1 0. 86 0. 95 0. 86 1 1 - 1 0. 74 0. 82 0. 87 0. 82 0. 86 0. 83 1. 00 0. 80 0. 99 0. 89 0. 98 0. 99 0. 86 0. 96 0. 92 0. 96 0. 82 0. 93 0. 94 0. 77 0. 93 0. 91 high How is good? 1. 00 1 1. 00
![Normalized Mutual information Kvalseth 1987 Data set KM Normalized Mutual Information NMI RKM KM Normalized Mutual information [Kvalseth, 1987] Data set KM Normalized Mutual Information (NMI) RKM KM++](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-27.jpg)
Normalized Mutual information [Kvalseth, 1987] Data set KM Normalized Mutual Information (NMI) RKM KM++ XM AC RS GKM Bridge 0. 77 0. 78 House 0. 80 Miss America 0. 64 GA 0. 78 0. 77 0. 80 0. 83 0. 82 1. 00 0. 81 0. 82 0. 81 0. 83 0. 84 1. 00 0. 63 0. 64 0. 66 1. 00 House 0. 81 0. 82 - 1. 00 Birch 1 Birch 2 Birch 3 S 1 S 2 S 3 S 4 0. 95 0. 97 0. 99 0. 96 0. 98 1. 00 - 1. 00 0. 96 0. 97 0. 99 0. 97 1. 00 - 1. 00 0. 94 0. 93 0. 96 - 1. 00 0. 93 1. 00 1. 00 0. 99 0. 95 0. 99 0. 93 0. 99 0. 92 0. 97 0. 94 0. 97 0. 88 0. 94 0. 95 0. 85 0. 94
![Normalized Van Dongen Kvalseth 1987 Data set Bridge House Miss America House Birch 1 Normalized Van Dongen [Kvalseth, 1987] Data set Bridge House Miss America House Birch 1](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-28.jpg)
Normalized Van Dongen [Kvalseth, 1987] Data set Bridge House Miss America House Birch 1 Birch 2 Birch 3 S 1 S 2 S 3 S 4 Normalized Van Dongen (NVD) KM RKM KM++ XM AC RS GKM GA 0. 45 0. 44 0. 60 0. 42 0. 43 0. 60 0. 43 0. 40 0. 61 0. 46 0. 37 0. 59 0. 38 0. 40 0. 57 0. 32 0. 33 0. 55 0. 33 0. 31 0. 53 0. 00 0. 40 0. 37 0. 34 0. 39 0. 34 - 0. 00 0. 09 0. 12 0. 19 0. 09 0. 11 0. 08 0. 11 0. 04 0. 08 0. 12 0. 00 0. 02 0. 04 0. 01 0. 03 0. 10 0. 00 0. 02 0. 04 0. 06 0. 02 0. 09 0. 00 0. 13 0. 00 0. 06 0. 01 0. 02 0. 05 0. 03 0. 13 0. 00 0. 06 -is Lower 0. 00 better 0. 04 0. 00 0. 02 0. 04
![Centroid Similarity Index Fränti Rezaei Zhao 2014 Centroid Similarity Index CSI Data set Bridge Centroid Similarity Index [Fränti, Rezaei, Zhao, 2014] Centroid Similarity Index (CSI) Data set Bridge](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-29.jpg)
Centroid Similarity Index [Fränti, Rezaei, Zhao, 2014] Centroid Similarity Index (CSI) Data set Bridge House Miss America House Birch 1 Birch 2 Birch 3 S 1 S 2 S 3 S 4 KM RKM KM++ XM AC RS GKM GA 0. 47 0. 49 0. 32 0. 51 0. 50 0. 32 0. 49 0. 54 0. 32 0. 45 0. 57 0. 33 0. 57 0. 55 0. 38 0. 62 0. 63 0. 40 0. 63 0. 66 0. 42 1. 00 0. 54 0. 57 0. 63 0. 54 0. 57 0. 62 --- 1. 00 0. 87 0. 76 0. 71 0. 83 0. 82 0. 89 0. 87 0. 94 0. 82 1. 00 0. 99 0. 98 0. 94 0. 87 1. 00 0. 99 0. 98 0. 93 0. 81 1. 00 0. 91 0. 99 1. 00 0. 86 1. 00 0. 98 0. 85 1. 00 --1. 00 Ok but---lacks 0. 93 --1. 00 threshold 1. 00 0. 99 0. 98
![Centroid Index Fränti Rezaei Zhao 2014 CIndex CI 2 Data set Bridge House Miss Centroid Index [Fränti, Rezaei, Zhao, 2014] C-Index (CI 2) Data set Bridge House Miss](https://slidetodoc.com/presentation_image_h/007dac8918ec369f68b0dde4ee185988/image-30.jpg)
Centroid Index [Fränti, Rezaei, Zhao, 2014] C-Index (CI 2) Data set Bridge House Miss America House Birch 1 Birch 2 Birch 3 S 1 S 2 S 3 S 4 KM RKM KM++ XM AC RS GKM GA 74 56 88 63 45 91 58 40 67 81 37 88 33 31 38 33 22 43 35 20 36 0 0 0 43 39 22 47 26 23 --- 0 7 18 23 2 2 1 1 3 11 11 0 0 1 4 7 0 0 4 12 10 0 1 0 0 7 0 0 0 1 0 0 2 0 0 ------0 0 0

Going deeper…

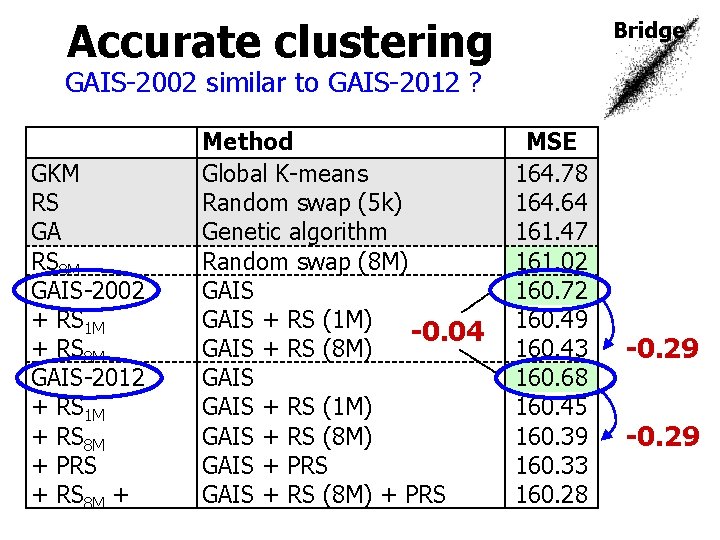
Accurate clustering Bridge GAIS-2002 similar to GAIS-2012 ? GKM RS GA RS 8 M GAIS-2002 + RS 1 M + RS 8 M GAIS-2012 + RS 1 M + RS 8 M + PRS + RS 8 M + Method Global K-means Random swap (5 k) Genetic algorithm Random swap (8 M) GAIS + RS (1 M) -0. 04 GAIS + RS (8 M) GAIS + RS (1 M) GAIS + RS (8 M) GAIS + PRS GAIS + RS (8 M) + PRS MSE 164. 78 164. 64 161. 47 161. 02 160. 72 160. 49 160. 43 160. 68 160. 45 160. 39 160. 33 160. 28 -0. 29

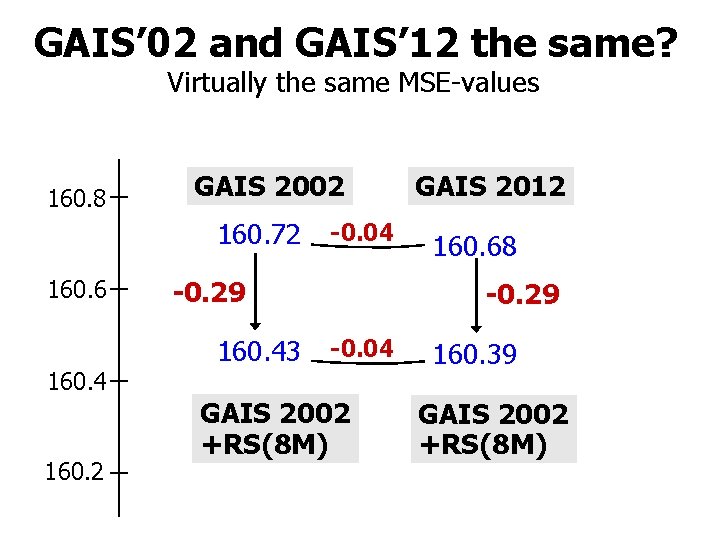
GAIS’ 02 and GAIS’ 12 the same? Virtually the same MSE-values 160. 8 GAIS 2002 160. 72 160. 6 160. 4 160. 2 -0. 04 -0. 29 160. 43 GAIS 2012 160. 68 -0. 29 -0. 04 GAIS 2002 +RS(8 M) 160. 39 GAIS 2002 +RS(8 M)

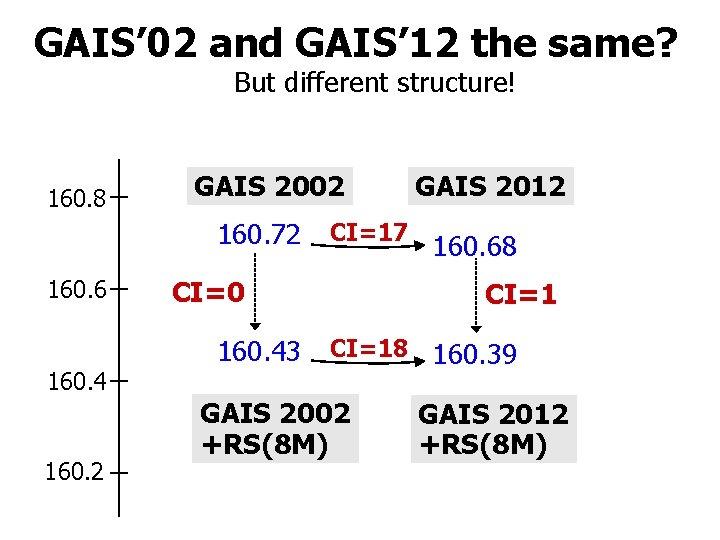
GAIS’ 02 and GAIS’ 12 the same? But different structure! 160. 8 GAIS 2002 160. 72 160. 6 160. 4 160. 2 CI=17 CI=0 160. 43 GAIS 2012 160. 68 CI=18 GAIS 2002 +RS(8 M) 160. 39 GAIS 2012 +RS(8 M)

Seemingly the same solutions Same structure “same family” Main RS 8 M algorithm: + Tuning 1 × + Tuning 2 × RS 8 M --GAIS (2002) 23 + RS 1 M 23 + RS 8 M 23 GAIS (2012) 25 + RS 1 M 25 + RS 8 M 25 + PRS 25 + RS 8 M + PRS 24 GAIS 2002 × × 19 --0 0 17 17 17 RS 1 M RS 8 M × × 19 19 0 0 --18 18 18 GAIS 2012 × × 23 14 14 14 --1 1 RS 1 M RS 8 M × × 24 24 15 15 15 1 1 --0 0 1 1 × 23 14 14 14 1 0 0 --1 RS 8 M 22 16 13 13 1 1 ---

But why? • • Real cluster structure missing Clusters allocated like well optimized “grid” Several grids results different allocation Overall clustering quality can still be the same

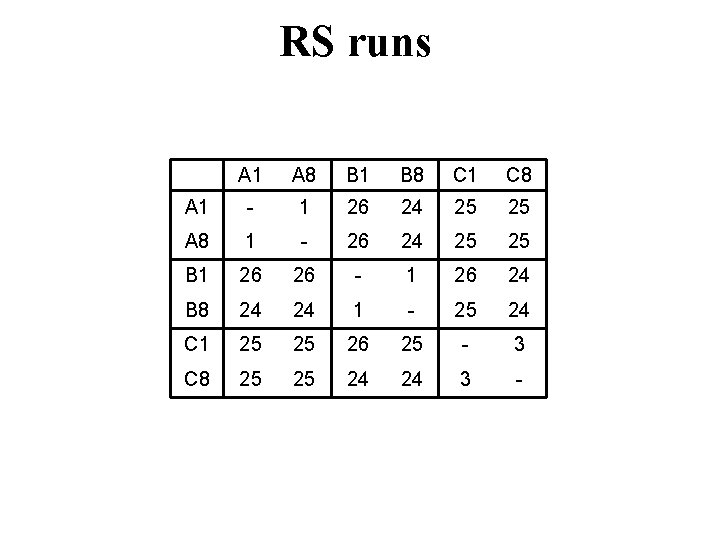
RS runs A 1 A 8 B 1 B 8 C 1 C 8 A 1 - 1 26 24 25 25 A 8 1 - 26 24 25 25 B 1 26 26 - 1 26 24 B 8 24 24 1 - 25 24 C 1 25 25 26 25 - 3 C 8 25 25 24 24 3 -

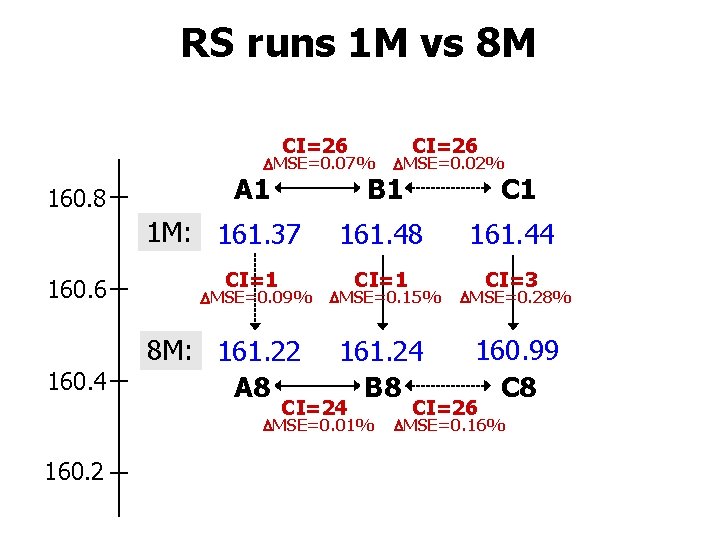
RS runs 1 M vs 8 M CI=26 MSE=0. 07% 160. 8 160. 6 160. 4 CI=1 MSE=0. 09% 8 M: 161. 22 A 8 C 1 161. 48 161. 44 CI=1 CI=3 MSE=0. 15% MSE=0. 28% 161. 24 B 8 160. 99 C 8 CI=24 MSE=0. 01% 160. 2 MSE=0. 02% B 1 A 1 1 M: 161. 37 CI=26 MSE=0. 16%

Generalization

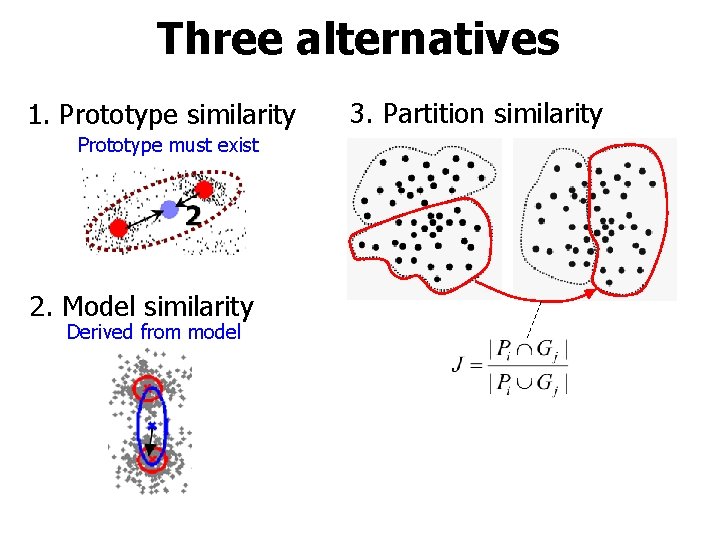
Three alternatives 1. Prototype similarity Prototype must exist 2. Model similarity Derived from model 3. Partition similarity

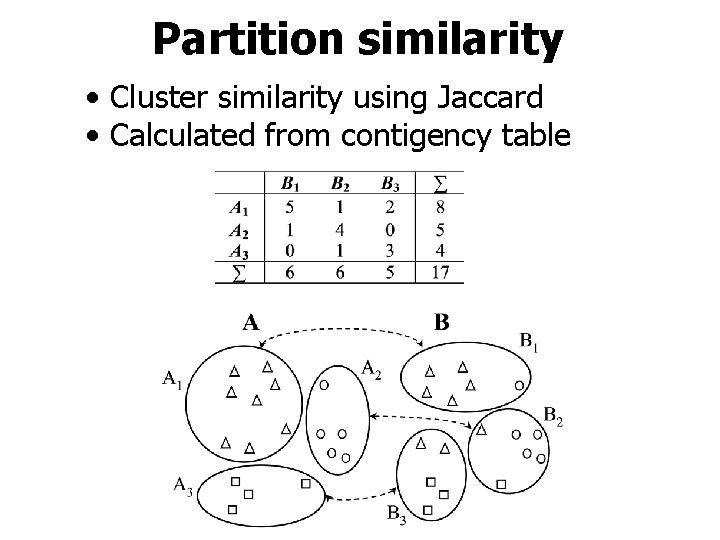
Partition similarity • Cluster similarity using Jaccard • Calculated from contigency table

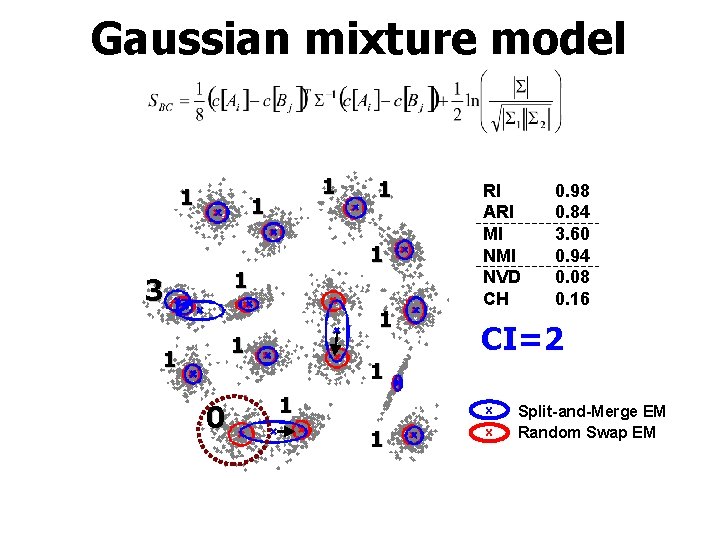
Gaussian mixture model 1 1 1 3 1 1 1 0 1 RI ARI MI NVD CH 0. 98 0. 84 3. 60 0. 94 0. 08 0. 16 CI=2 1 1 x Split-and-Merge EM Random Swap EM

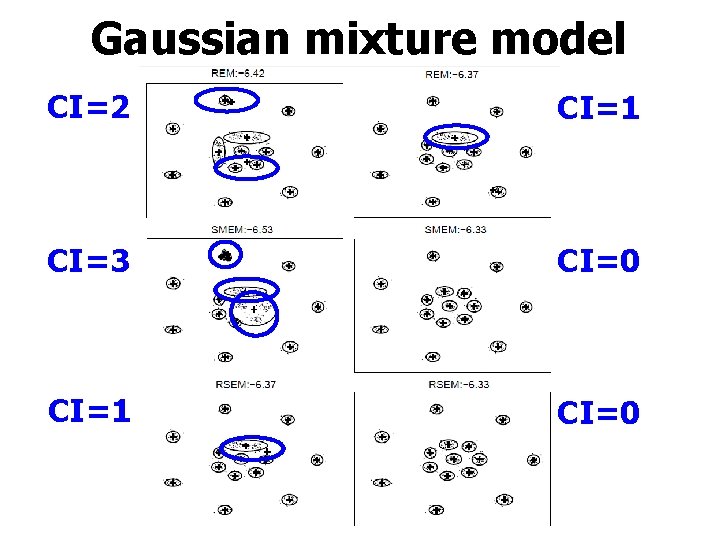
Gaussian mixture model CI=2 CI=1 CI=3 CI=0 CI=1 CI=0

Arbitrary-shape data

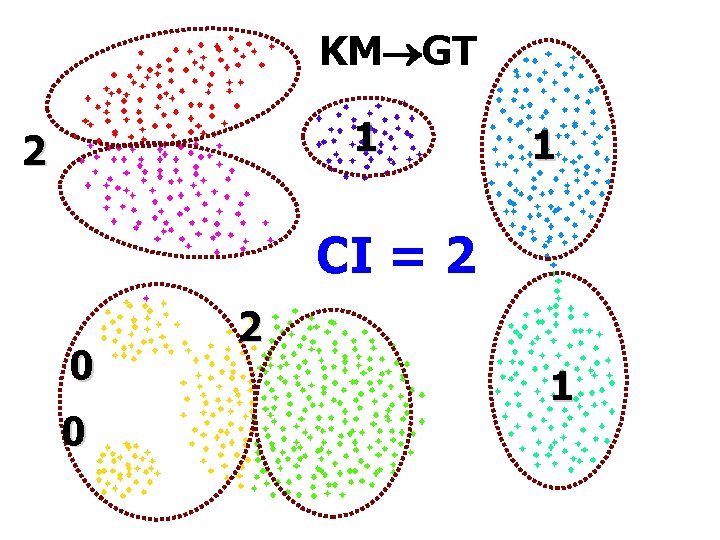
KM GT 1 2 1 CI = 2 0 0 2 1

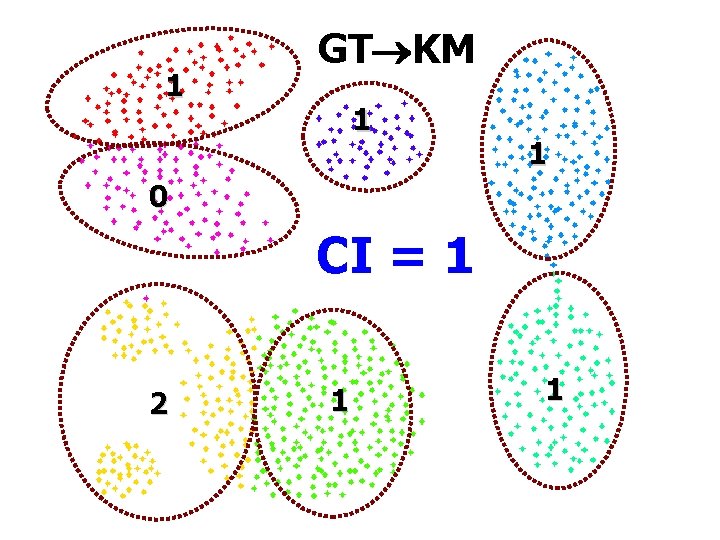
1 GT KM 1 1 0 CI = 1 2 1 1

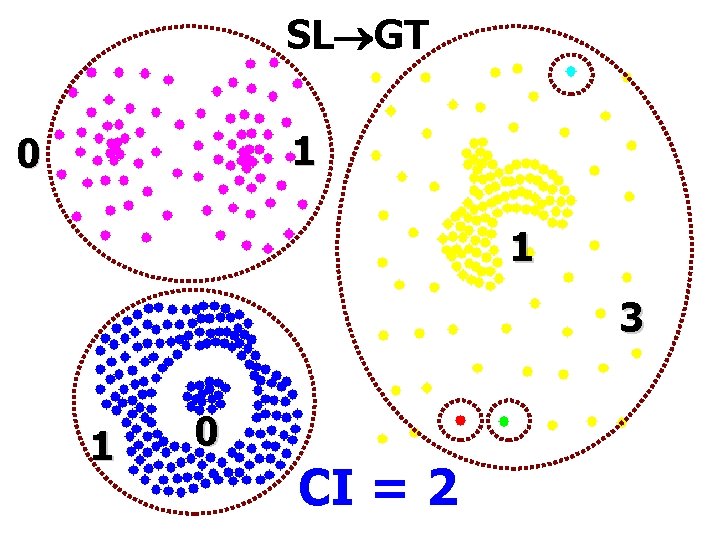
SL GT 1 0 1 3 1 0 CI = 2

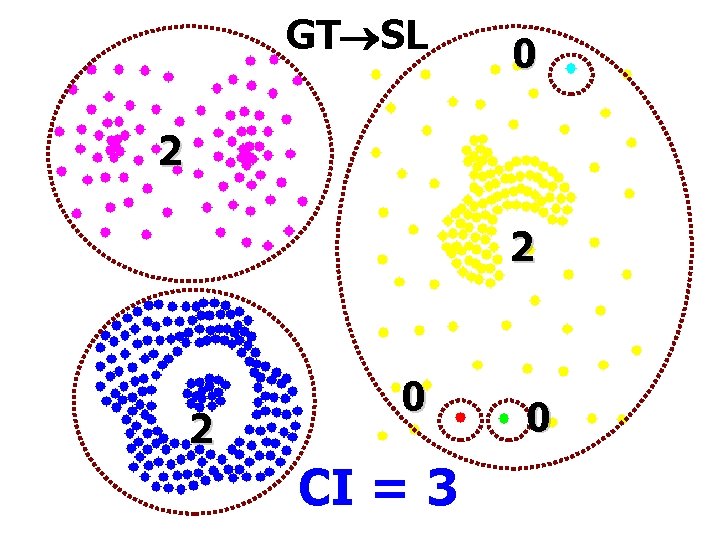
GT SL 0 2 2 2 0 CI = 3 0

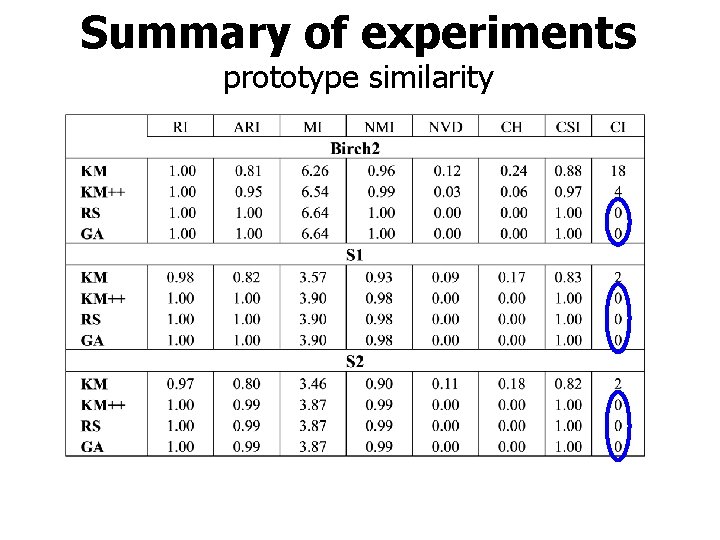
Summary of experiments prototype similarity

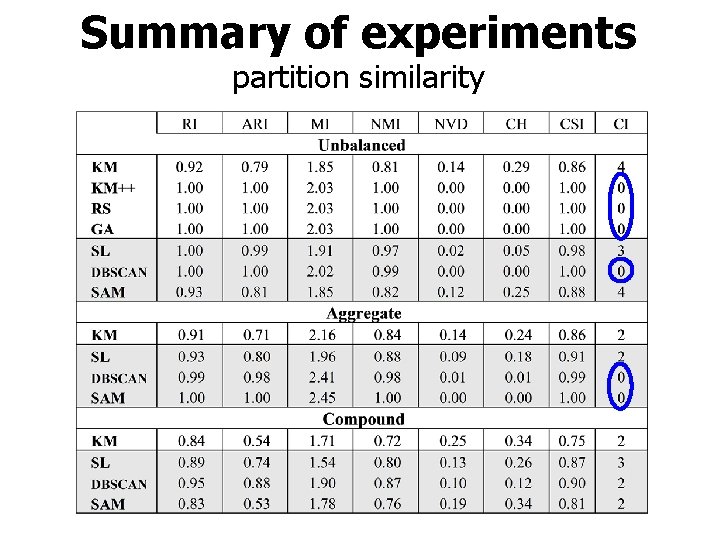
Summary of experiments partition similarity

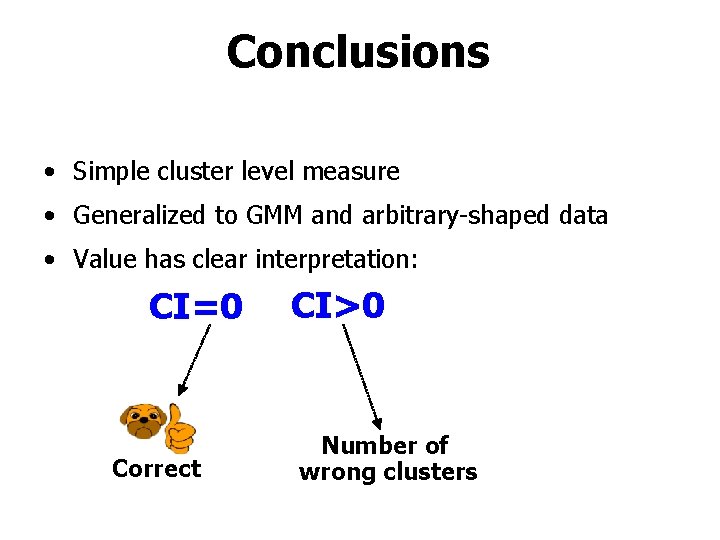
Conclusions • Simple cluster level measure • Generalized to GMM and arbitrary-shaped data • Value has clear interpretation: CI=0 Correct CI>0 Number of wrong clusters
 Compare india and sri lanka on the basis of hdi
Compare india and sri lanka on the basis of hdi Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Pasi fränti
Pasi fränti Frnti
Frnti Pasi hongisto
Pasi hongisto Frnti
Frnti Cluster validation methods
Cluster validation methods Frnti
Frnti Frnti
Frnti Frnti
Frnti Frnti
Frnti Regula celor 8 secunde baschet
Regula celor 8 secunde baschet Pasi kaipainen
Pasi kaipainen Lidheza tek
Lidheza tek Pasi fränti
Pasi fränti Sfs-en 1838
Sfs-en 1838 Pasi kaipainen
Pasi kaipainen Typasi
Typasi Un pasi
Un pasi Pasi puranen
Pasi puranen Structura repetitiva cu numar cunoscut de pasi
Structura repetitiva cu numar cunoscut de pasi Sehr geehrte frau
Sehr geehrte frau Nicoletta pasi
Nicoletta pasi Elenger marine
Elenger marine Pasi rantahalvari
Pasi rantahalvari Pasi fränti
Pasi fränti Gibbons jacobean city comedy download
Gibbons jacobean city comedy download Hail
Hail What does the simpson diversity index measure
What does the simpson diversity index measure Measure page quality
Measure page quality Data quality dimensions
Data quality dimensions Diff between step index and graded index fiber
Diff between step index and graded index fiber Dense secondary index
Dense secondary index Leprosy
Leprosy Mode theory of circular waveguide
Mode theory of circular waveguide Consistency index and liquidity index
Consistency index and liquidity index Clustered index và non clustered index
Clustered index và non clustered index Air quality index calculation
Air quality index calculation Zhou wang
Zhou wang Proxy for institutional quality
Proxy for institutional quality Dr zhou wang
Dr zhou wang Quality control and quality assurance
Quality control and quality assurance Pmp quality vs grade
Pmp quality vs grade Pmp gold plating
Pmp gold plating Ana quality assurance model
Ana quality assurance model Compliance vs quality
Compliance vs quality Qa basic concepts
Qa basic concepts Which one is jurans three role model
Which one is jurans three role model