Applications of Voronoi tessellations in protein structure prediction















- Slides: 47

Applications of Voronoi tessellations in protein structure prediction and analysis Brendan Mc. Conkey Department of Biology University of Waterloo

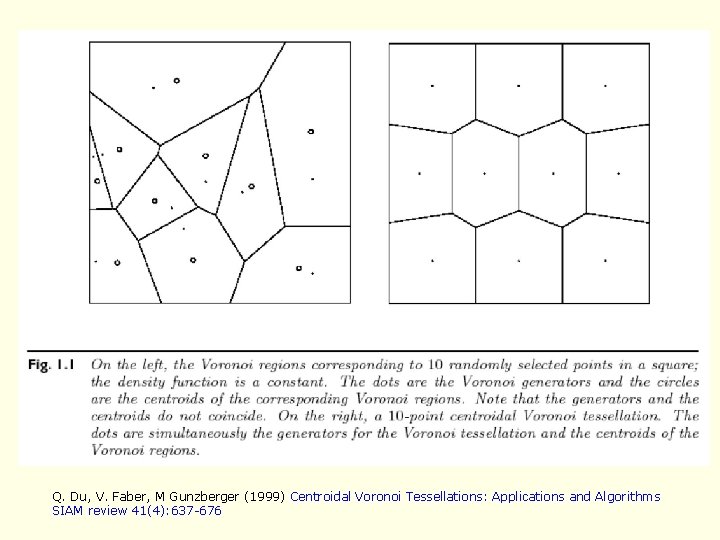
Q. Du, V. Faber, M Gunzberger (1999) Centroidal Voronoi Tessellations: Applications and Algorithms SIAM review 41(4): 637 -676

Gravitational influence of stars. Descartes. 1644. http: //www. snibbe. com/scott

Wigner-Seitz Cells Soap Bubbles in Frame. Fig. 52 from Soap Bubbles, Their Colors and Forces which Mold Them. C. V. Boys. The distribution of Mc. Donald's Restaurants in San Francisco. http: //www. snibbe. com/scott http: //www. chembio. uoguelph. ca/educmat/chm 729/wscells/start. htm

Part of a dragonfly's wing. Fig. 162. From On Growth and Form. D'Arcy Thompson. "Reticulum Plasmatique. " Fig. 321. From On Growth and Form. D'Arcy Thompson. http: //www. snibbe. com/scott

Frogs' eggs showing various partitionings of first eight cells. Fig. 257. From On Growth and Form. D'Arcy Thompson. http: //www. snibbe. com/scott

Applications in protein structure analysis: - scoring functions for protein folding (statistical assessment of contacts within a protein) - generation of protein Voronoi contact maps (2 D targets for structure prediction) - calculating surfaces, areas, and volumes of atoms and amino acid residues

Structure prediction methods The structure prediction problem may be divided into two related tasks: 1. A search procedure - comparative modeling - ab initio prediction 2. An energetic or scoring function - physicochemical potentials - statistical potentials

What determines the structure of a protein? * energetics – structure should have a minimum energy - amino acid sequence - topology of the protein - environment (solvation, membrane interactions) - constitutive ligands (ions, heme groups, …) - interactions with other proteins, cofactors, ligands - folding of cytosolic proteins is largely driven by desolvation That an amino acid sequence can spontaneously form a functional protein implies that the structure is robust to small changes (structure is in a low energy conformation, and will return to this conformation if perturbed)

Protein folding energy landscape protein energy landscape iscomplex, with many local minima believed to have a funnel-likeshape, with global minimum representing native structure image from http: //bioinfo. mshri. on. ca/

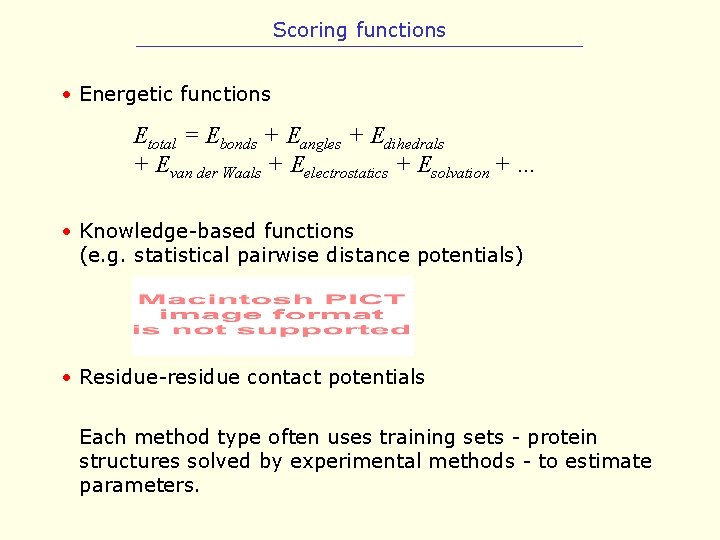
Scoring functions • Energetic functions Etotal = Ebonds + Eangles + Edihedrals + Evan der Waals + Eelectrostatics + Esolvation + … • Knowledge-based functions (e. g. statistical pairwise distance potentials) • Residue-residue contact potentials Each method type often uses training sets - protein structures solved by experimental methods - to estimate parameters.

Development of an atom-atom contact scoring function Advantages of contact-based scoring: • can treat the solvent accessible surface as an atomic contact, eliminating the need to add corrective terms • solvation energy is proportional to the solvent contact area (Eisenberg, 1986) • hydrophobic interactions are largely due to desolvation, so are correlated to loss of solvent contact area • knowledge-based statistical methodology may be applied to contact areas as well as inter-atomic distances * contact scores require a reliable quantification of inter-atomic contacts. This can be done using a Voronoi tessellation.

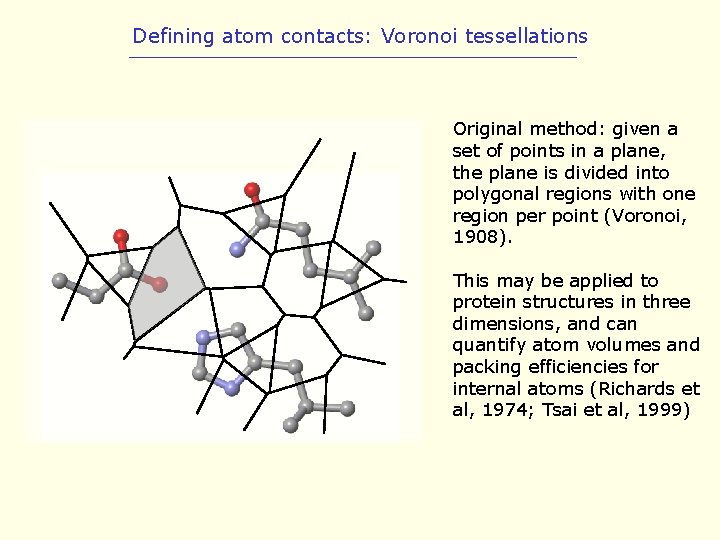
Defining atom contacts: Voronoi tessellations Original method: given a set of points in a plane, the plane is divided into polygonal regions with one region per point (Voronoi, 1908). This may be applied to protein structures in three dimensions, and can quantify atom volumes and packing efficiencies for internal atoms (Richards et al, 1974; Tsai et al, 1999)

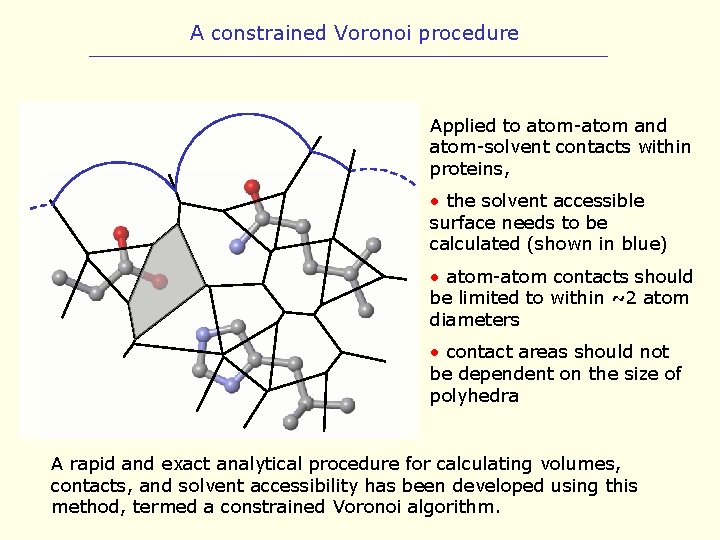
A constrained Voronoi procedure Applied to atom-atom and atom-solvent contacts within proteins, • the solvent accessible surface needs to be calculated (shown in blue) • atom-atom contacts should be limited to within ~2 atom diameters • contact areas should not be dependent on the size of polyhedra A rapid and exact analytical procedure for calculating volumes, contacts, and solvent accessibility has been developed using this method, termed a constrained Voronoi algorithm.

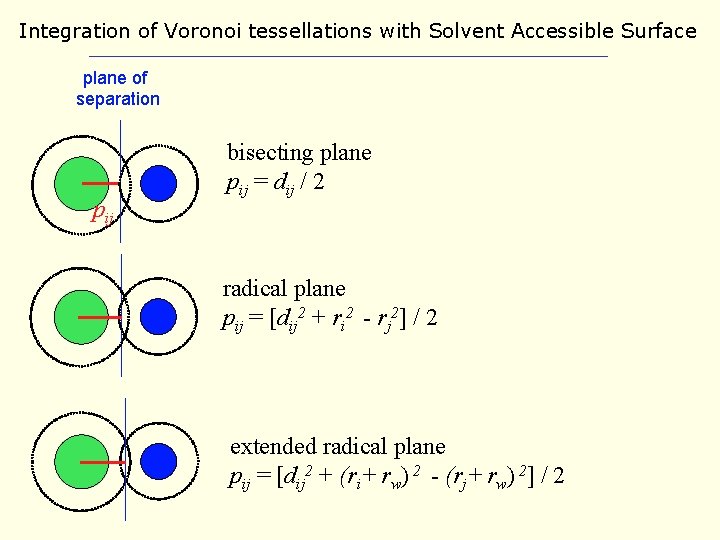
Integration of Voronoi tessellations with Solvent Accessible Surface plane of separation pij bisecting plane pij = dij / 2 radical plane pij = [dij 2 + ri 2 - rj 2] / 2 extended radical plane pij = [dij 2 + (ri+ rw) 2 - (rj+ rw) 2] / 2

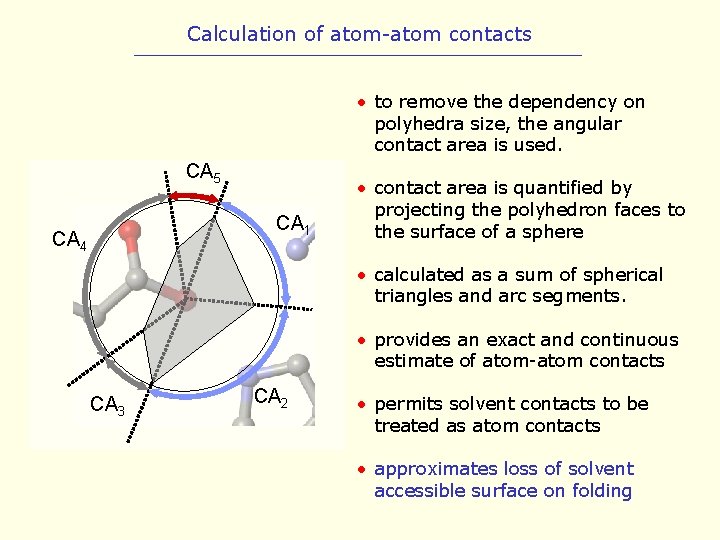
Calculation of atom-atom contacts • to remove the dependency on polyhedra size, the angular contact area is used. CA 5 CA 1 CA 4 • contact area is quantified by projecting the polyhedron faces to the surface of a sphere • calculated as a sum of spherical triangles and arc segments. • provides an exact and continuous estimate of atom-atom contacts CA 3 CA 2 • permits solvent contacts to be treated as atom contacts • approximates loss of solvent accessible surface on folding

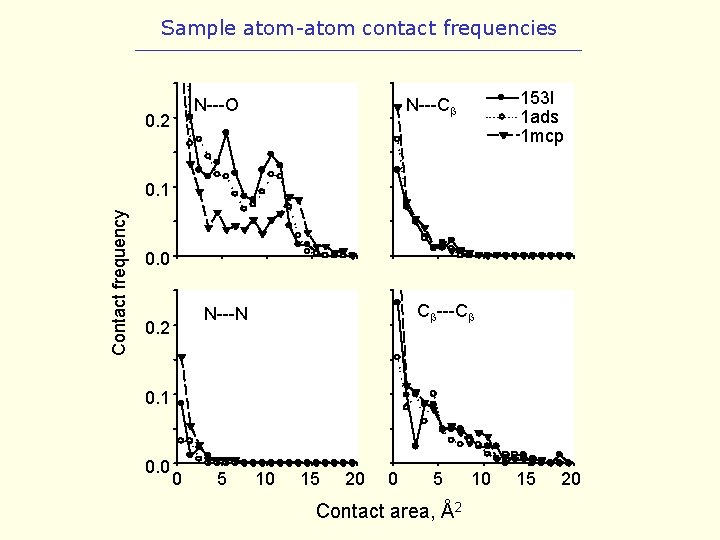
Sample atom-atom contact frequencies N---O 0. 2 153 l 1 ads 1 mcp N---Cb Contact frequency 0. 1 0. 0 Cb---Cb N---N 0. 2 0. 1 0. 0 0 5 10 15 20 0 5 Contact area, Å2 10 15 20

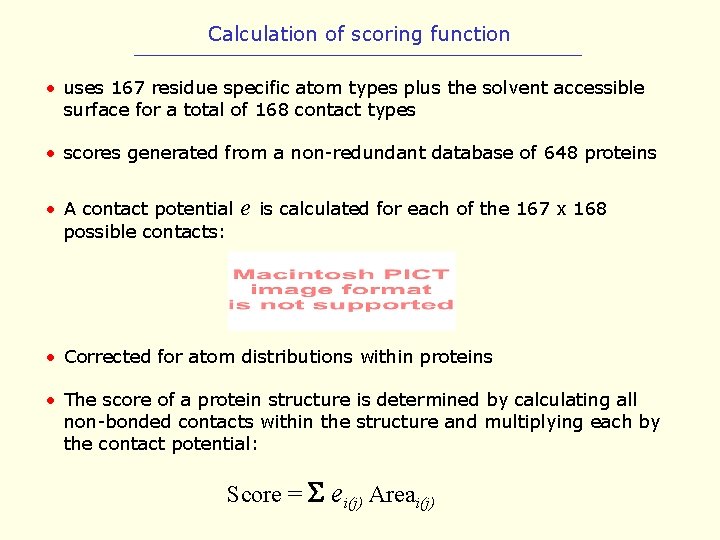
Calculation of scoring function • uses 167 residue specific atom types plus the solvent accessible surface for a total of 168 contact types • scores generated from a non-redundant database of 648 proteins • A contact potential possible contacts: e is calculated for each of the 167 x 168 • Corrected for atom distributions within proteins • The score of a protein structure is determined by calculating all non-bonded contacts within the structure and multiplying each by the contact potential: Score = ei(j) Areai(j)

A few words on the reference state. . . • reference states (expected distributions) have a large influence on statistical scoring functions • here, the unfolded protein (maximum possible solvent contact) is used as a reference state • results in a closed system, with fixed amount of solvent • statistics independent of size of protein • consistent with the idea that protein folding is largely due to hydrophobic interactions

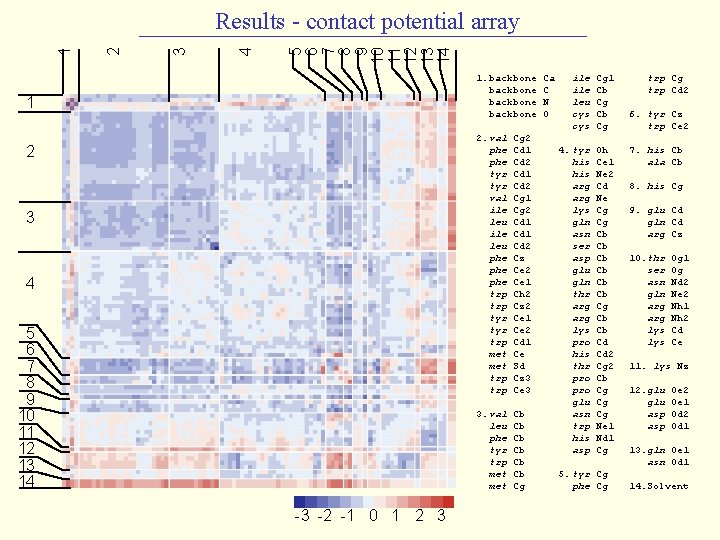
5 6 7 8 9 10 11 12 13 14 4 3 2 1 Results - contact potential array 1. backbone 1 2 3 4 5 6 7 8 9 10 11 12 13 14 -3 -2 -1 0 1 2 3 2. val phe tyr val ile leu phe phe trp tyr trp met trp Cg 2 Cd 1 Cd 2 Cg 1 Cg 2 Cd 1 Cd 2 Cz Ce 2 Ce 1 Ch 2 Cz 2 Ce 1 Ce 2 Cd 1 Ce Sd Cz 3 Ce 3 3. val leu phe tyr trp met Cb Cb Cb Cg Ca C N O ile leu cys Cg 1 Cb Cg 4. tyr his arg lys gln asn ser asp glu gln thr arg lys pro his thr pro glu asn trp his asp Oh Ce 1 Ne 2 Cd Ne Cg Cg Cb Cb Cb Cg Cb Cb Cd Cd 2 Cg 2 Cb Cg Cg Cg Ne 1 Nd 1 Cg 5. tyr Cg phe Cg trp Cd 2 6. tyr Cz trp Ce 2 7. his Cb ala Cb 8. his Cg 9. glu Cd gln Cd arg Cz 10. thr ser asn gln arg lys Og 1 Og Nd 2 Ne 2 Nh 1 Nh 2 Cd Ce 11. lys Nz 12. glu asp Oe 2 Oe 1 Od 2 Od 1 13. gln Oe 1 asn Od 1 14. Solvent

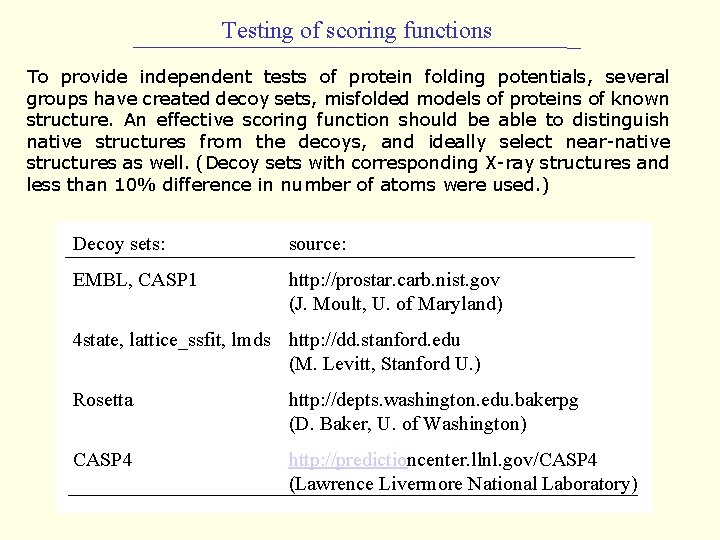
Testing of scoring functions To provide independent tests of protein folding potentials, several groups have created decoy sets, misfolded models of proteins of known structure. An effective scoring function should be able to distinguish native structures from the decoys, and ideally select near-native structures as well. (Decoy sets with corresponding X-ray structures and less than 10% difference in number of atoms were used. ) Decoy sets: source: EMBL, CASP 1 http: //prostar. carb. nist. gov (J. Moult, U. of Maryland) 4 state, lattice_ssfit, lmds http: //dd. stanford. edu (M. Levitt, Stanford U. ) Rosetta http: //depts. washington. edu. bakerpg (D. Baker, U. of Washington) CASP 4 http: //predictioncenter. llnl. gov/CASP 4 (Lawrence Livermore National Laboratory)

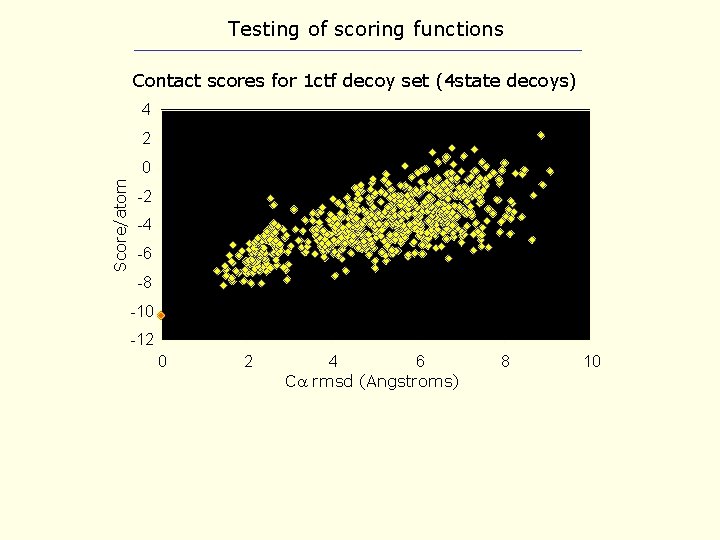
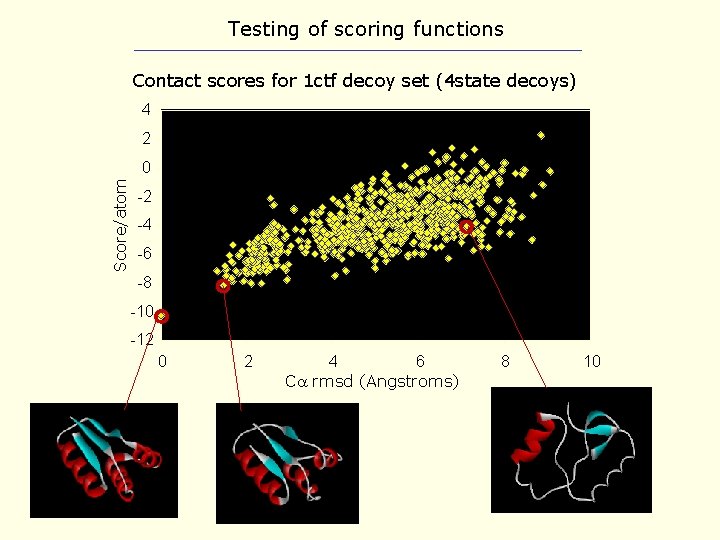
Testing of scoring functions Contact scores for 1 ctf decoy set (4 state decoys) 4 2 Score/atom 0 -2 -4 -6 -8 -10 -12 0 2 4 6 C rmsd (Angstroms) 8 10

Testing of scoring functions Contact scores for 1 ctf decoy set (4 state decoys) 4 2 Score/atom 0 -2 -4 -6 -8 -10 -12 0 2 4 6 C rmsd (Angstroms) 8 10

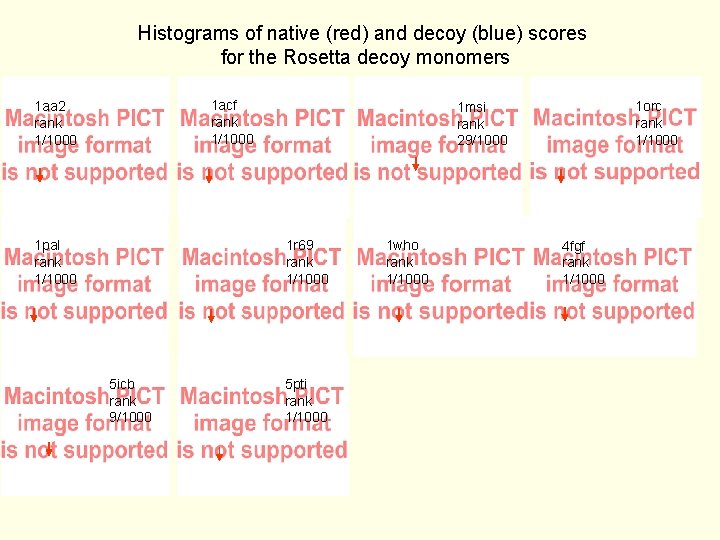
Histograms of native (red) and decoy (blue) scores for the Rosetta decoy monomers 1 acf rank 1/1000 1 aa 2 rank 1/1000 1 pal rank 1/1000 1 r 69 rank 1/1000 5 icb rank 9/1000 1 orc rank 1/1000 1 msi rank 29/1000 5 pti rank 1/1000 1 who rank 1/1000 4 fgf rank 1/1000

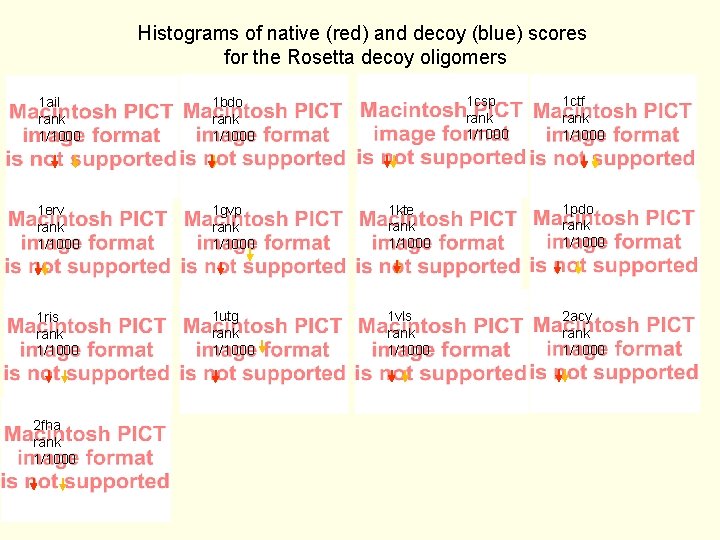
Histograms of native (red) and decoy (blue) scores for the Rosetta decoy oligomers 1 csp rank 1/1000 1 ctf rank 1/1000 1 ail rank 1/1000 1 bdo rank 1/1000 1 erv rank 1/1000 1 gvp rank 1/1000 1 kte rank 1/1000 1 pdo rank 1/1000 1 ris rank 1/1000 1 utg rank 1/1000 1 vls rank 1/1000 2 acy rank 1/1000 2 fha rank 1/1000

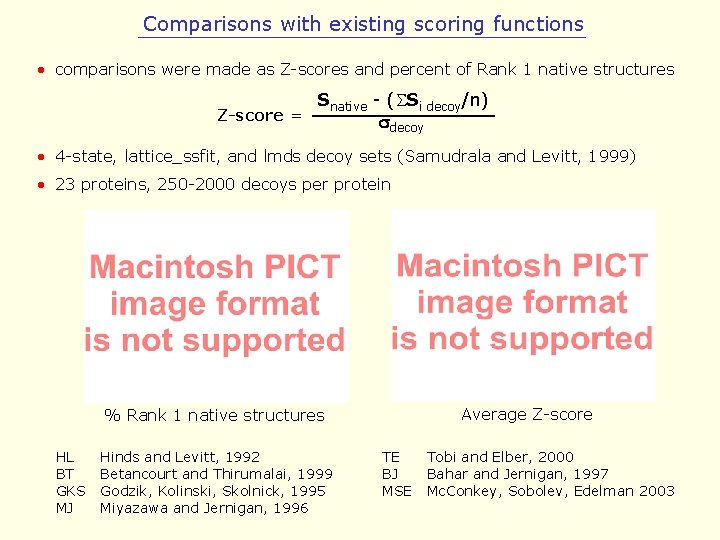
Comparisons with existing scoring functions • comparisons were made as Z-scores and percent of Rank 1 native structures Snative - (SSi decoy/n) Z-score = sdecoy • 4 -state, lattice_ssfit, and lmds decoy sets (Samudrala and Levitt, 1999) • 23 proteins, 250 -2000 decoys per protein Average Z-score % Rank 1 native structures HL BT GKS MJ Hinds and Levitt, 1992 Betancourt and Thirumalai, 1999 Godzik, Kolinski, Skolnick, 1995 Miyazawa and Jernigan, 1996 TE BJ MSE Tobi and Elber, 2000 Bahar and Jernigan, 1997 Mc. Conkey, Sobolev, Edelman 2003

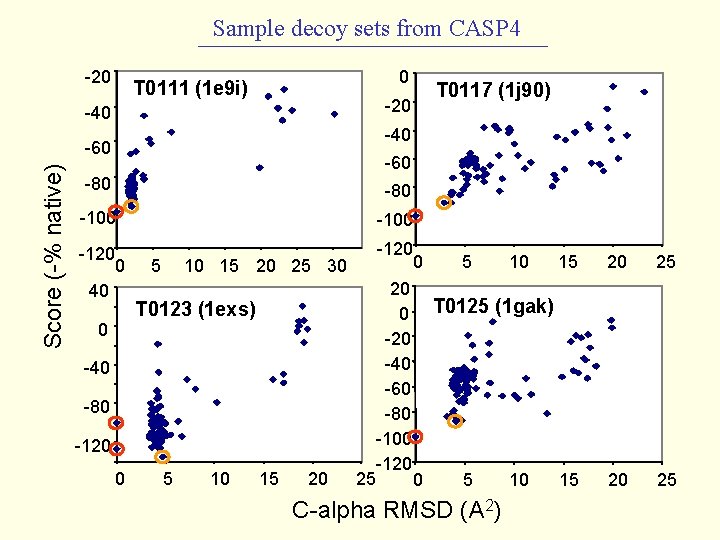
Sample decoy sets from CASP 4 -20 0 T 0111 (1 e 9 i) -20 -40 -60 Score (-% native) T 0117 (1 j 90) -60 -80 -100 -120 0 40 -120 0 0 5 10 15 20 25 30 T 0123 (1 exs) -40 -80 -120 0 5 10 15 20 5 15 20 25 20 0 T 0125 (1 gak) -20 -40 -60 -80 -100 -120 25 0 5 10 15 20 25 C-alpha RMSD (A 2) 10

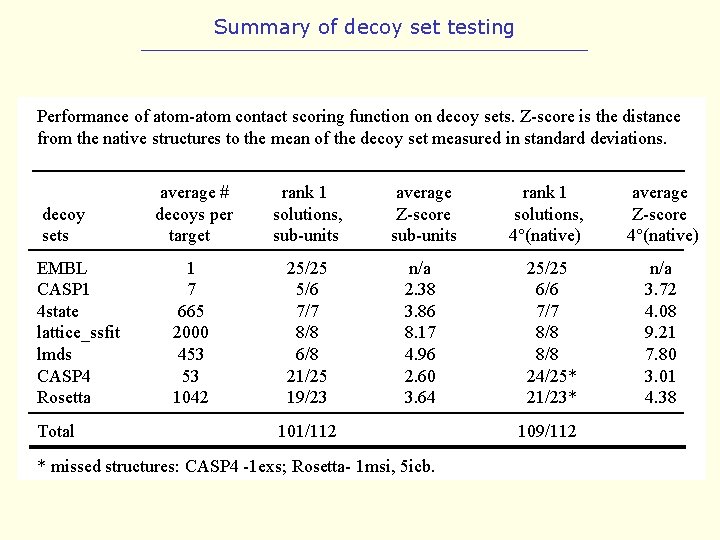
Summary of decoy set testing Performance of atom-atom contact scoring function on decoy sets. Z-score is the distance from the native structures to the mean of the decoy set measured in standard deviations. decoy sets EMBL CASP 1 4 state lattice_ssfit lmds CASP 4 Rosetta Total average # decoys per target rank 1 solutions, sub-units average Z-score sub-units 1 7 665 2000 453 53 1042 25/25 5/6 7/7 8/8 6/8 21/25 19/23 n/a 2. 38 3. 86 8. 17 4. 96 2. 60 3. 64 101/112 * missed structures: CASP 4 -1 exs; Rosetta- 1 msi, 5 icb. rank 1 solutions, 4°(native) 25/25 6/6 7/7 8/8 24/25* 21/23* 109/112 average Z-score 4°(native) n/a 3. 72 4. 08 9. 21 7. 80 3. 01 4. 38

Summary of atom-atom contact scoring • the Voronoi tessellation permits a precise and continuous quantification of atom-atom contacts • the contact scoring function qualitatively resembles energetic interaction potentials • the scoring function has a very high success rate for recognition of correctly folded protein structures, and has greater accuracy than other currently available scoring functions • Native protein structures could be identified in 97% of the decoy sets tested

Observations from all-atom potential • backbone atoms behave similarly, independent of residue type • statistical potential less accurate for backbone atoms due to severe topology constraints (e. g. C --N interaction) • backbone N and O are almost always H-bonded or solvent exposed • there is an reasonably strong effect of neighboring atoms on the potential (e. g. Lysine NZ and Lysine CE)

But. . . • contact potential is still an all-atom potential • requires all atoms to be positioned for a structure to be scored • does not readily permit simplification of folding algorithms • a simplified potential would be useful in initial stages of protein folding. the same methodology for creating the all atom potential has been used to create a folding potential.

First attempt at simplification: • reduce number of contact types from 168 to less than 30 • use residue types to define united atom types • assume backbone atoms behave similarly • GLY is treated as part of backbone • implicitly includes interactions with solvent • initial function remains area dependent

One possibility is a residue-residue potential: A beads-on-a-string model of amino acid chain Unfortunately, this approach has had only moderate success in the past.

A variation of beads on a string: A united-atom model of the amino acid chain backbone interactions are ignored • (assumed to be hydrogen bonded) approximates a residue-residue contact potential •

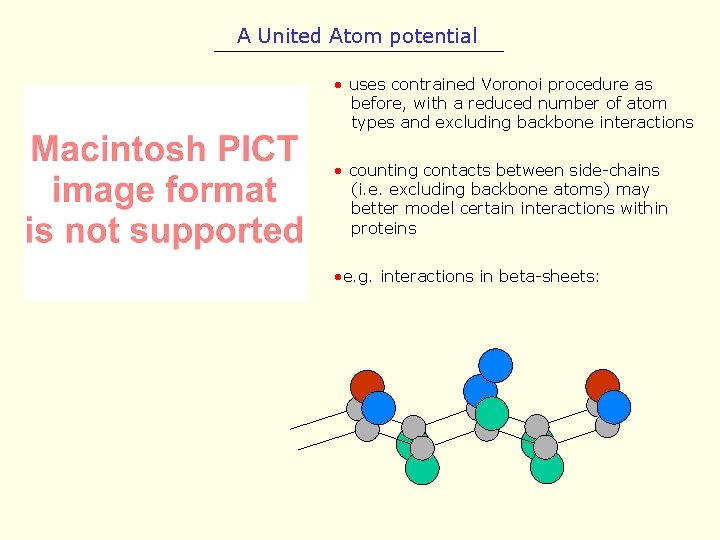
A United Atom potential • uses contrained Voronoi procedure as before, with a reduced number of atom types and excluding backbone interactions • counting contacts between side-chains (i. e. excluding backbone atoms) may better model certain interactions within proteins • e. g. interactions in beta-sheets:

United Atom potential #1 the UA potential was compared to the all-atom potential • using the Rosetta decoy set Area dependence was used: 19/23 Score = ei(j) Areai(j) • 21/23 17/23 19/23

United Atom potentials the initial United Atom function is still dependent on • calculating contact areas between amino acid residues, so relies on knowledge of the position of side chains a binary potential (residues in contact or not) would • be more useful, as it doesn’t require coordinate information A binary contact potential was developed, where side- • chains were considered in contact if they shared > 8 Å 2 contact area. Solvent contact was also enumerated, with 10 -30 Å2 = 1 • contact, 30 -50 Å2 = 2 contacts, etc. binary potential tested using Rosetta decoys •

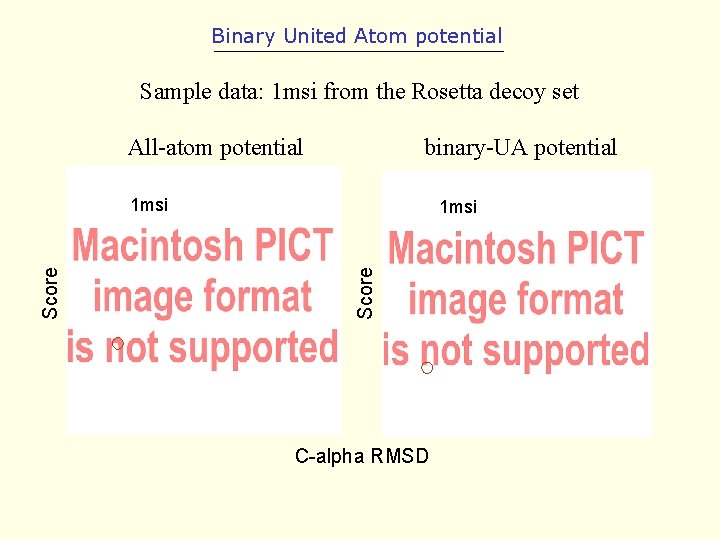
Binary United Atom potential Sample data: 1 msi from the Rosetta decoy set All-atom potential binary-UA potential 1 msi Score 1 msi C-alpha RMSD

Binary United Atom potential 19/23 22/23 21/23 17/23 19/23 20/23 The binary UA potential recognized the native structure in • all test sets except one (5 pti, rank 2/1000)

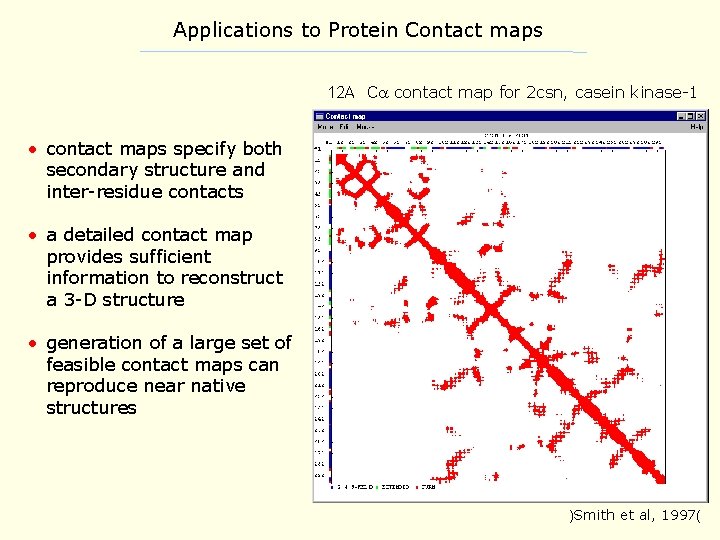
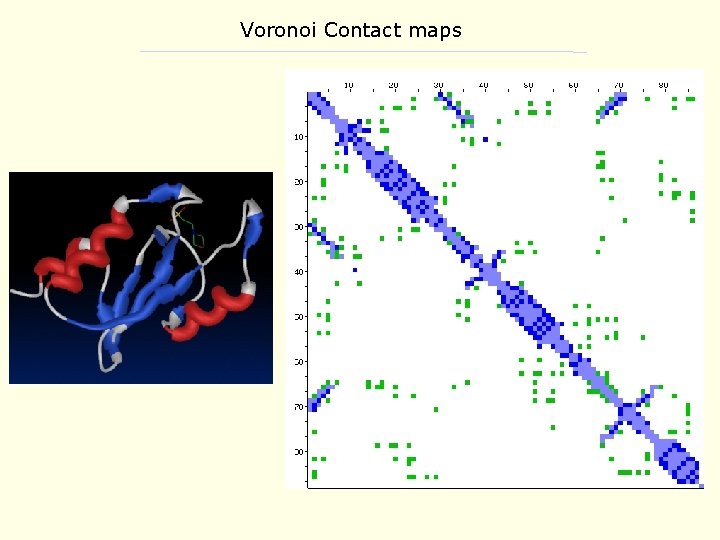
Applications to Protein Contact maps 12 A C contact map for 2 csn, casein kinase-1 • contact maps specify both secondary structure and inter-residue contacts • a detailed contact map provides sufficient information to reconstruct a 3 -D structure • generation of a large set of feasible contact maps can reproduce near native structures )Smith et al, 1997(

Protein structure prediction: Contact maps Some issues with distance based contact maps: • typically use C -C distances - dependent on appropriate choice of cutoff • short cutoff distance biases map towards contacts within secondary structures • longer cutoff distance results in more contacts, and a noisy data set • C atoms in close proximity may have little interaction - e. g. n, n+2 residues in an alpha helix • contact with solvent not readily integrated A tessellation procedure based on residue sidechains can circumvent some of these issues

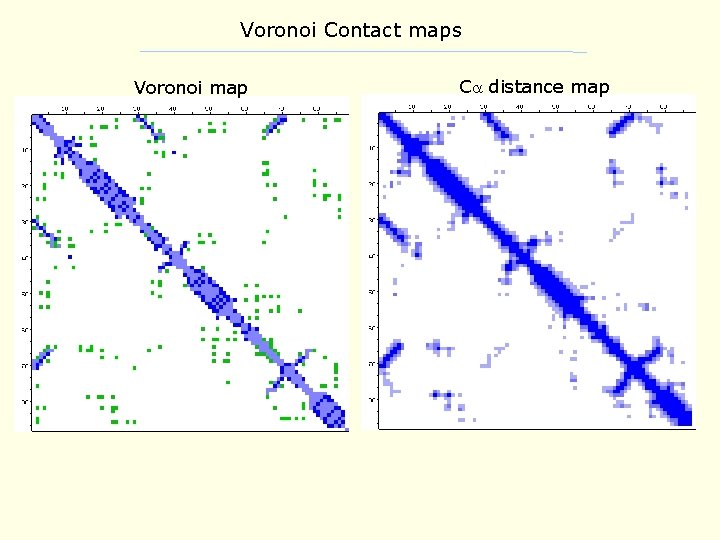
Voronoi Contact maps • similar to C -C distance-based maps • uses a tesselation procedure to determine if residues are in contact • contacts can be subdivided by type: - sidechain contacts - backbone contacts - both sidechain and backbone • results in recognizable patterns of interaction for within and between secondary structures • it is possible to integrate solvent contact into this scheme as well

Voronoi Contact maps

Voronoi Contact maps Voronoi map C distance map

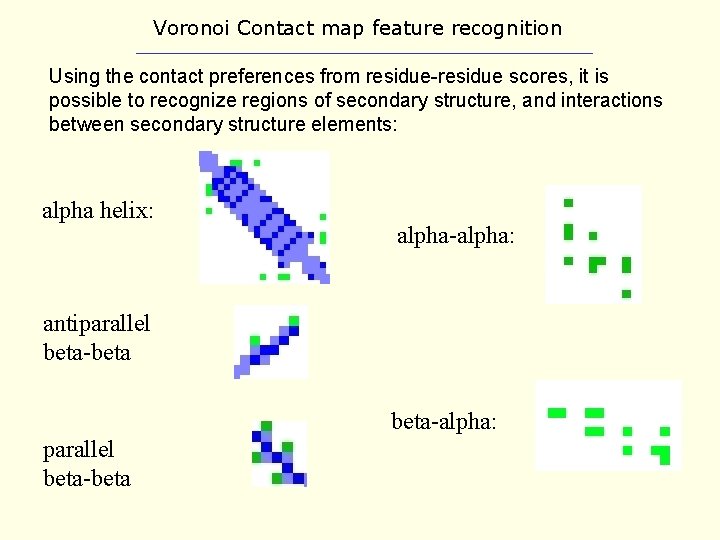
Voronoi Contact map feature recognition Using the contact preferences from residue-residue scores, it is possible to recognize regions of secondary structure, and interactions between secondary structure elements: alpha helix: alpha-alpha: antiparallel beta-beta-alpha: parallel beta-beta

Future work • Further refinement of binary contact scoring functions - incorporate different contact types - beta sheet vs. alpha helix • Development of search procedures to explore contact map space • Other unrelated stuff - proteomics - gene expression and divergence - physicochemical pattern recognition

Thanks!. . questions?
 Toxic waste dump problem voronoi
Toxic waste dump problem voronoi Voronoi diagram tutorial
Voronoi diagram tutorial Voronoi diagram
Voronoi diagram Voroneho diagram
Voroneho diagram Voronoi cella
Voronoi cella Protein function prediction via graph kernels
Protein function prediction via graph kernels Types of tessellations
Types of tessellations Demi regular tessellation patterns
Demi regular tessellation patterns Mc escher nationality
Mc escher nationality Escher ghosts
Escher ghosts Examples of tessellations in nature
Examples of tessellations in nature Carrier vs channel proteins
Carrier vs channel proteins Protein-protein docking
Protein-protein docking Void volume
Void volume Phd secondary structure prediction
Phd secondary structure prediction Rna secondary structure prediction
Rna secondary structure prediction Rna secondary structure prediction
Rna secondary structure prediction Super secondary structure of protein
Super secondary structure of protein Peptide bonds in primary structure of protein
Peptide bonds in primary structure of protein Super secondary structure of protein
Super secondary structure of protein Secondary structure of protein
Secondary structure of protein Primary secondary and tertiary protein structure
Primary secondary and tertiary protein structure Protein primary structure
Protein primary structure Example of quaternary
Example of quaternary Protein monomer
Protein monomer Primary secondary and tertiary protein structure
Primary secondary and tertiary protein structure Adenine thymine cytosine and guanine
Adenine thymine cytosine and guanine วสท.
วสท. Quaternary structure of protein
Quaternary structure of protein Protein tertiary structure bonds
Protein tertiary structure bonds Protein structure
Protein structure Hierarchy of protein structure
Hierarchy of protein structure Protein structure determination
Protein structure determination 64 amino acids
64 amino acids Super secondary structure of protein
Super secondary structure of protein Super secondary structure of protein
Super secondary structure of protein Hierarchy of protein structure
Hierarchy of protein structure Amino acid structures
Amino acid structures Leucine polar or nonpolar
Leucine polar or nonpolar Protein unfolding
Protein unfolding Nnuuu
Nnuuu Primary secondary and tertiary structure of protein
Primary secondary and tertiary structure of protein Protein structure
Protein structure Protein structure
Protein structure Will won't may might for predictions
Will won't may might for predictions Championship branch prediction
Championship branch prediction Corner prediction
Corner prediction The hunger games chapter 24 questions and answers
The hunger games chapter 24 questions and answers