Profile Hidden Markov Models Mark Stamp PHMM 1













- Slides: 49

Profile Hidden Markov Models Mark Stamp PHMM 1

Hidden Markov Models q Here, we assume you know about HMMs o If not, see “A revealing introduction to hidden Markov models” q Executive summary of HMMs HMM is a machine learning technique… …and a discrete hill climb technique Train model based on observation sequence Score any given sequence to determine how closely it matches the model o Efficient algorithms and many useful apps o o PHMM 2

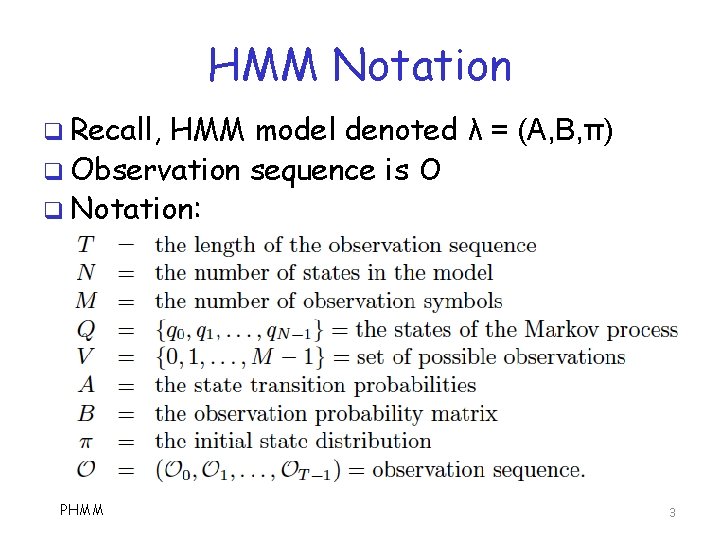
HMM Notation q Recall, HMM model denoted λ = (A, B, π) q Observation sequence is O q Notation: PHMM 3

Hidden Markov Models q Among the many uses for HMMs… q Speech analysis q Music search engine q Malware detection q Intrusion detection systems (IDS) q And more all the time PHMM 4

Limitations of HMMs q Positional information not considered o HMM has no “memory” beyond previous state o Higher order models have more “memory” o But no explicit use of positional information q q With HMM, no insertions or deletions These limitations are serious problems in some applications o In bioinformatics string comparison, sequence alignment is critical o Also, insertions and deletions can occur PHMM 5

Profile HMM q Profile HMM (PHMM) designed to overcome limitations on previous slide o In some ways, PHMM easier than HMM o In some ways, PHMM more complex q The basic idea of PHMM ? o Define multiple B matrices o Almost like having an HMM for each position in sequence PHMM 6

PHMM q In bioinformatics, begin by aligning multiple related sequences o Multiple sequence alignment (MSA) o Analogous to training phase for HMM q Generate PHMM based on MSA o This is easy, once MSA is known o Again, hard part is generating MSA q Then can score sequences using PHMM o Use forward algorithm, similar to HMM PHMM 7

Training: PHMM vs HMM q Training PHMM o Determine MSA challenging o Determine PHMM matrices easy q Training HMM o Append training sequences trivial o Determine HMM matrices challenging q PHMM and HMM are, in this sense, almost opposites… PHMM 8

Generic View of PHMM q Have delete, insert, and match states o Match states correspond to HMM states q Arrows are possible transitions o Each transition has a probability q Transition probabilities are A matrix q Emission probabilities are B matrices o In PHMM, observations called emissions o Match and insert states have emissions PHMM 9

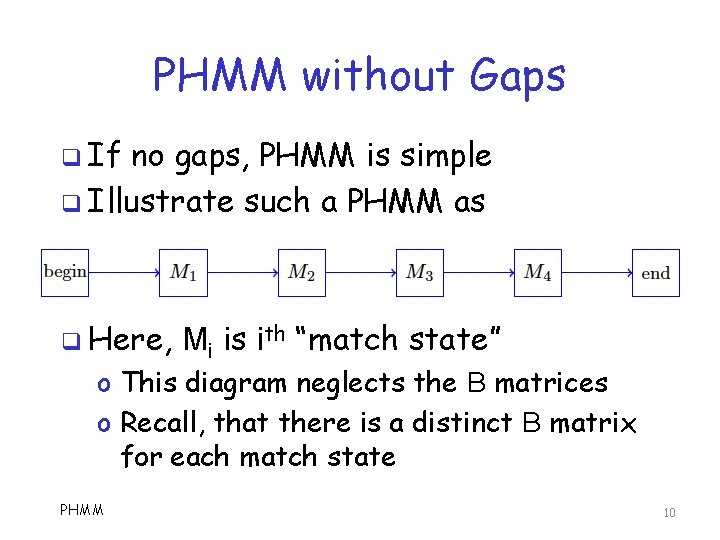
PHMM without Gaps q If no gaps, PHMM is simple q Illustrate such a PHMM as q Here, Mi is ith “match state” o This diagram neglects the B matrices o Recall, that there is a distinct B matrix for each match state PHMM 10

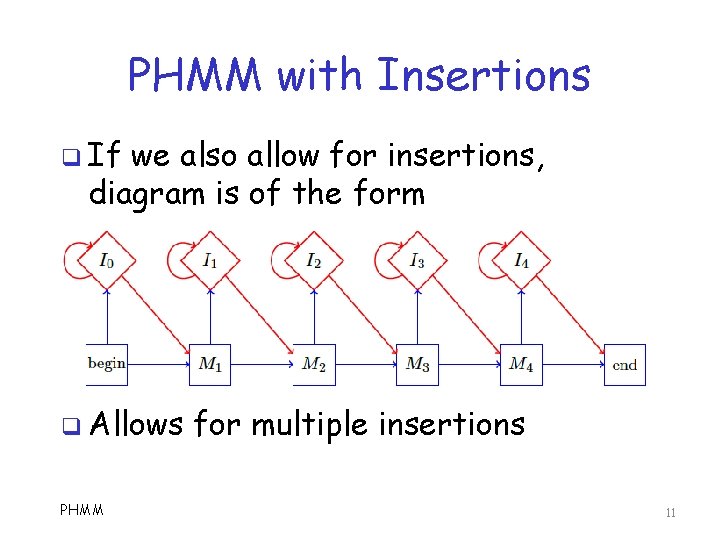
PHMM with Insertions q If we also allow for insertions, diagram is of the form q Allows PHMM for multiple insertions 11

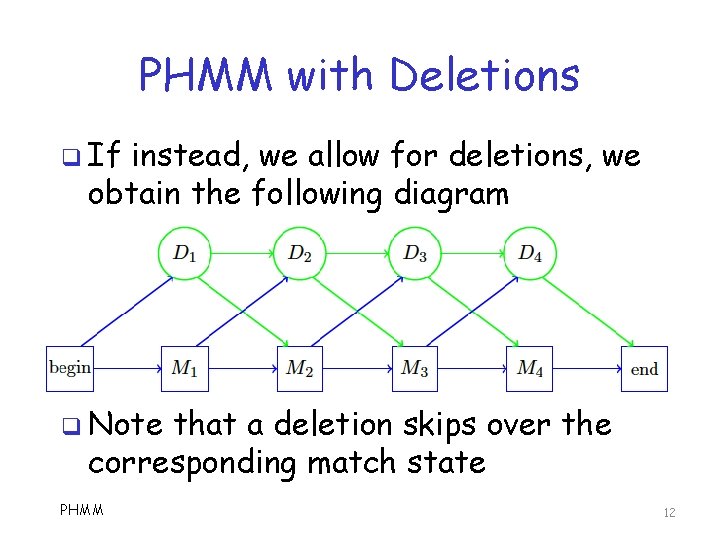
PHMM with Deletions q If instead, we allow for deletions, we obtain the following diagram q Note that a deletion skips over the corresponding match state PHMM 12

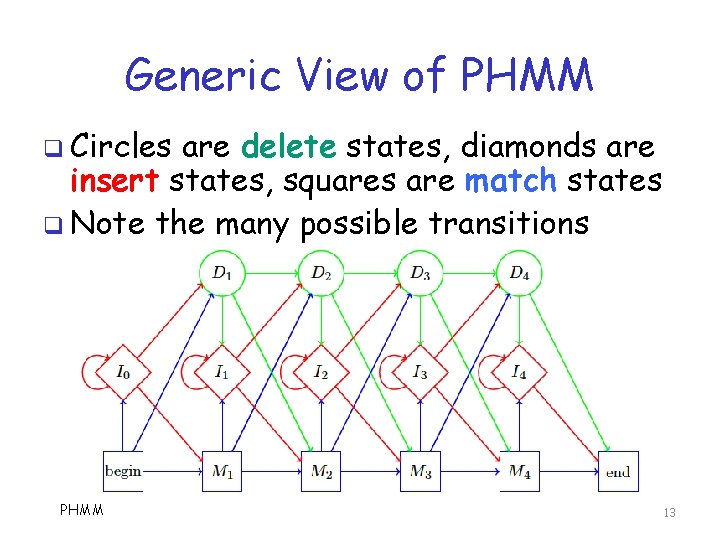
Generic View of PHMM q Circles are delete states, diamonds are insert states, squares are match states q Note the many possible transitions PHMM 13

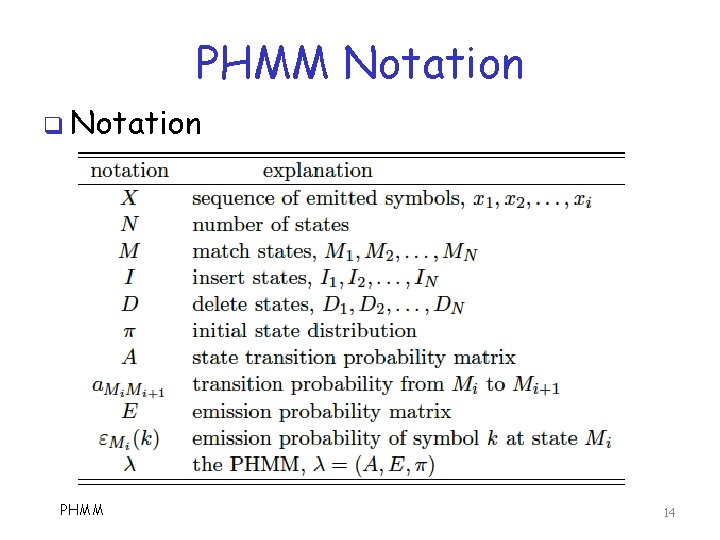
PHMM Notation q Notation PHMM 14

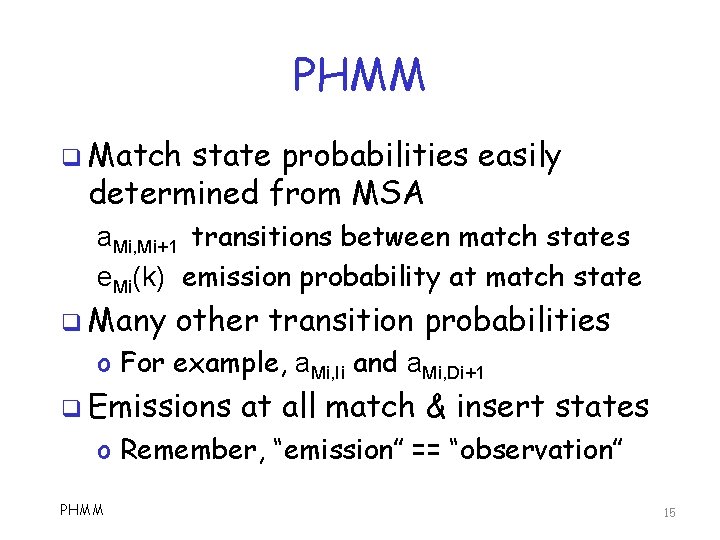
PHMM q Match state probabilities easily determined from MSA a. Mi, Mi+1 transitions between match states e. Mi(k) emission probability at match state q Many other transition probabilities o For example, a. Mi, Ii and a. Mi, Di+1 q Emissions at all match & insert states o Remember, “emission” == “observation” PHMM 15

Multiple Sequence Alignment q First we show MSA construction q Then construct PHMM from MSA o This is the difficult part o Lots of ways to do this o “Best” way depends on specific problem o This is the easy part o Standard algorithm to generate PHMM q How to score a sequence? o Forward algorithm, similar to HMM PHMM 16

MSA q How to construct MSA? o Construct pairwise alignments o Combine pairwise alignments into MSA q Allow gaps to be inserted o To make better matches q Gaps tend to weaken PHMM scoring o So, tradeoff between number of gaps and strength of score PHMM 17

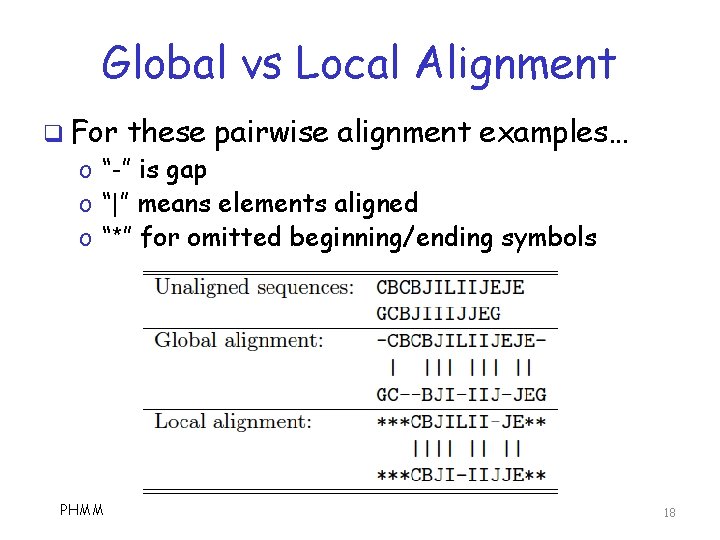
Global vs Local Alignment q For these pairwise alignment examples… o “-” is gap o “|” means elements aligned o “*” for omitted beginning/ending symbols PHMM 18

Global vs Local Alignment q Global o o alignment is lossless But gaps tend to proliferate And gaps increase when we do MSA More gaps, more random sequences match… …and result is less useful for scoring q We usually only consider local alignment o That is, omit ends for better alignment q For simplicity, we do global alignment in examples presented here PHMM 19

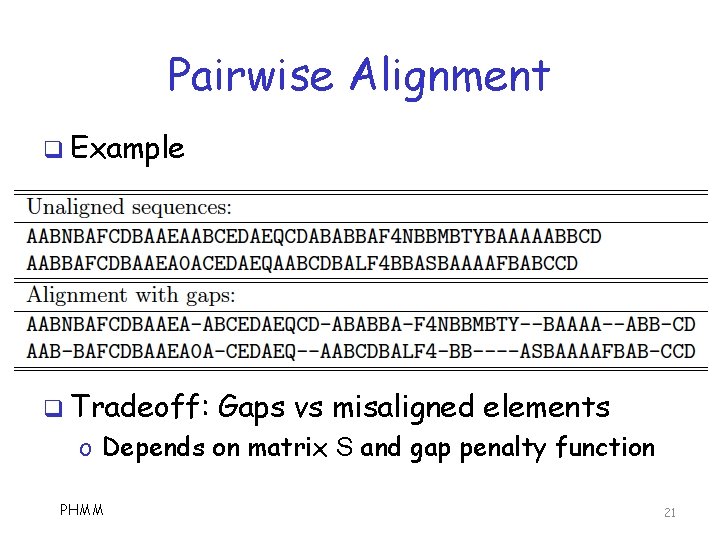
Pairwise Alignment q Allow gaps when aligning q How to score an alignment? o Based on n x n substitution matrix S o Where n is number of symbols q What algorithm(s) to align sequences? q Local alignment? Additional issues arise… o Usually, dynamic programming o Sometimes, HMM is used o Other? PHMM 20

Pairwise Alignment q Example q Tradeoff: Gaps vs misaligned elements o Depends on matrix S and gap penalty function PHMM 21

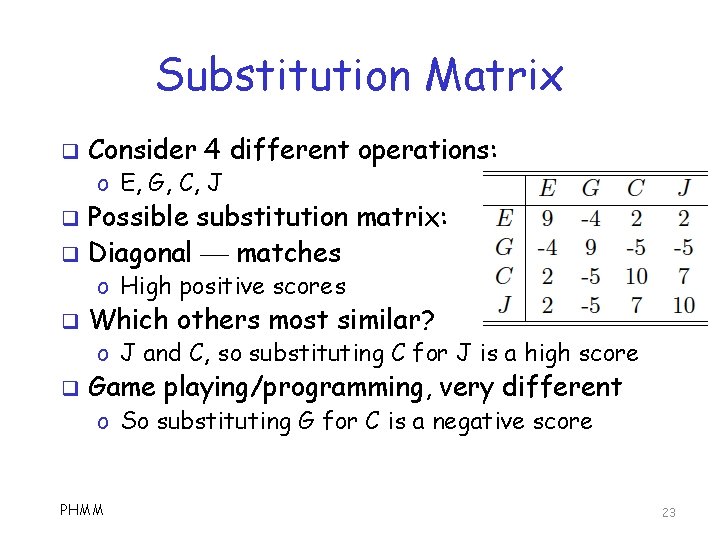
Substitution Matrix q For example, masquerade detection o Detect imposter using computer account q Consider o o E == send email G == play games C == C programming J == Java programming q How PHMM 4 different operations similar are these to each other? 22

Substitution Matrix q Consider 4 different operations: o E, G, C, J q q Possible substitution matrix: Diagonal matches o High positive scores q Which others most similar? o J and C, so substituting C for J is a high score q Game playing/programming, very different o So substituting G for C is a negative score PHMM 23

Substitution Matrix q Depending on problem, might be easy or very difficult to find useful S matrix q Consider masquerade detection based on UNIX commands o Sometimes difficult to say how “close” 2 commands are q Suppose instead, aligning DNA sequences o Biologically valid reasons for S matrix PHMM 24

Gap Penalty q Generally must allow gaps to be inserted q But gaps make alignment more generic o Less useful for scoring, so we penalize gaps q How to penalize gaps? Two common ways q Linear gap penalty function: g(x) = ax (constant penalty for every gap) q Affine gap penalty function g(x) = a + b(x – 1) o Gap opening penalty a and constant penalty of b for each extension of existing gap PHMM 25

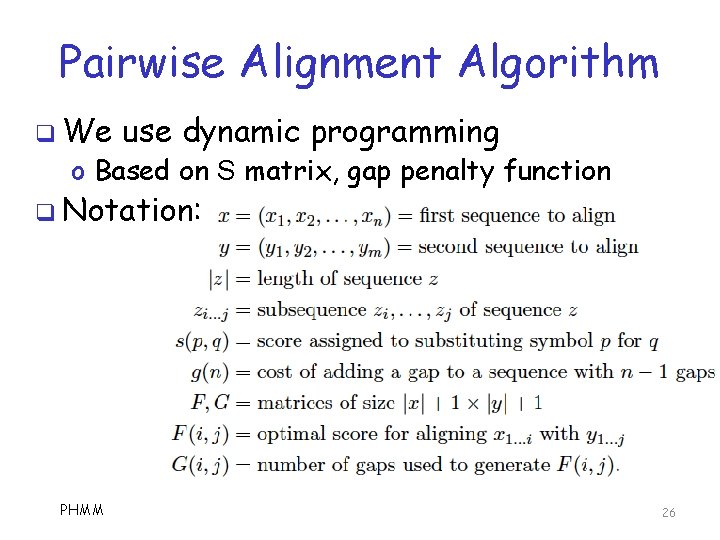
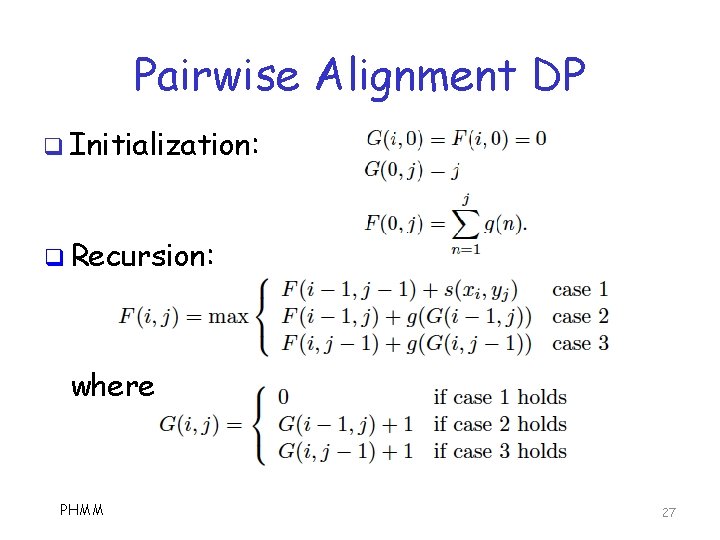
Pairwise Alignment Algorithm q We use dynamic programming o Based on S matrix, gap penalty function q Notation: PHMM 26

Pairwise Alignment DP q Initialization: q Recursion: where PHMM 27

MSA from Pairwise Alignments q Given pairwise alignments… q How to construct MSA? q Generally use “progressive alignment” o Select one pairwise alignment o Select another and combine with first o Continue to add more until all are used q Relatively easy (good) q Gaps proliferate, and it’s unstable (bad) PHMM 28

MSA from Pairwise Alignments q Lots of ways to improve on generic progressive alignment o Here, we mention one such approach o Not necessarily “best” or most popular q Feng-Dolittle progressive alignment o Compute scores for all pairs of n sequences o Select n-1 alignments that a) “connect” all sequences and b) maximize pairwise scores o Then generate a minimum spanning tree o For MSA, add sequences in the order that they appear in the spanning tree PHMM 29

MSA Construction q Create pairwise alignments o Generate substitution matrix S o Dynamic program for pairwise alignments q Use pairwise alignments to make MSA o Use pairwise alignments to construct spanning tree (e. g. , Prim’s Algorithm) o Add sequences in spanning tree order (from high score, insert gaps as needed) o Note: gap penalty is used here PHMM 30

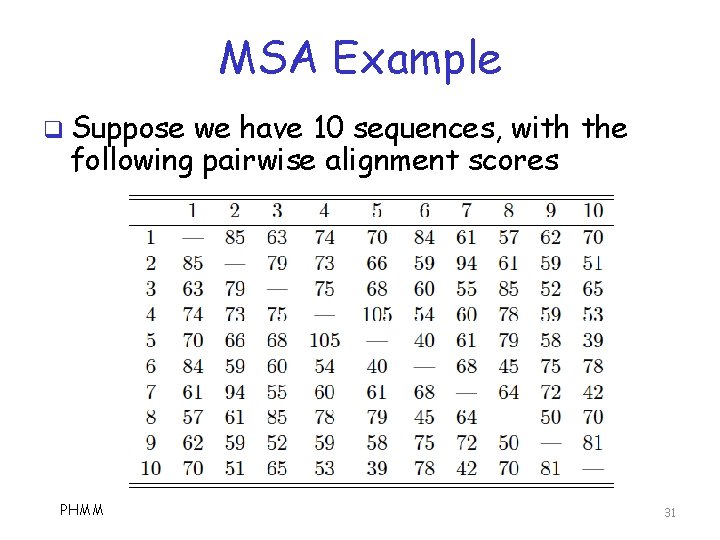
MSA Example q Suppose we have 10 sequences, with the following pairwise alignment scores PHMM 31

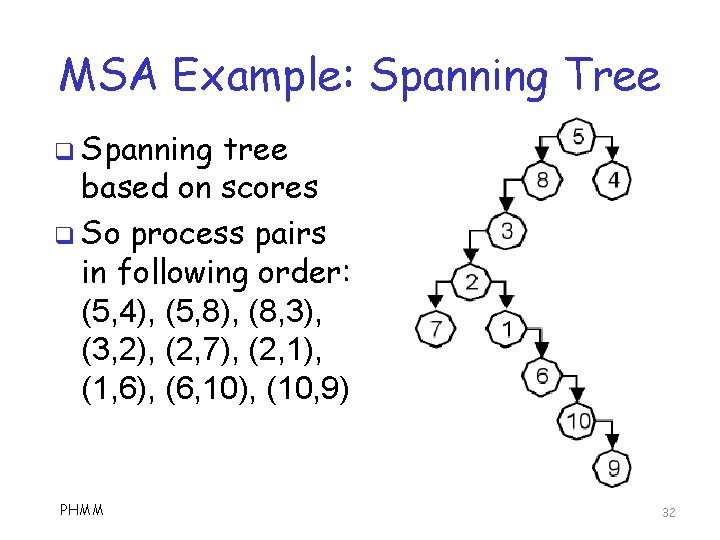
MSA Example: Spanning Tree q Spanning tree based on scores q So process pairs in following order: (5, 4), (5, 8), (8, 3), (3, 2), (2, 7), (2, 1), (1, 6), (6, 10), (10, 9) PHMM 32

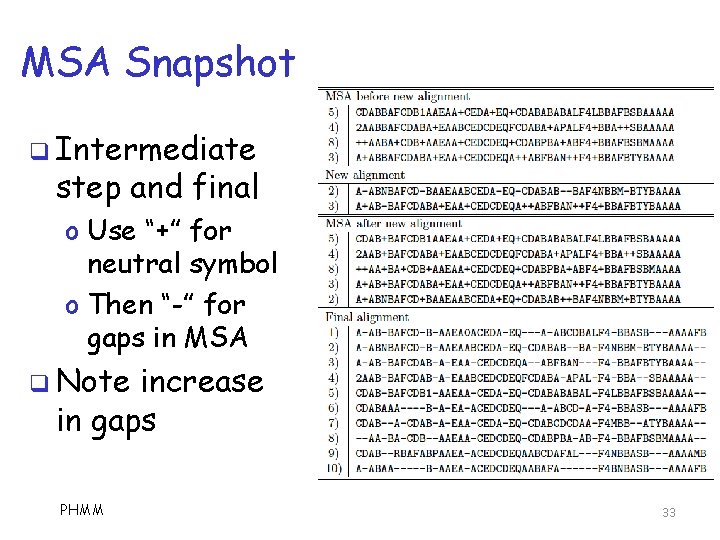
MSA Snapshot q Intermediate step and final o Use “+” for neutral symbol o Then “-” for gaps in MSA q Note increase in gaps PHMM 33

PHMM from MSA q In PHMM, determine match and insert states & probabilities from MSA q “Conservative” columns are match states o Half or less of symbols are gaps q Other columns are insert states o Majority of symbols are gaps q Delete PHMM states are a separate issue 34

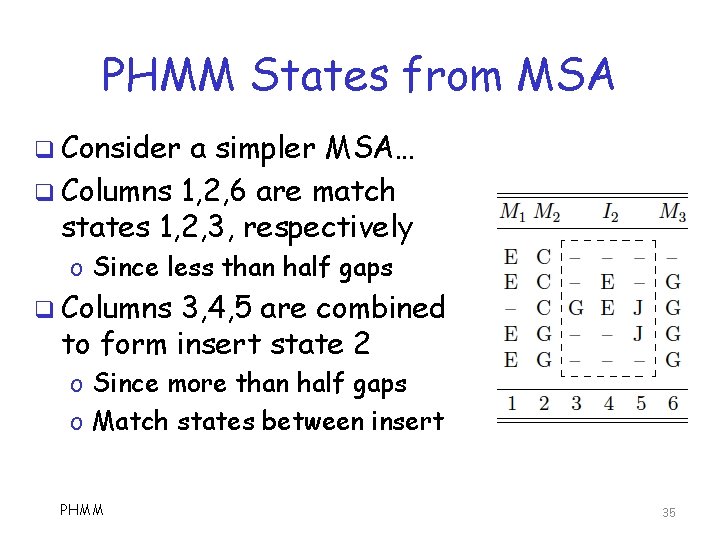
PHMM States from MSA q Consider a simpler MSA… q Columns 1, 2, 6 are match states 1, 2, 3, respectively o Since less than half gaps q Columns 3, 4, 5 are combined to form insert state 2 o Since more than half gaps o Match states between insert PHMM 35

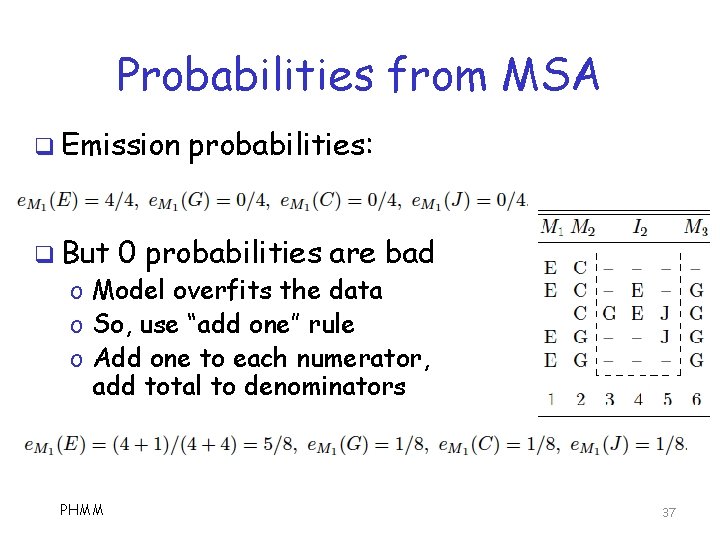
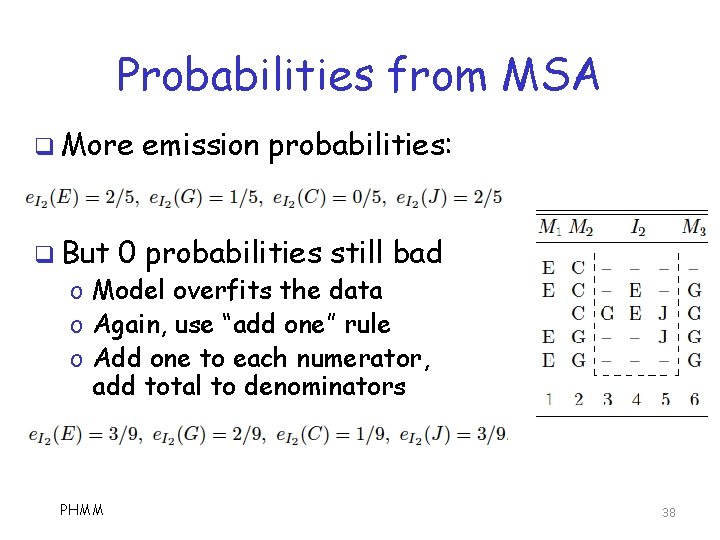
Probabilities from MSA q Emission probabilities o Based on symbol distribution in match and insert states q State transition probs o Based on transitions in the MSA PHMM 36

Probabilities from MSA q Emission q But probabilities: 0 probabilities are bad o Model overfits the data o So, use “add one” rule o Add one to each numerator, add total to denominators PHMM 37

Probabilities from MSA q More q But emission probabilities: 0 probabilities still bad o Model overfits the data o Again, use “add one” rule o Add one to each numerator, add total to denominators PHMM 38

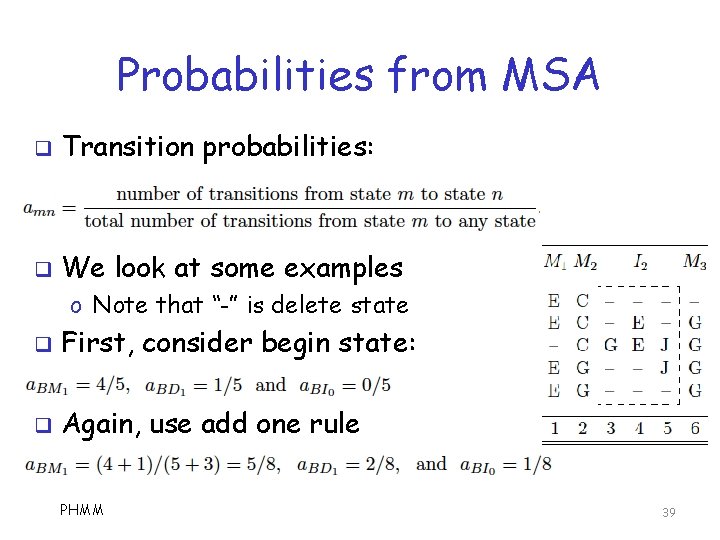
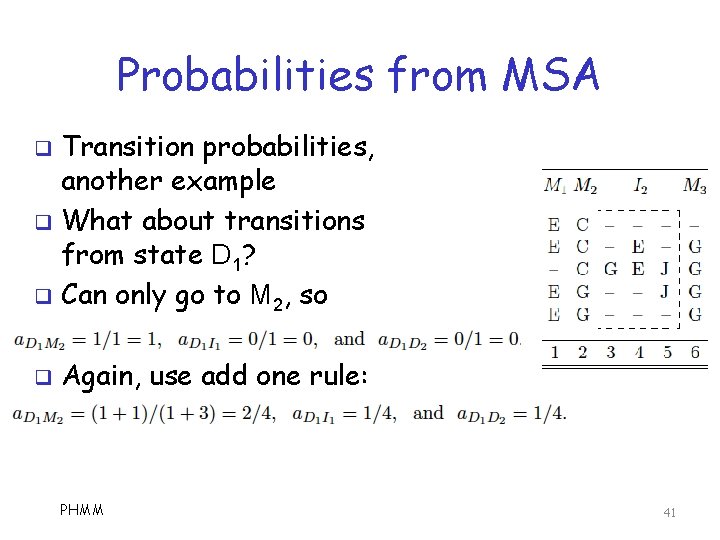
Probabilities from MSA q Transition probabilities: q We look at some examples o Note that “-” is delete state q First, consider begin state: q Again, use add one rule PHMM 39

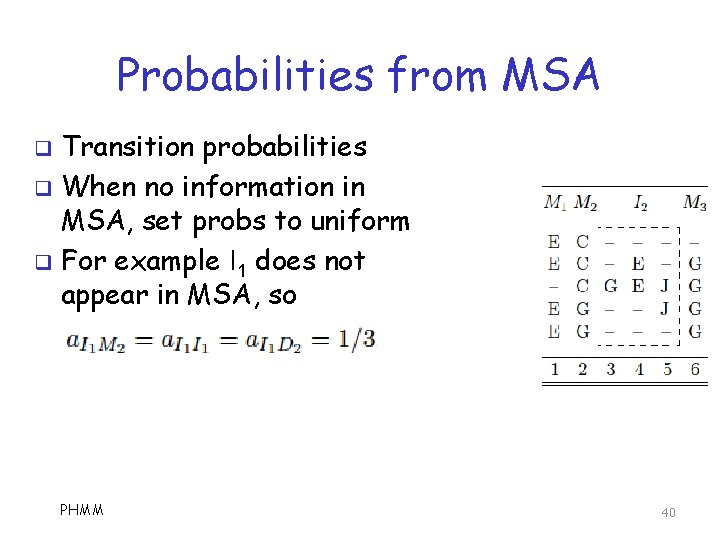
Probabilities from MSA Transition probabilities q When no information in MSA, set probs to uniform q For example I 1 does not appear in MSA, so q PHMM 40

Probabilities from MSA Transition probabilities, another example q What about transitions from state D 1? q Can only go to M 2, so q q Again, use add one rule: PHMM 41

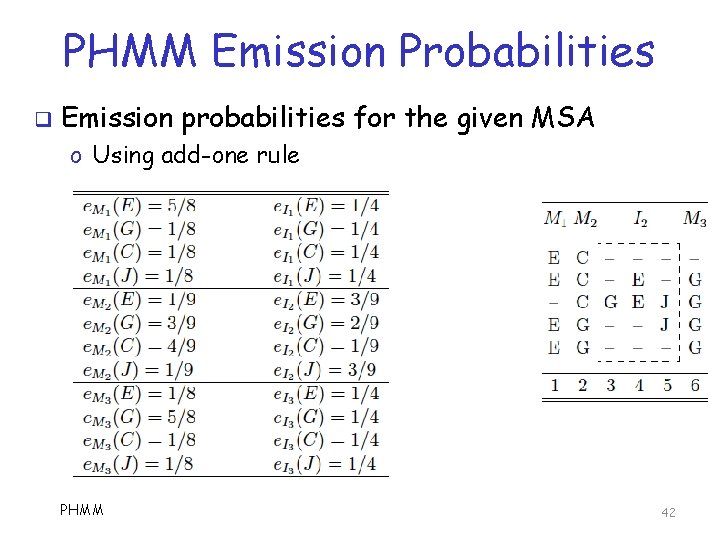
PHMM Emission Probabilities q Emission probabilities for the given MSA o Using add-one rule PHMM 42

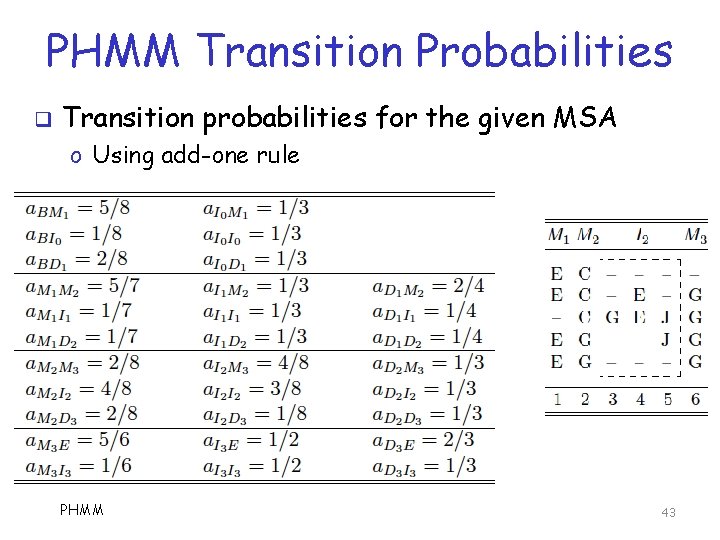
PHMM Transition Probabilities q Transition probabilities for the given MSA o Using add-one rule PHMM 43

PHMM Summary q Construct pairwise alignments o Usually, use dynamic programming q Use these to construct MSA o Lots of ways to do this q Using MSA, determine probabilities q Then we have trained a PHMM o Emission probabilities o State transition probabilities o Now what? ? ? PHMM 44

PHMM Scoring q Want to score sequences to see how closely they match PHMM q How did we score using HMM? o Forward algorithm q How to score sequences with PHMM? o Forward algorithm (surprised? ) q But, algorithm is a little more complex o Due to more complex state transitions PHMM 45

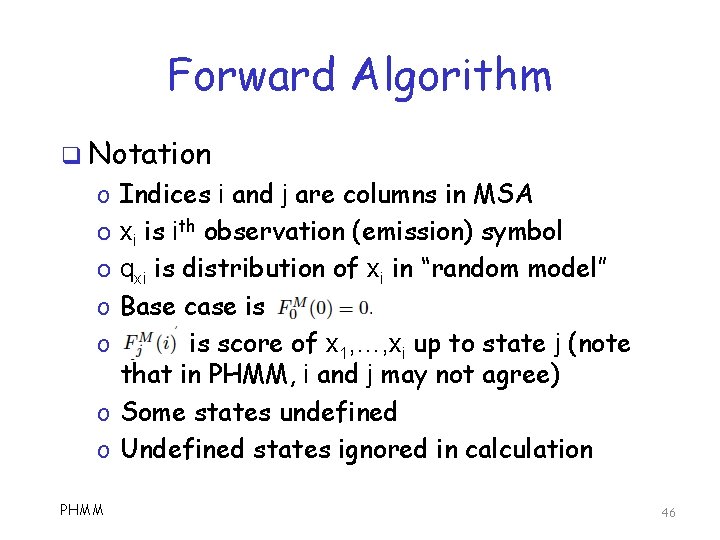
Forward Algorithm q Notation Indices i and j are columns in MSA xi is ith observation (emission) symbol qxi is distribution of xi in “random model” Base case is is score of x 1, …, xi up to state j (note that in PHMM, i and j may not agree) o Some states undefined o Undefined states ignored in calculation o o o PHMM 46

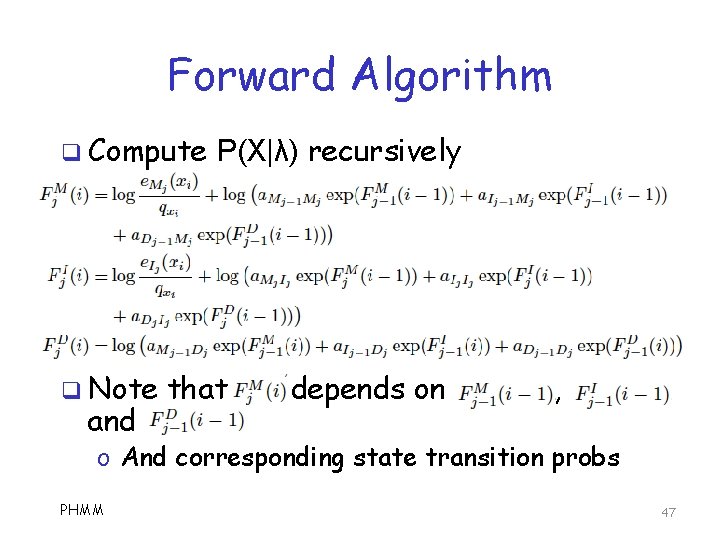
Forward Algorithm q Compute q Note and P(X|λ) recursively that depends on , o And corresponding state transition probs PHMM 47

PHMM q We will see examples of PHMM used in security application later q In particular, o Malware detection based on opcodes o Masquerade detection based on UNIX commands o Malware detection based on dynamically extracted API calls PHMM 48

References q q q Durbin, et al, Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids L. Huang and M. Stamp, Masquerade detection using profile hidden Markov models, Computers & Security, 30(8): 732 -747, 2011 S. Attaluri, S. Mc. Ghee, and M. Stamp, Profile hidden Markov models for metamorphic virus detection, Journal in Computer Virology, 5(2): 151 -169, 2009 PHMM 49