Hidden Markov Models A firstorder Hidden Markov Model







- Slides: 24

Hidden Markov Models A first-order Hidden Markov Model is completely defined by: • A set of states. • An alphabet of symbols. • A transition probability matrix T=(tij) • An emission probability matrix E=(ei. X)

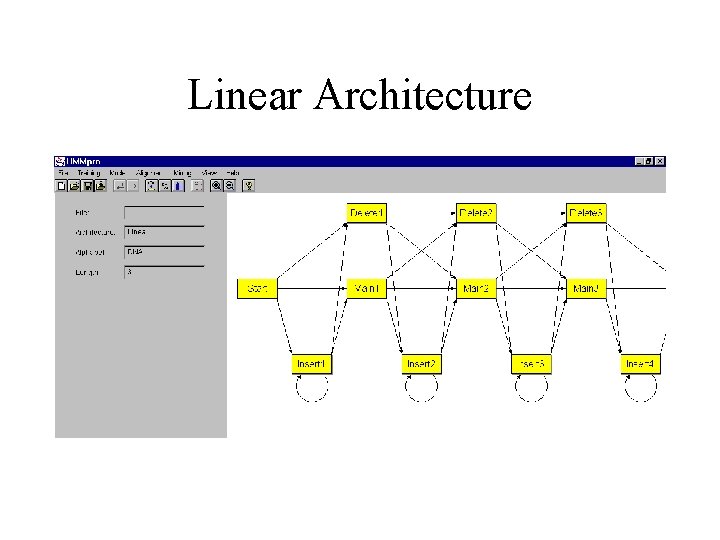
Linear Architecture

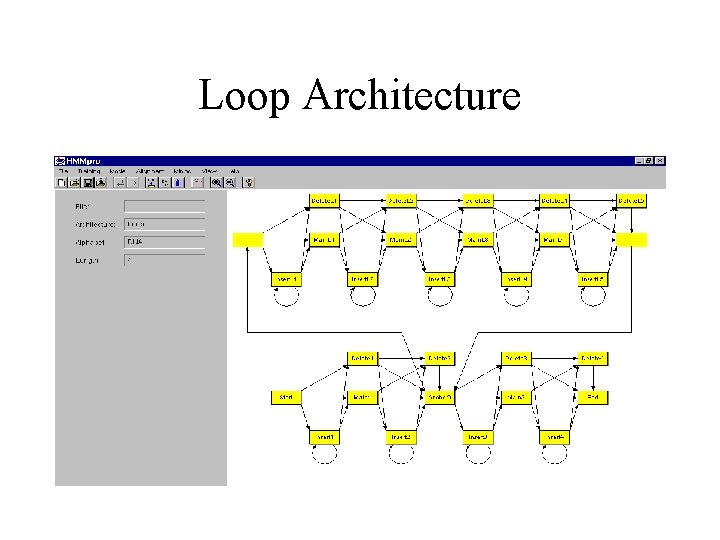
Loop Architecture

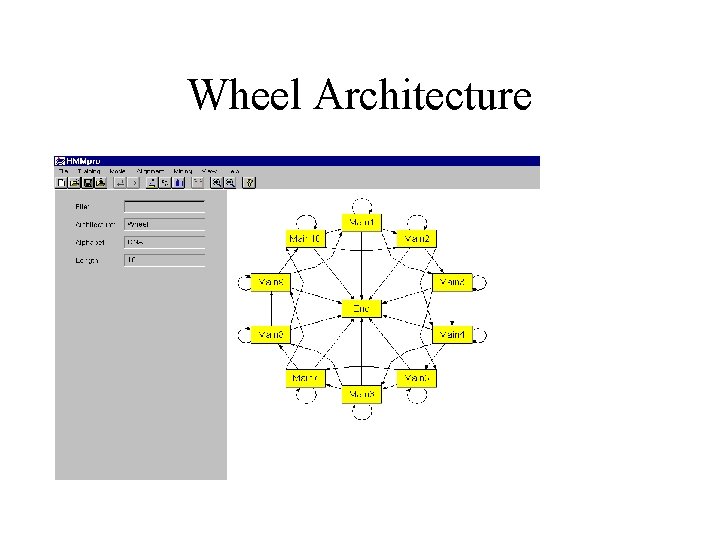
Wheel Architecture

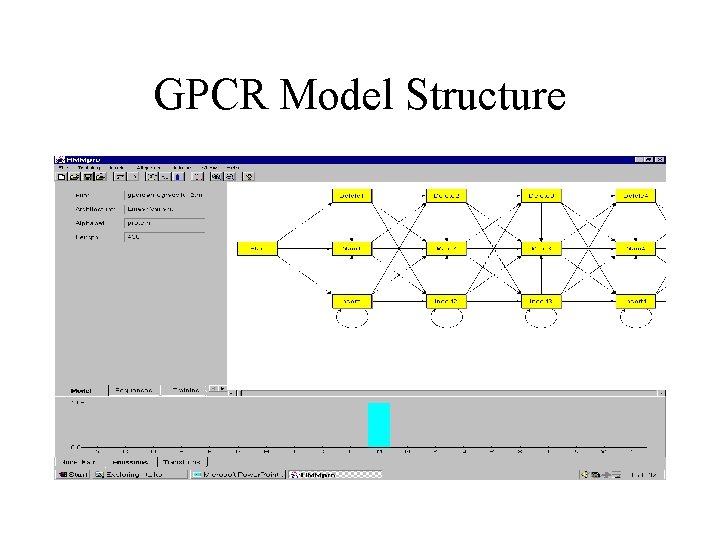
Basic Ideas • As in speech recognition, use Hidden Markov Models (HMM) to model a family of related primary sequences. • As in speech recognition, in general use a left to right HMM: once the system leaves a state it can never reenter it. The basic architecture consists of a main backbone chain of main states, and two side chains of insert and delete states. • The parameters of the model are the transition and emission probabilities. These parameters are adjusted during training from examples. • After learning, the model can be used in a variety of tasks including: multiple alignments, detection of motifs, classification, data base searches.

HMM APPLICATIONS • MULTIPLE ALIGNMENTS • DATA BASE SEARCHES AND DISCRIMINATION/CLASSIFICATION • STRUCTURAL ANALYSIS AND PATTERN DISCOVERY

Multiple Alignments • No precise definition of what a good alignment is (low entropy, detection of motifs). • The multiple alignment problem is NP complete (finding longest subsequence). • Pairwise alignment can be solved efficiently by dynamic programming in O(N 2) steps. • For K sequences of average length N, dynamic programming scales like O(NK), exponentially in the number of sequences. • Problem of variable scores and gap penalties.

HMMs of Protein Families • • • Globins Immunoglobulins Kinases G-Protein-Coupled Receptors Pfam is a data base of protein domains

HMMs of DNA • • coding/non-coding regions (E. Coli) exons/introns/acceptor sites promoter regions gene finding

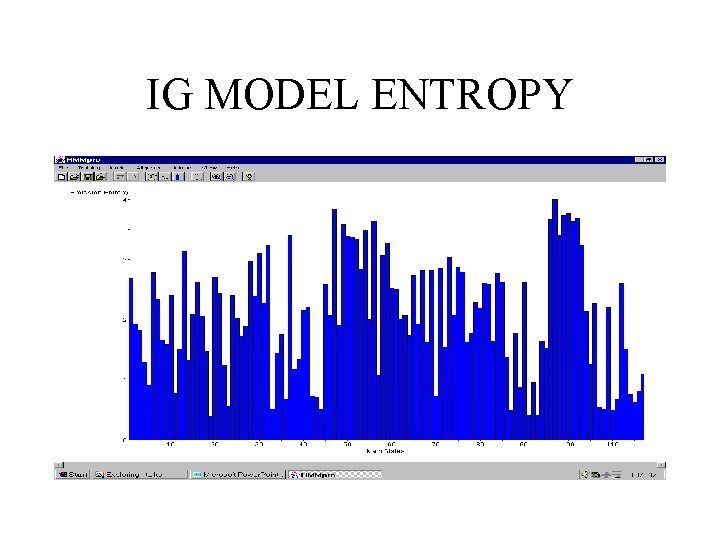
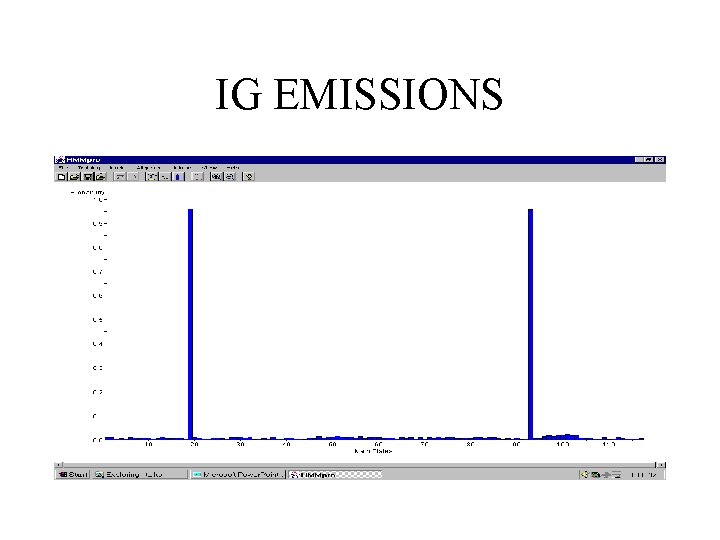
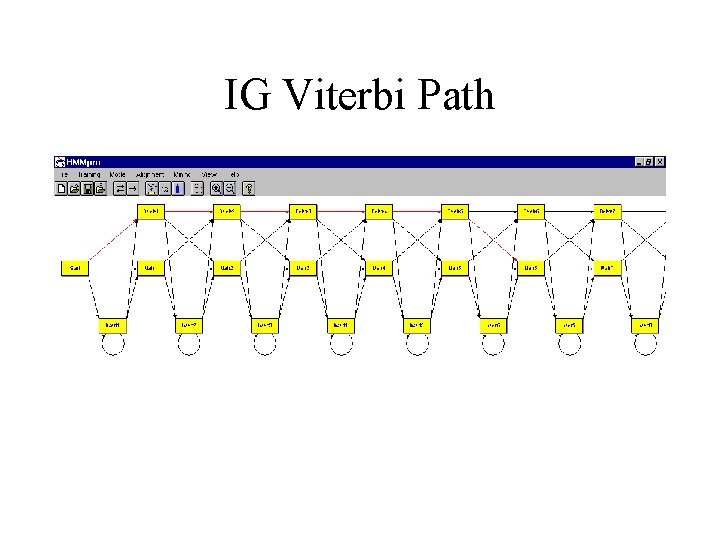
IMMUNOGLOBULINS • 294 sequences (V regions) with minimum length 90, average length 117, and maximal length 254 • linear model of length 117 trained with a random subset of 150 sequences

IG MODEL ENTROPY

IG EMISSIONS

IG Viterbi Path

IG MULTIPLE ALIGNMENT

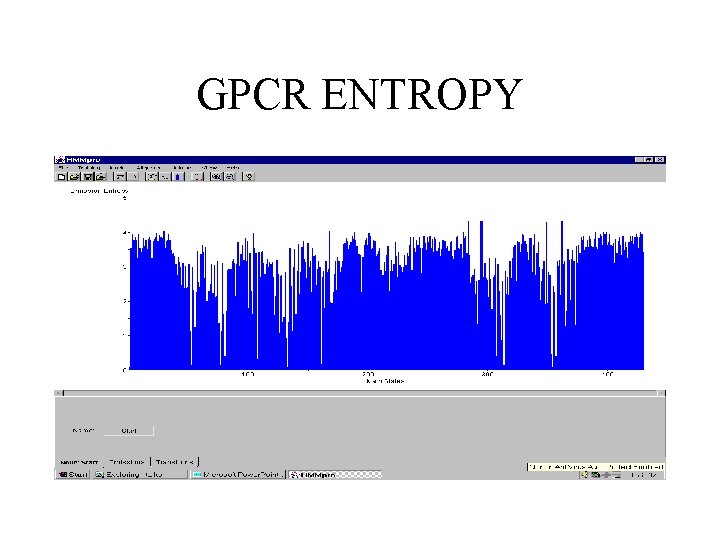
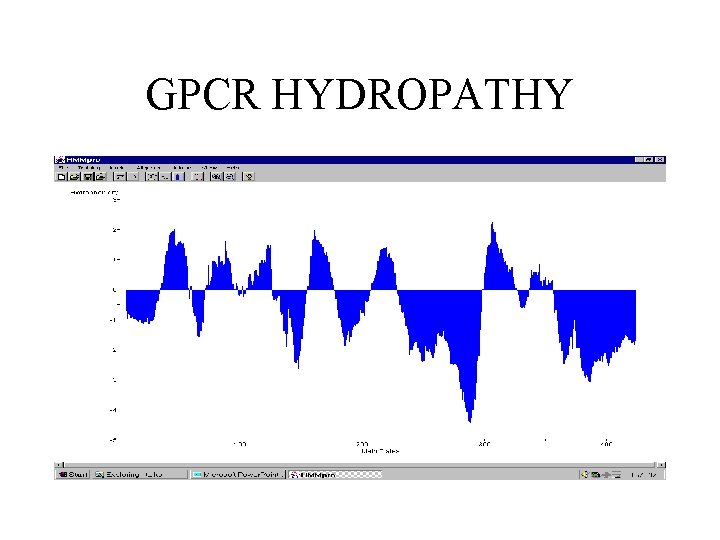
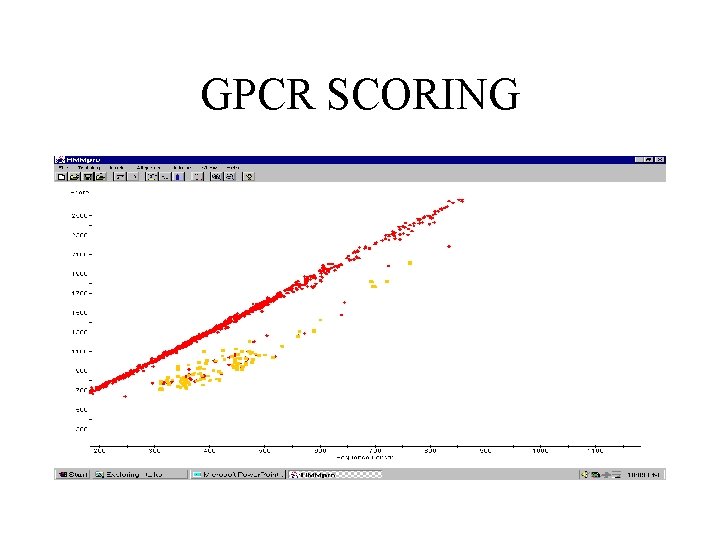
G-PROTEIN-COUPLED RECEPTORS • 145 sequences with minimum length 310, average length 430, and maximal length 764. • Model trained with 143 sequences (3 sequences contained undefined symbols) using Viterbi learning.

GPCR ENTROPY

GPCR HYDROPATHY

GPCR Model Structure

GPCR SCORING

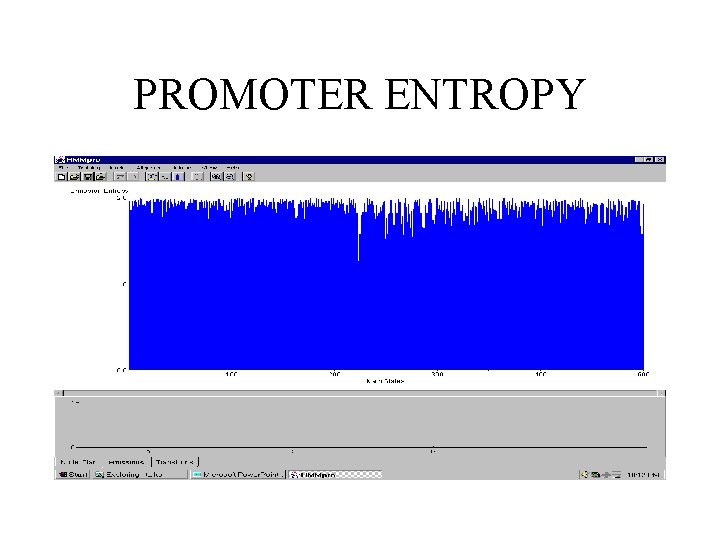
PROMOTER ENTROPY

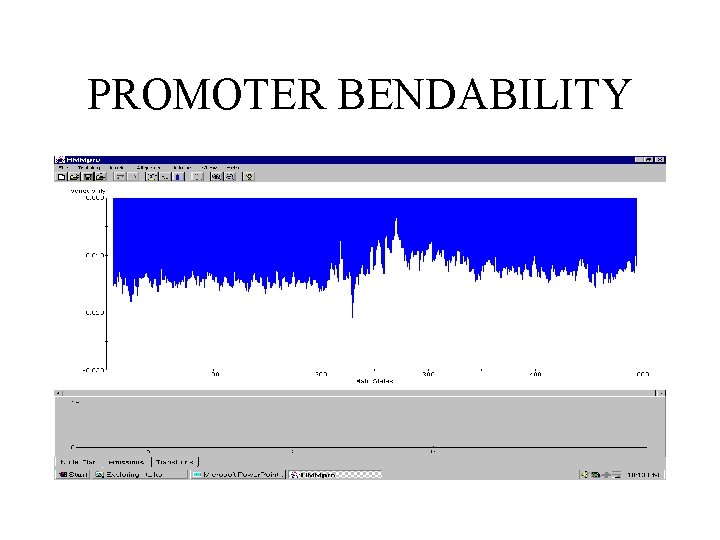
PROMOTER BENDABILITY

PROMOTER PROPELLER TWIST

SOFTWARE STRUCTURE • OBJECT-ORIENTED LIBRARY FOR MACHINE LEARNING • ENGINE IN C++ • GRAPHICAL USER INTERFACE IN JAVA • RUNS UNDER WINDOWS NT AND UNIX (SOLARIS, IRIX)

INFORMATION • ADDITIONAL INFORMATION, POINTERS, REFERENCES, AND SOFTWARE DOWNLOAD: WWW. NETID. COM