Nucleic acid structure and chemistry Andy Howard Introductory
![Base composition for DNA n n As noted, [A]=[T], [C]=[G] because of base pairing Base composition for DNA n n As noted, [A]=[T], [C]=[G] because of base pairing](https://slidetodoc.com/presentation_image_h2/6ae00e10cf9930f88d034d98b98e7755/image-28.jpg)

- Slides: 68

Nucleic acid structure and chemistry Andy Howard Introductory Biochemistry Monday 20 October 2014 Nucleic Acid Structure and Chemistry 10/20/2014

Nucleic acid structure and function n Nucleic acid bases, nucleosides, and nucleotides have characteristic structural and chemical properties, both on their own and as polymers 10/20/2014 Nucleic Acid Structure and Chemistry p. 2 of 68

What we’ll discuss n n Nucleosides and deoxynucleosides Oligomers and polymers Duplexes & helicity DNA structure n n Characterizations B, A, and Z-DNA Dynamics Function n n DNA sequencing RNA: structure & types n n m. RNA t. RNA r. RNA Small RNAs 10/20/2014 Nucleic Acid Structure and Chemistry p. 3 of 68

i. Clicker quiz, 1 st question 1. Which type of ion-channel gating have we not discussed? n (a) ligand-gating n (b) substrate-gating n (c) magnetic-field gating n (d) voltage-gating n (e) we discussed all of these 10/20/2014 Nucleic Acid Structure and Chemistry p. 4 of 68

i. Clicker quiz, question 2 2. We would expect the central pore of a cation channel to be lined with n (a) hydrophobic residues n (b) asp and glu residues n (c) lys and arg residues n (d) cofactors n (e) none of the above. 10/20/2014 Nucleic Acid Structure and Chemistry p. 5 of 68

i. Clicker quiz, question 3 3. Which of the following atom types are absent in nucleotides? n (a) P and S n (b) S and K n (c) N and O n (d) P and H n (e) all of these atom types are found in nucleotides. 10/20/2014 Nucleic Acid Structure and Chemistry p. 6 of 68

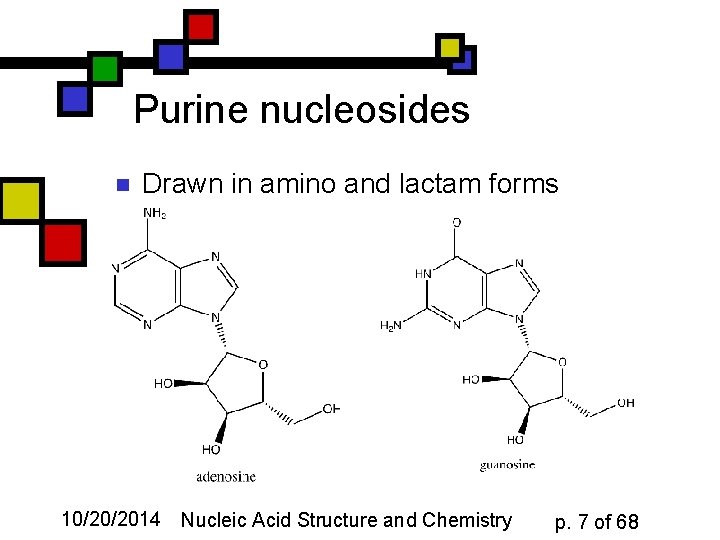
Purine nucleosides n Drawn in amino and lactam forms 10/20/2014 Nucleic Acid Structure and Chemistry p. 7 of 68

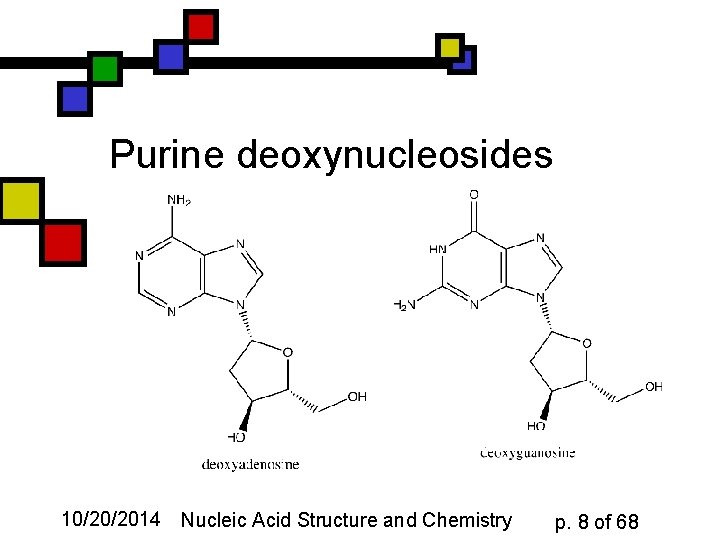
Purine deoxynucleosides 10/20/2014 Nucleic Acid Structure and Chemistry p. 8 of 68

Conformations around the glycosidic bond n n Rotation of the base around the glycosidic bond is sterically hindered In the syn conformation there would be some interference between the sugar ring and the 2 -position of the base Therefore pyrimidines are always anti, and purines are usually anti Furanose and base rings are roughly perpendicular 10/20/2014 Nucleic Acid Structure and Chemistry p. 9 of 68

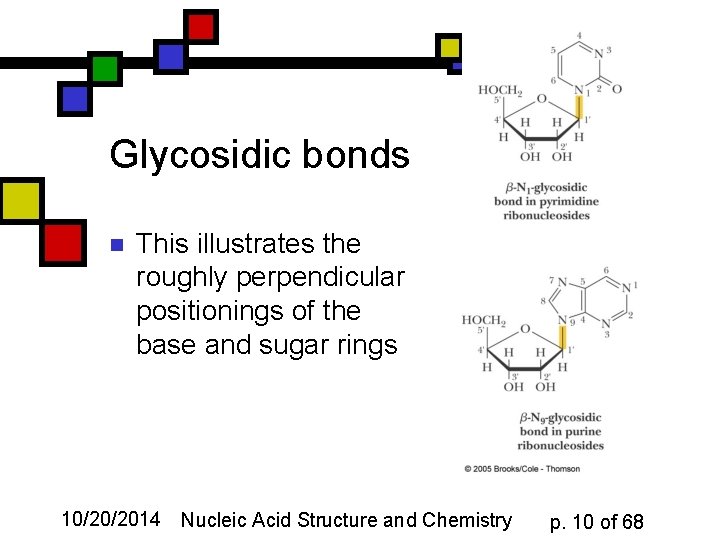
Glycosidic bonds n This illustrates the roughly perpendicular positionings of the base and sugar rings 10/20/2014 Nucleic Acid Structure and Chemistry p. 10 of 68

Solubility of nucleosides and lability of glycosidic linkages n n n The sugar makes nucleosides more soluble than the free bases Nucleosides are generally stable to basic hydrolysis at the glycosidic bond Acid hydrolysis: n n Purines: glycosidic bond fairly readily hydrolyzed Pyrimidines: resistant to acid hydrolysis 10/20/2014 Nucleic Acid Structure and Chemistry p. 11 of 68

Chirality in nucleic acids n n n Bases themselves are achiral Four asymmetric centers in ribofuranose, counting the glycosidic bond. Three in deoxyribofuranose Glycosidic bond is one of those 4 or 3. Same for nucleotides: phosphates don’t add asymmetries 10/20/2014 Nucleic Acid Structure and Chemistry p. 12 of 68

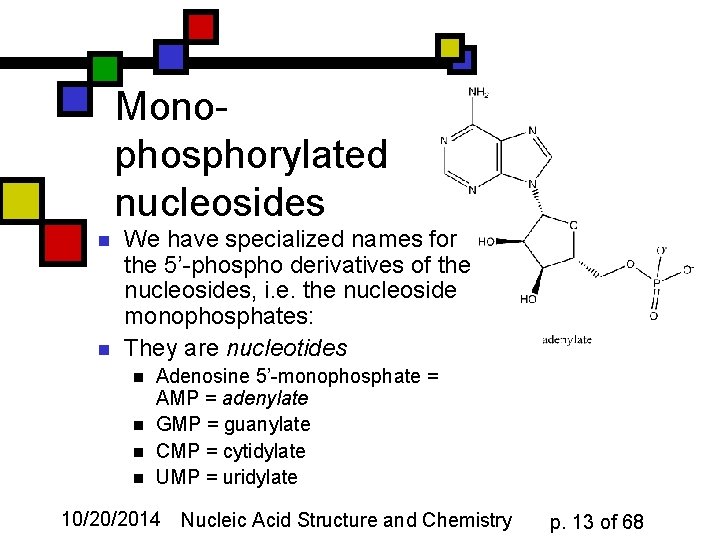
Monophosphorylated nucleosides n n We have specialized names for the 5’-phospho derivatives of the nucleosides, i. e. the nucleoside monophosphates: They are nucleotides n n Adenosine 5’-monophosphate = AMP = adenylate GMP = guanylate CMP = cytidylate UMP = uridylate 10/20/2014 Nucleic Acid Structure and Chemistry p. 13 of 68

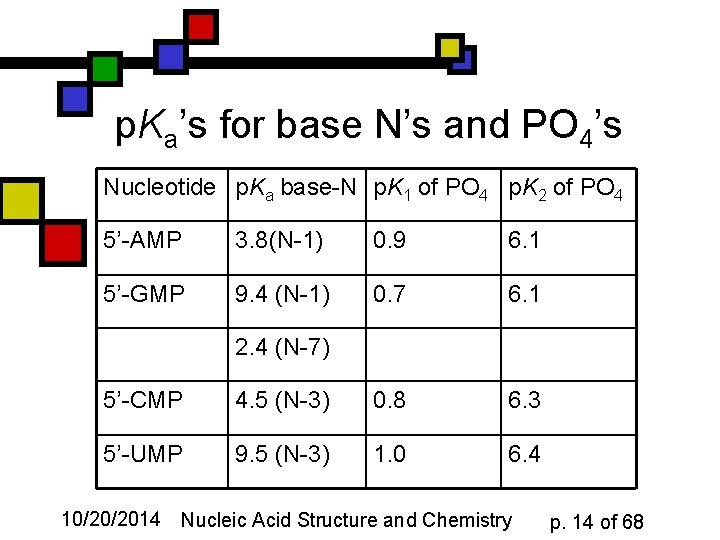
p. Ka’s for base N’s and PO 4’s Nucleotide p. Ka base-N p. K 1 of PO 4 p. K 2 of PO 4 5’-AMP 3. 8(N-1) 0. 9 6. 1 5’-GMP 9. 4 (N-1) 0. 7 6. 1 2. 4 (N-7) 5’-CMP 4. 5 (N-3) 0. 8 6. 3 5’-UMP 9. 5 (N-3) 1. 0 6. 4 10/20/2014 Nucleic Acid Structure and Chemistry p. 14 of 68

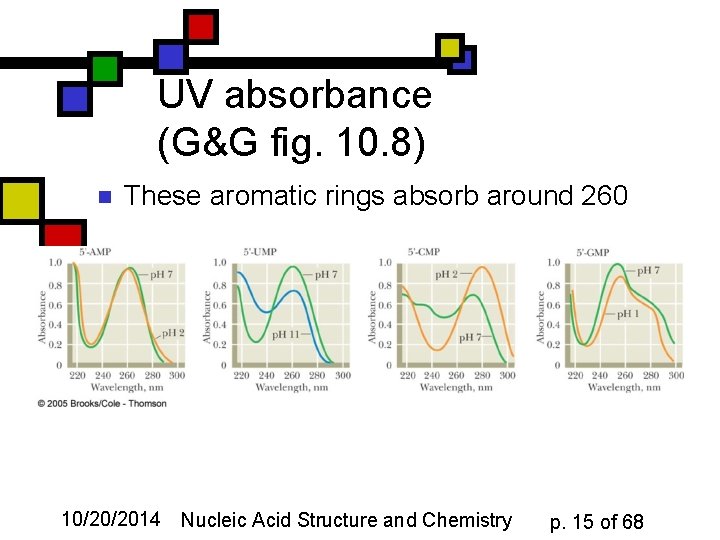
UV absorbance (G&G fig. 10. 8) n These aromatic rings absorb around 260 10/20/2014 Nucleic Acid Structure and Chemistry p. 15 of 68

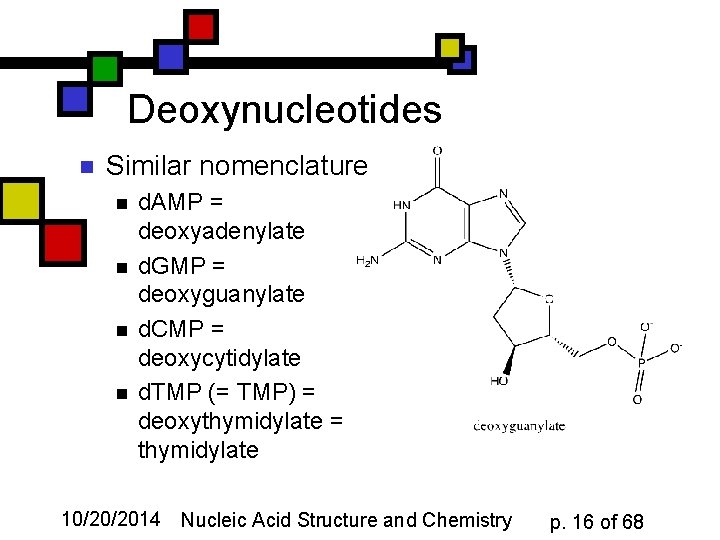
Deoxynucleotides n Similar nomenclature n n d. AMP = deoxyadenylate d. GMP = deoxyguanylate d. CMP = deoxycytidylate d. TMP (= TMP) = deoxythymidylate = thymidylate 10/20/2014 Nucleic Acid Structure and Chemistry p. 16 of 68

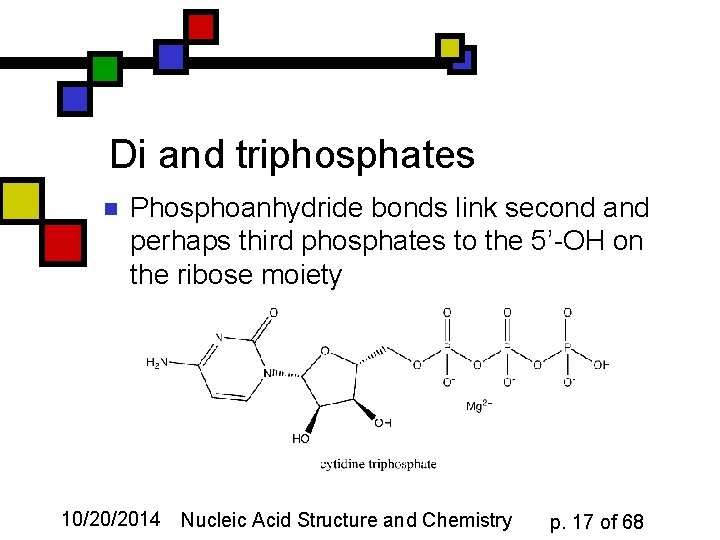
Di and triphosphates n Phosphoanhydride bonds link second and perhaps third phosphates to the 5’-OH on the ribose moiety 10/20/2014 Nucleic Acid Structure and Chemistry p. 17 of 68

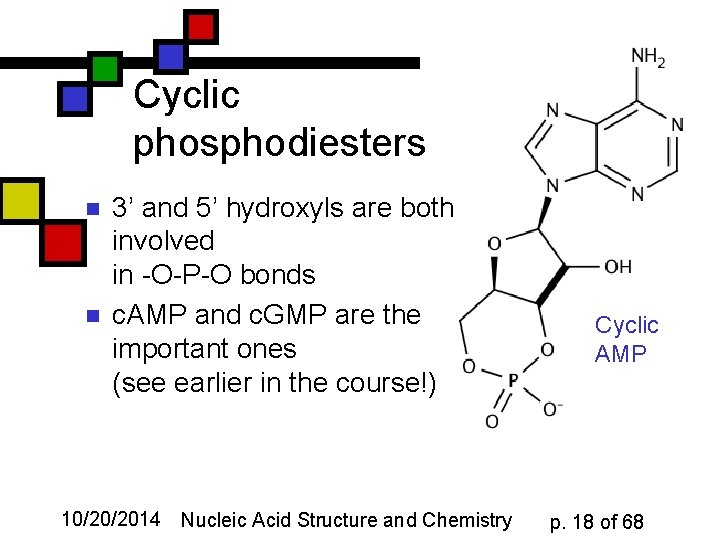
Cyclic phosphodiesters n n 3’ and 5’ hydroxyls are both involved in -O-P-O bonds c. AMP and c. GMP are the important ones (see earlier in the course!) 10/20/2014 Nucleic Acid Structure and Chemistry Cyclic AMP p. 18 of 68

Oligomers and Polymers n n n Monomers are nucleotides or deoxynucleotides Linkages are phosphodiester linkages between 3’ of one ribose and 5’ of the next ribose It’s logical to start from the 5’ end for synthetic reasons 10/20/2014 Nucleic Acid Structure and Chemistry p. 19 of 68

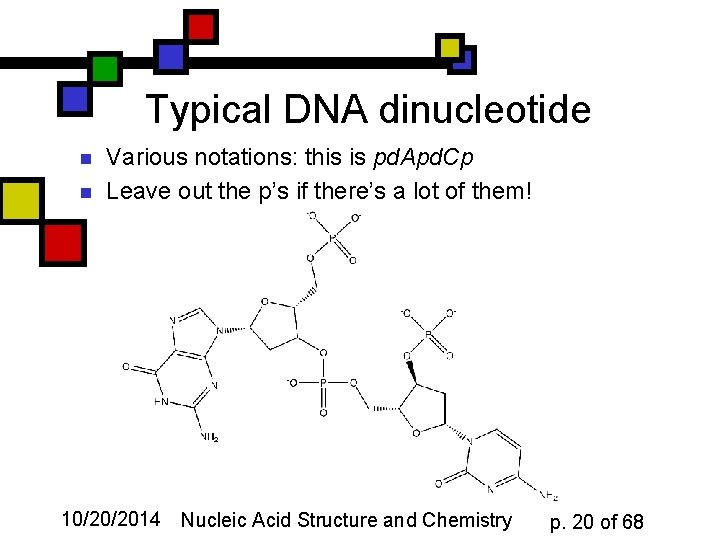
Typical DNA dinucleotide n n Various notations: this is pd. Apd. Cp Leave out the p’s if there’s a lot of them! 10/20/2014 Nucleic Acid Structure and Chemistry p. 20 of 68

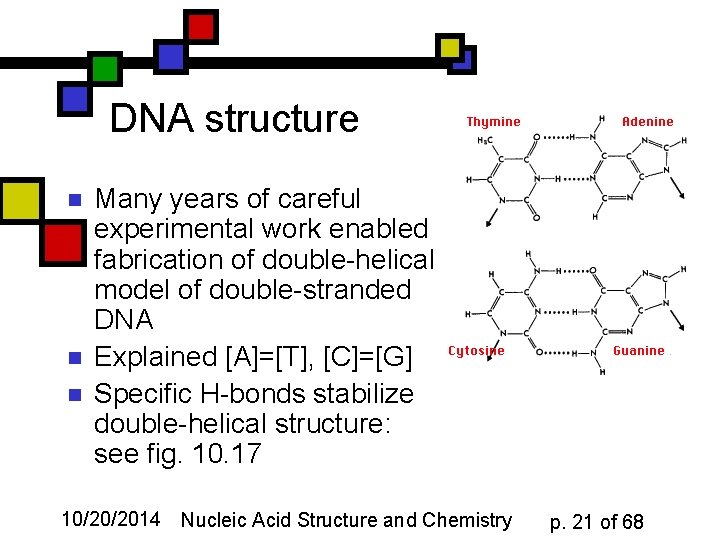
DNA structure n n n Many years of careful experimental work enabled fabrication of double-helical model of double-stranded DNA Explained [A]=[T], [C]=[G] Specific H-bonds stabilize double-helical structure: see fig. 10. 17 10/20/2014 Nucleic Acid Structure and Chemistry p. 21 of 68

What does double-stranded DNA really look like? n n Picture on previous slide emphasizes only the H-bond interactions; it ignores the orientation of the sugars, which are actually tilted relative to the helix axis Planes of the bases are almost perpendicular to the helical axes on both sides of the double helix 10/20/2014 Nucleic Acid Structure and Chemistry p. 22 of 68

Sizes (cf. fig. 11. 8, table 11. 1) n n Diameter of the double helix: 2. 37 nm Length along one full turn: 10. 4 base pairs = pitch = 3. 40 nm Distance between stacked base pairs = rise = 0. 33 nm Major groove is wider and shallower; minor groove is narrower and deeper 10/20/2014 Nucleic Acid Structure and Chemistry p. 23 of 68

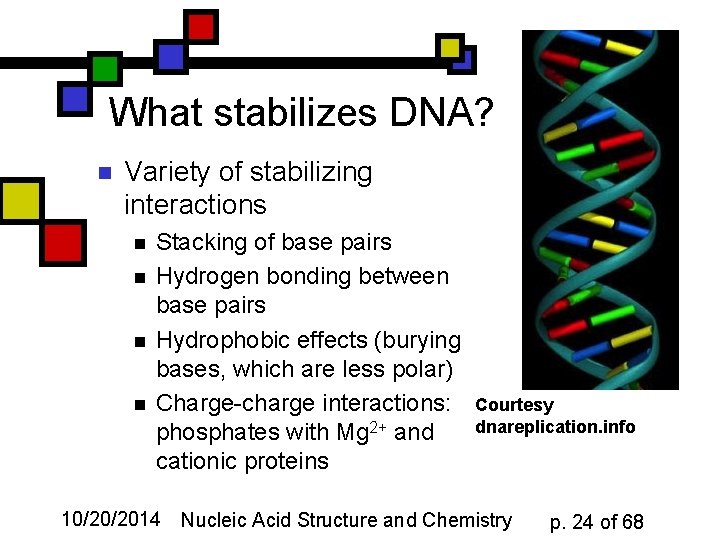
What stabilizes DNA? n Variety of stabilizing interactions n n Stacking of base pairs Hydrogen bonding between base pairs Hydrophobic effects (burying bases, which are less polar) Charge-charge interactions: phosphates with Mg 2+ and cationic proteins Courtesy dnareplication. info 10/20/2014 Nucleic Acid Structure and Chemistry p. 24 of 68

How close to instability is it? n n Pretty close. Heating DNA makes it melt: fig. 11. 20 p. H > 10 separates strands too The more GC pairs, the harder it is to melt DNA thermally n n Weaker stacking interactions in A-T One more H-bond per GC than per AT 10/20/2014 Nucleic Acid Structure and Chemistry p. 25 of 68

Do the differences between RNA and DNA matter? Yes! n n Deoxy sugars don’t degrade as readily! DNA has deoxythymidine, RNA has uridine: n n cytidine spontaneously degrades to uridine d. C spontaneously degrades to d. U The only d. U found in DNA is there because of degradation: d. T goes with d. A So when a cell finds d. U in its DNA, it knows it should replace it with d. C or else synthesize d. G opposite the d. U instead of d. A 10/20/2014 Nucleic Acid Structure and Chemistry p. 26 of 68

Ribose vs. deoxyribose n n n Presence of -OH on 2’ position makes the 3’ position in RNA more susceptible to nonenzymatic cleavage than the 3’ in DNA The ribose vs. deoxyribose distinction also influences enzymatic degradation of nucleic acids I can carry DNA in my shirt pocket, but not RNA 10/20/2014 Nucleic Acid Structure and Chemistry p. 27 of 68
![Base composition for DNA n n As noted AT CG because of base pairing Base composition for DNA n n As noted, [A]=[T], [C]=[G] because of base pairing](https://slidetodoc.com/presentation_image_h2/6ae00e10cf9930f88d034d98b98e7755/image-28.jpg)
Base composition for DNA n n As noted, [A]=[T], [C]=[G] because of base pairing [A]/[C] etc. not governed by base pairing n n Can vary considerably (table 10. 3) E. coli : [A], [C] about equal Mycobacterium tuberculosis: [C] > 2*[A] Mammals: [C] < 0. 74*[A] 10/20/2014 Nucleic Acid Structure and Chemistry p. 28 of 68

DNA secondary structures n If double-stranded DNA were simply a straightlegged ladder: n n n Base pairs would be 0. 6 nm apart Watson-Crick base-pairs have very uniform dimensions because the H-bonds are fixed lengths But water could get to the apolar bases So, in fact, the ladder gets twisted into a helix. The most common helix is B-DNA, but there are others. B-DNA’s properties include: n n Sugar-sugar distance is still 0. 6 nm Helix repeats itself every 3. 4 nm, i. e. 10 bp 10/20/2014 Nucleic Acid Structure and Chemistry p. 29 of 68

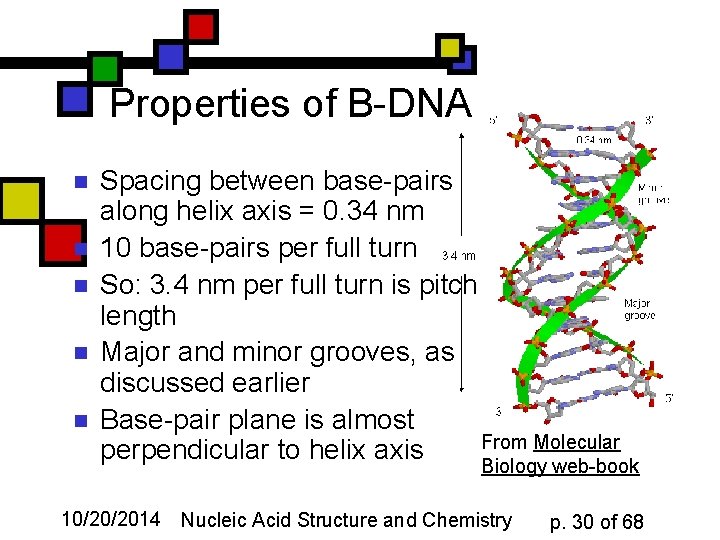
Properties of B-DNA n n n Spacing between base-pairs along helix axis = 0. 34 nm 10 base-pairs per full turn So: 3. 4 nm per full turn is pitch length Major and minor grooves, as discussed earlier Base-pair plane is almost From Molecular perpendicular to helix axis Biology web-book 10/20/2014 Nucleic Acid Structure and Chemistry p. 30 of 68

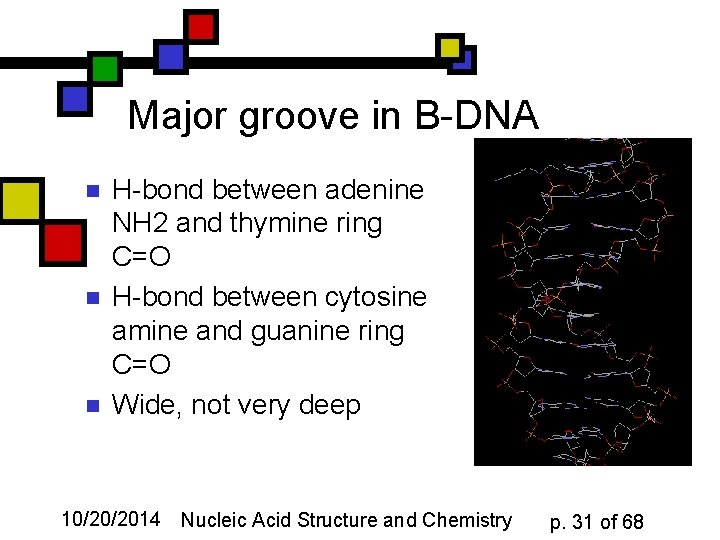
Major groove in B-DNA n n n H-bond between adenine NH 2 and thymine ring C=O H-bond between cytosine amine and guanine ring C=O Wide, not very deep 10/20/2014 Nucleic Acid Structure and Chemistry p. 31 of 68

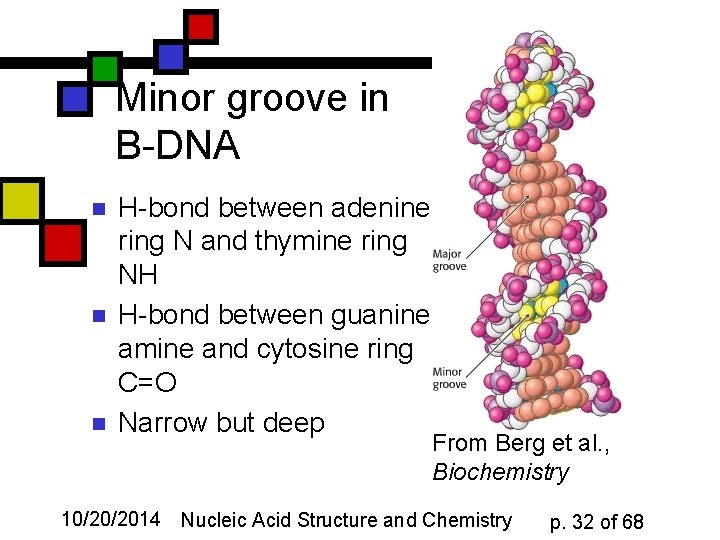
Minor groove in B-DNA n n n H-bond between adenine ring N and thymine ring NH H-bond between guanine amine and cytosine ring C=O Narrow but deep From Berg et al. , Biochemistry 10/20/2014 Nucleic Acid Structure and Chemistry p. 32 of 68

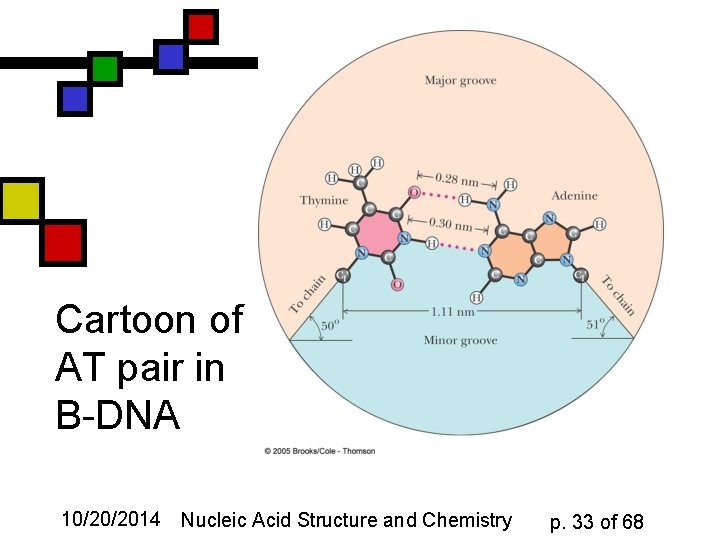
Cartoon of AT pair in B-DNA 10/20/2014 Nucleic Acid Structure and Chemistry p. 33 of 68

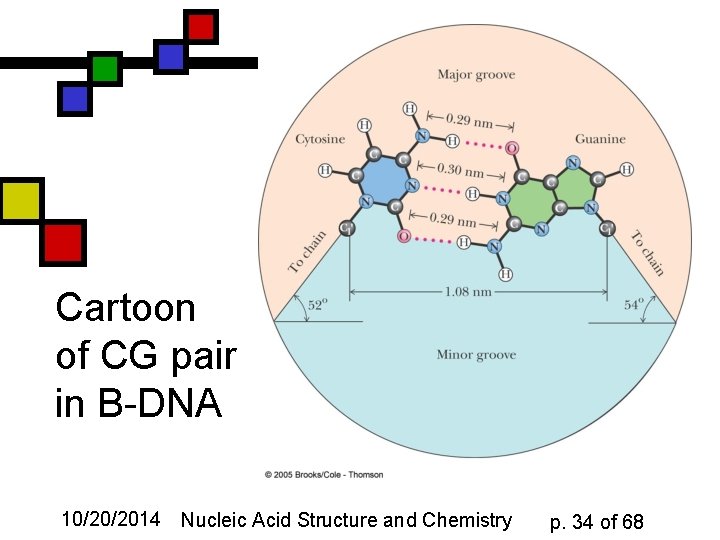
Cartoon of CG pair in B-DNA 10/20/2014 Nucleic Acid Structure and Chemistry p. 34 of 68

What holds duplex B-DNA together? n n n H-bonds (but just barely) Electrostatics: Mg 2+ –HPO 4 van der Waals interactions n - interactions in bases n n Solvent exclusion Recognize role of grooves in defining DNA-protein interactions 10/20/2014 Nucleic Acid Structure and Chemistry p. 35 of 68

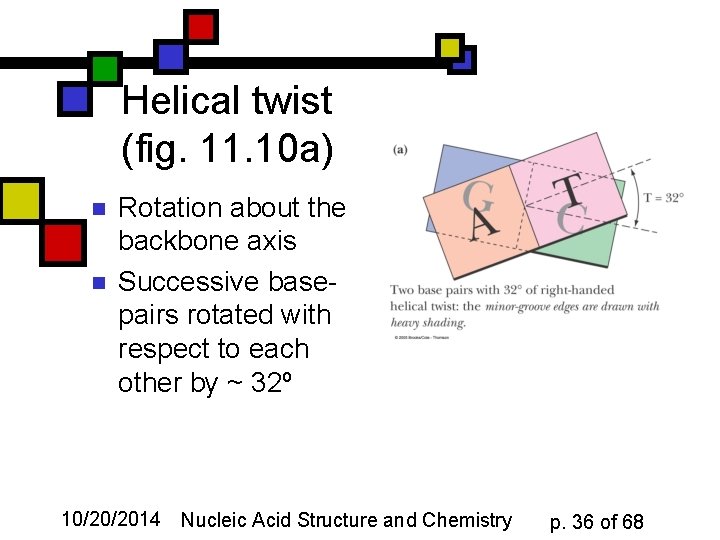
Helical twist (fig. 11. 10 a) n n Rotation about the backbone axis Successive basepairs rotated with respect to each other by ~ 32º 10/20/2014 Nucleic Acid Structure and Chemistry p. 36 of 68

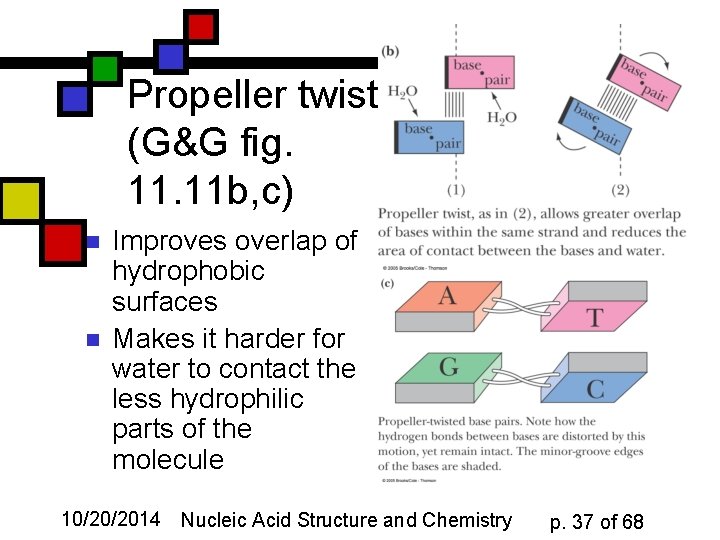
Propeller twist (G&G fig. 11 b, c) n n Improves overlap of hydrophobic surfaces Makes it harder for water to contact the less hydrophilic parts of the molecule 10/20/2014 Nucleic Acid Structure and Chemistry p. 37 of 68

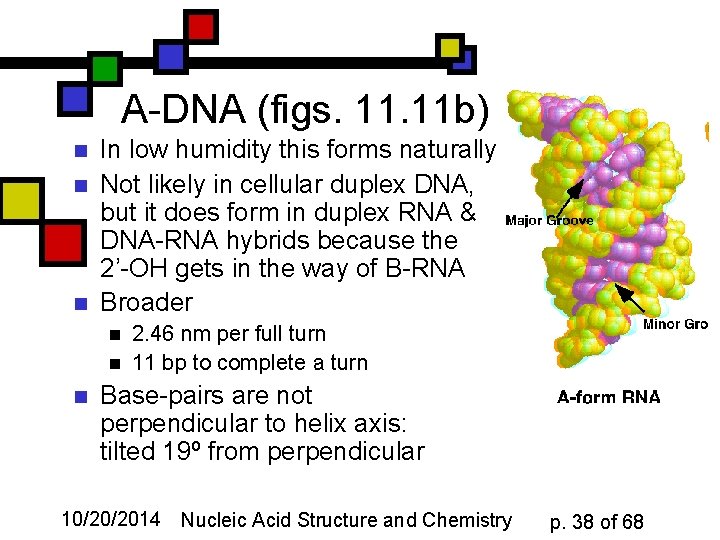
A-DNA (figs. 11 b) n n n In low humidity this forms naturally Not likely in cellular duplex DNA, but it does form in duplex RNA & DNA-RNA hybrids because the 2’-OH gets in the way of B-RNA Broader n n n 2. 46 nm per full turn 11 bp to complete a turn Base-pairs are not perpendicular to helix axis: tilted 19º from perpendicular 10/20/2014 Nucleic Acid Structure and Chemistry p. 38 of 68

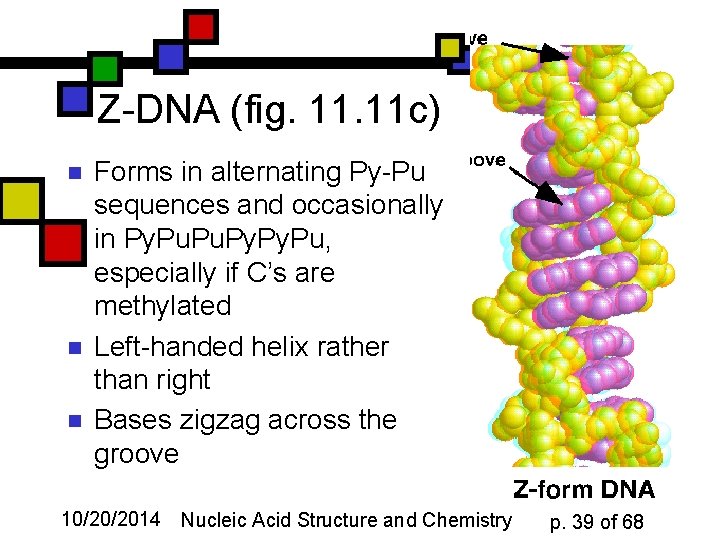
Z-DNA (fig. 11 c) n n n Forms in alternating Py-Pu sequences and occasionally in Py. Pu, especially if C’s are methylated Left-handed helix rather than right Bases zigzag across the groove 10/20/2014 Nucleic Acid Structure and Chemistry p. 39 of 68

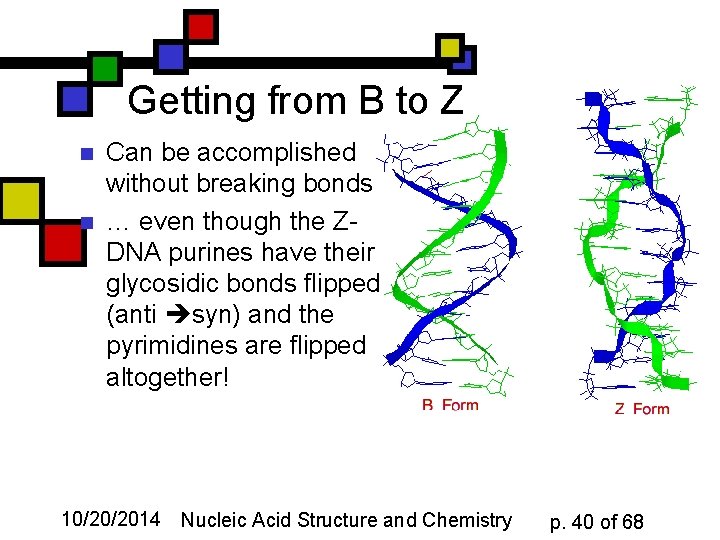
Getting from B to Z n n Can be accomplished without breaking bonds … even though the ZDNA purines have their glycosidic bonds flipped (anti syn) and the pyrimidines are flipped altogether! 10/20/2014 Nucleic Acid Structure and Chemistry p. 40 of 68

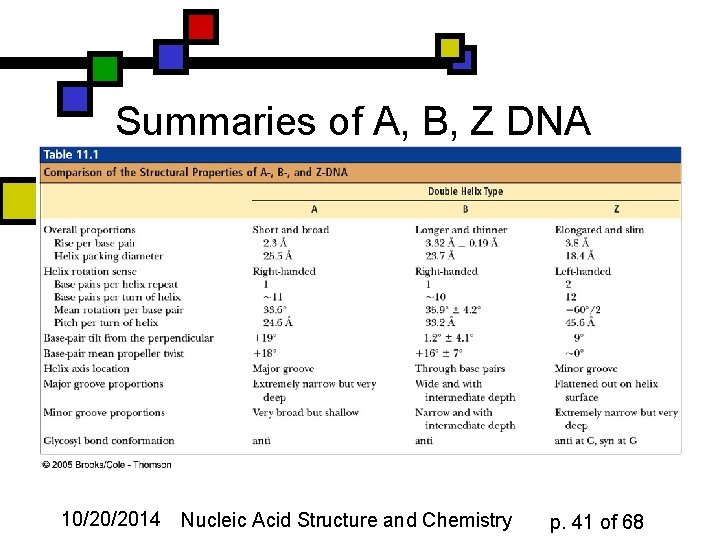
Summaries of A, B, Z DNA 10/20/2014 Nucleic Acid Structure and Chemistry p. 41 of 68

DNA is dynamic n n n Don’t think of these diagrams as static The H-bonds stretch and the torsions allow some rotations, so the ropes can form roughly spherical shapes when not constrained by histones Shape is sequence-dependent, which influences protein-DNA interactions 10/20/2014 Nucleic Acid Structure and Chemistry p. 42 of 68

What does DNA do? n Serve as the storehouse and the propagator of genetic information: That means that it’s made up of genes n n n Some code for m. RNAs that code for protein Others code for other types of RNA Genes contain non-coding segments (introns) But it also contains stretches that are not parts of genes at all and are serving controlling or structural roles Avoid the term junk DNA! 10/20/2014 Nucleic Acid Structure and Chemistry p. 43 of 68

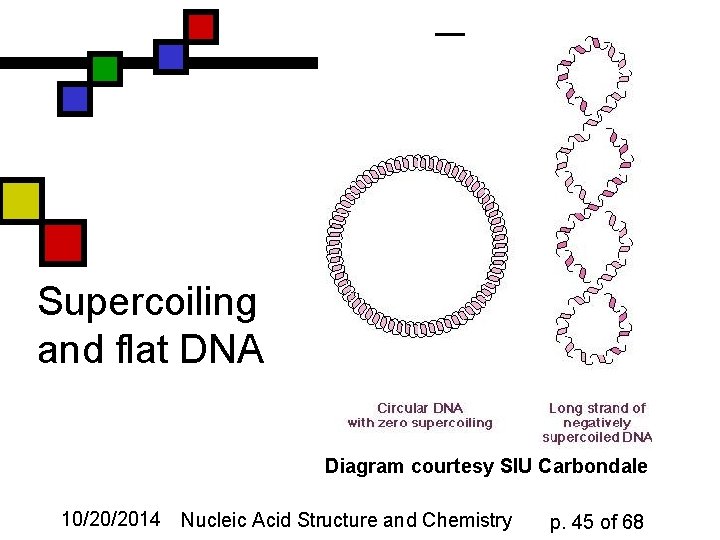
Supercoiling n n Refers to levels of organization of DNA beyond the immediate double-helix We describe circular DNA as relaxed if the closed double helix could lie flat It’s underwound or overwound if the ends are broken, twisted, and rejoined. Supercoils restore 10. 4 bp/turn relation upon rejoining 10/20/2014 Nucleic Acid Structure and Chemistry p. 44 of 68

Supercoiling and flat DNA Diagram courtesy SIU Carbondale 10/20/2014 Nucleic Acid Structure and Chemistry p. 45 of 68

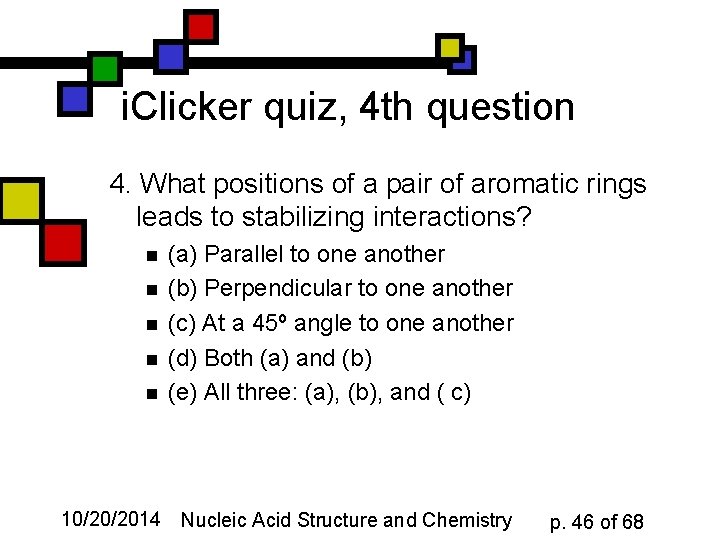
i. Clicker quiz, 4 th question 4. What positions of a pair of aromatic rings leads to stabilizing interactions? n n n (a) Parallel to one another (b) Perpendicular to one another (c) At a 45º angle to one another (d) Both (a) and (b) (e) All three: (a), (b), and ( c) 10/20/2014 Nucleic Acid Structure and Chemistry p. 46 of 68

i. Clicker question 5 n 5. Which has the highest molecular mass among the compounds listed? n n n (a) cytidylate (b) thymidylate (c) adenylate (d) adenosine triphosphate (e) they’re all the same MW 10/20/2014 Nucleic Acid Structure and Chemistry p. 47 of 68

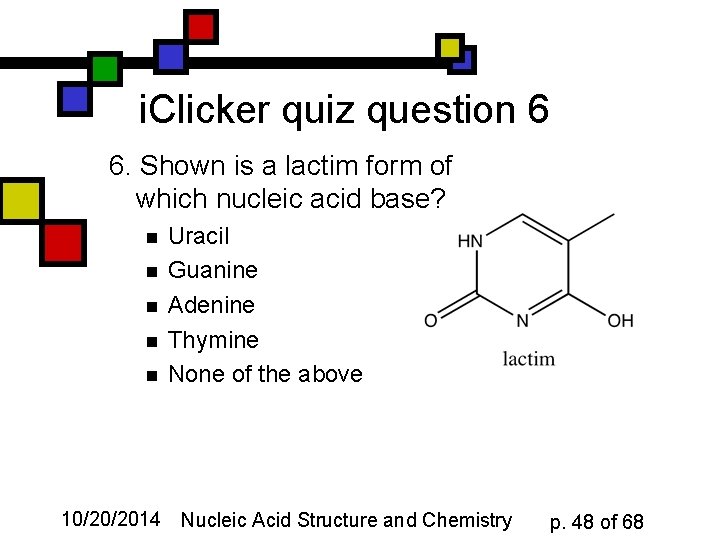
i. Clicker quiz question 6 6. Shown is a lactim form of which nucleic acid base? n n n Uracil Guanine Adenine Thymine None of the above 10/20/2014 Nucleic Acid Structure and Chemistry p. 48 of 68

i. Clicker quiz question 7 7. Suppose someone reports that he has characterized the genomic DNA of an organism as having 29% A and 22% T. How would you respond? n n (a) That’s a reasonable result (b) This result is unlikely because [A] ~ [T] in duplex DNA (c) That’s plausible if it’s a bacterium, but not if it’s a eukaryote (d) none of the above 10/20/2014 Nucleic Acid Structure and Chemistry p. 49 of 68

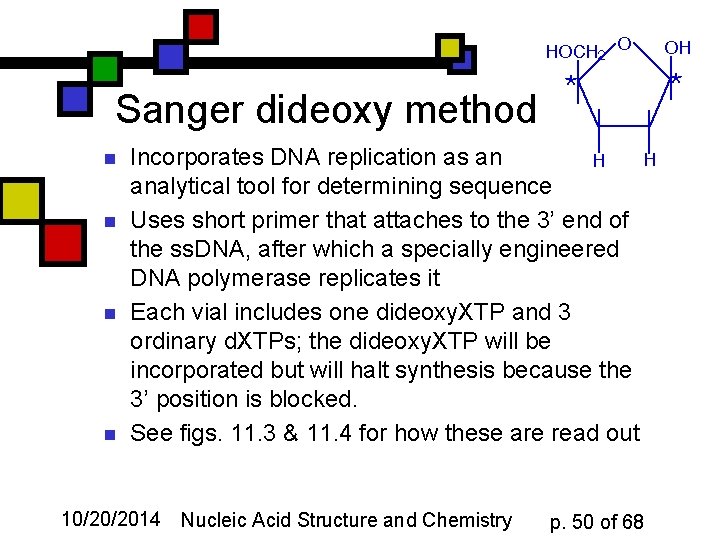
Sanger dideoxy method n n Incorporates DNA replication as an analytical tool for determining sequence Uses short primer that attaches to the 3’ end of the ss. DNA, after which a specially engineered DNA polymerase replicates it Each vial includes one dideoxy. XTP and 3 ordinary d. XTPs; the dideoxy. XTP will be incorporated but will halt synthesis because the 3’ position is blocked. See figs. 11. 3 & 11. 4 for how these are read out 10/20/2014 Nucleic Acid Structure and Chemistry p. 50 of 68

Automating dideoxy sequencing n n n Laser fluorescence detection allows for primer identification in real time An automated sequencing machine can handle 4500 bases/hour That’s one of the technologies that has made large-scale sequencing projects like the human genome project possible 10/20/2014 Nucleic Acid Structure and Chemistry p. 51 of 68

Ribonucleic acid n n We’re done with DNA for the moment. Let’s discuss RNA is generally, but not always, singlestranded The regions where localized base-pairing occurs (local double-stranded regions) often are of functional significance 10/20/2014 Nucleic Acid Structure and Chemistry p. 52 of 68

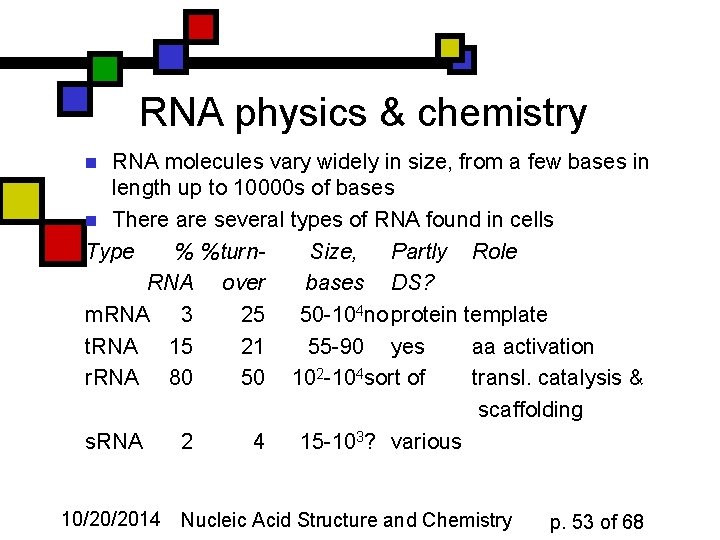
RNA physics & chemistry RNA molecules vary widely in size, from a few bases in length up to 10000 s of bases n There are several types of RNA found in cells Type % %turn. Size, Partly Role RNA over bases DS? m. RNA 3 25 50 -104 no protein template t. RNA 15 21 55 -90 yes aa activation r. RNA 80 50 102 -104 sort of transl. catalysis & scaffolding s. RNA 2 4 15 -103? various n 10/20/2014 Nucleic Acid Structure and Chemistry p. 53 of 68

Messenger RNA n n m. RNA: transcription vehicle DNA 5’-d. Ad. Cd. Gd. Td. Ad. Td. G-3’ RNA 3’- U G G C A U A C-5’ typical protein is ~500 amino acids; 3 m. RNA bases/aa: 1500 bases (after splicing) Additional noncoding regions (see later) brings it up to ~4000 bases = 4000*300 Da/base=1, 200, 000 Da Only about 3% of cellular RNA but unstable! 10/20/2014 Nucleic Acid Structure and Chemistry p. 54 of 68

Relative quantities n n Note that we said there wasn’t much m. RNA around at any given moment The amount synthesized is much greater because it has a much shorter lifetime than the others Ribonucleases act more avidly on it We need a mechanism for eliminating it because the cell wants to control concentrations of specific proteins 10/20/2014 Nucleic Acid Structure and Chemistry p. 55 of 68

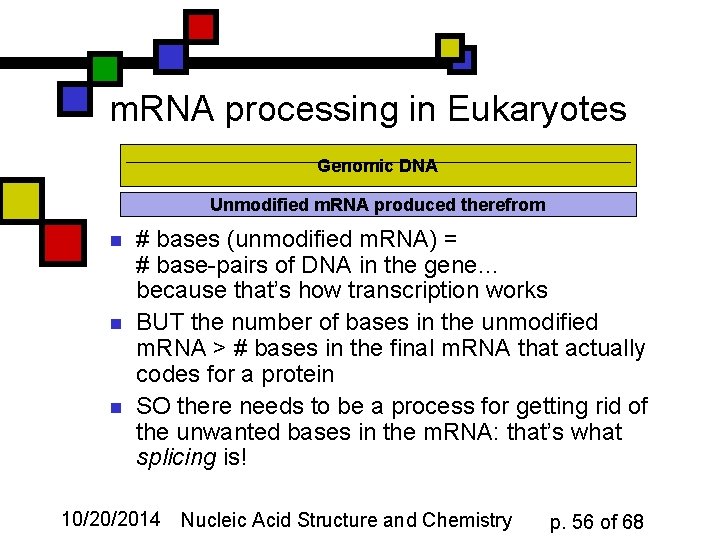
m. RNA processing in Eukaryotes Genomic DNA Unmodified m. RNA produced therefrom n n n # bases (unmodified m. RNA) = # base-pairs of DNA in the gene… because that’s how transcription works BUT the number of bases in the unmodified m. RNA > # bases in the final m. RNA that actually codes for a protein SO there needs to be a process for getting rid of the unwanted bases in the m. RNA: that’s what splicing is! 10/20/2014 Nucleic Acid Structure and Chemistry p. 56 of 68

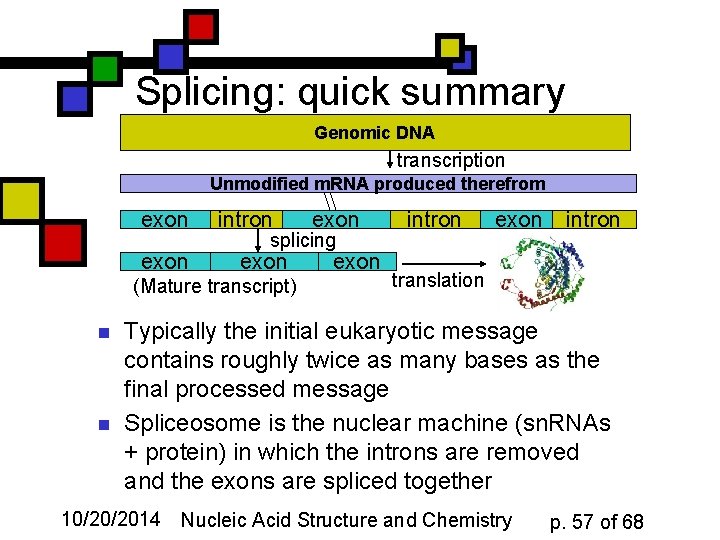
Splicing: quick summary Genomic DNA transcription Unmodified m. RNA produced therefrom exon intron splicing exon (Mature transcript) n n exon intron translation Typically the initial eukaryotic message contains roughly twice as many bases as the final processed message Spliceosome is the nuclear machine (sn. RNAs + protein) in which the introns are removed and the exons are spliced together 10/20/2014 Nucleic Acid Structure and Chemistry p. 57 of 68

Heterogeneity via spliceosomal flexibility n n Specific RNA sequences in the initial m. RNA signal where to start and stop each intron, but with some flexibility That flexibility enables a single gene to code for multiple mature RNAs and therefore multiple proteins 10/20/2014 Nucleic Acid Structure and Chemistry p. 58 of 68

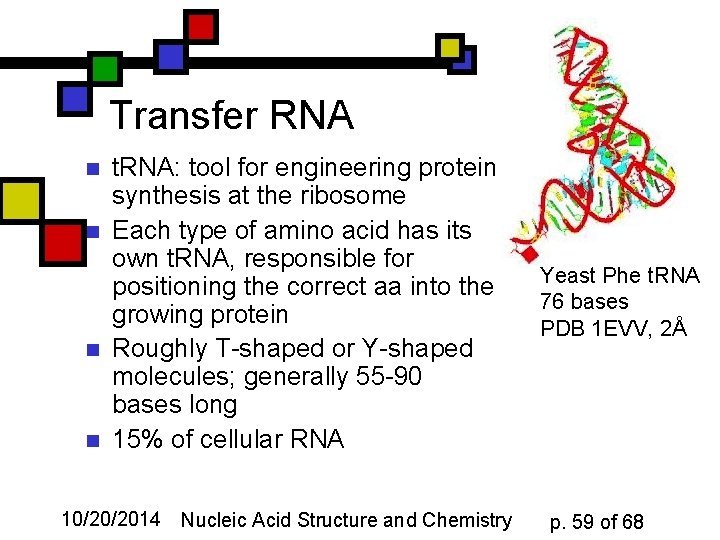
Transfer RNA n n t. RNA: tool for engineering protein synthesis at the ribosome Each type of amino acid has its own t. RNA, responsible for positioning the correct aa into the growing protein Roughly T-shaped or Y-shaped molecules; generally 55 -90 bases long 15% of cellular RNA 10/20/2014 Nucleic Acid Structure and Chemistry Yeast Phe t. RNA 76 bases PDB 1 EVV, 2Å p. 59 of 68

Secondary and Tertiary Structure of t. RNA n n Extensive H-bonding creates four double helical domains, three capped by loops, one by a stem Only one t. RNA structure (alone) is known Phenylalanine t. RNA is "L-shaped" Many non-canonical bases found in t. RNA 10/20/2014 Nucleic Acid Structure and Chemistry p. 60 of 68

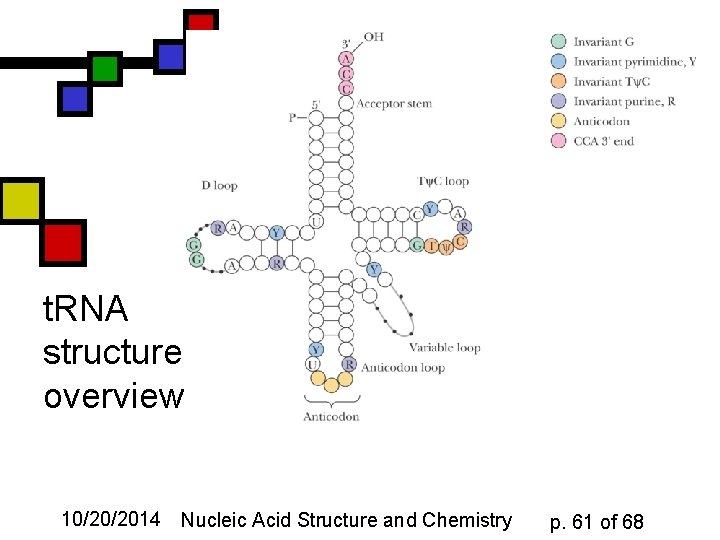
t. RNA structure: overview 10/20/2014 Nucleic Acid Structure and Chemistry p. 61 of 68

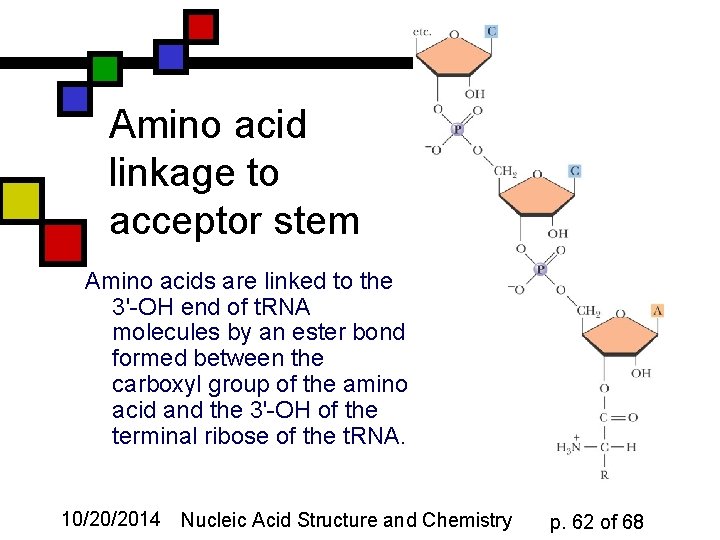
Amino acid linkage to acceptor stem Amino acids are linked to the 3'-OH end of t. RNA molecules by an ester bond formed between the carboxyl group of the amino acid and the 3'-OH of the terminal ribose of the t. RNA. 10/20/2014 Nucleic Acid Structure and Chemistry p. 62 of 68

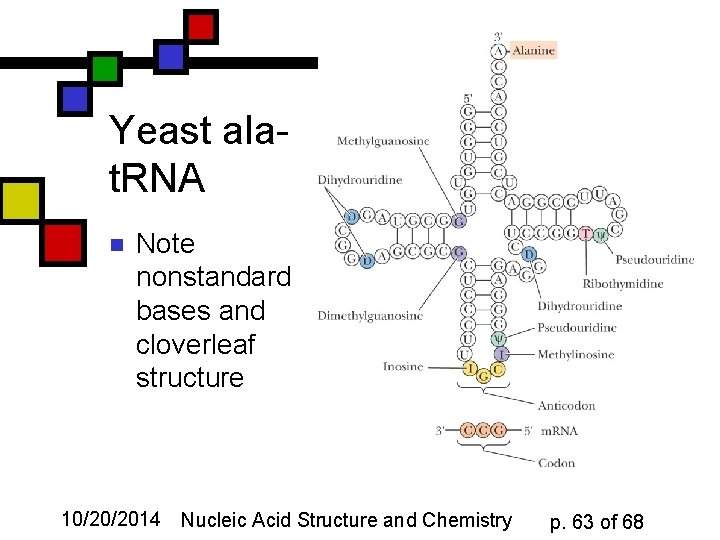
Yeast alat. RNA n Note nonstandard bases and cloverleaf structure 10/20/2014 Nucleic Acid Structure and Chemistry p. 63 of 68

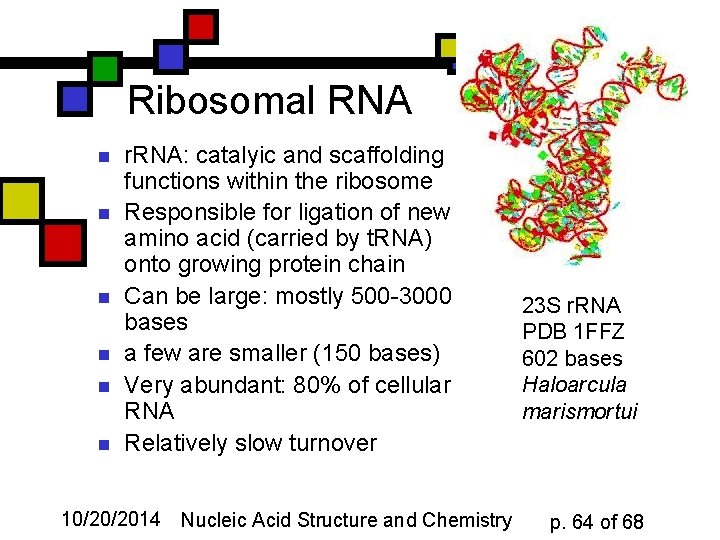
Ribosomal RNA n n n r. RNA: catalyic and scaffolding functions within the ribosome Responsible for ligation of new amino acid (carried by t. RNA) onto growing protein chain Can be large: mostly 500 -3000 bases a few are smaller (150 bases) Very abundant: 80% of cellular RNA Relatively slow turnover 10/20/2014 Nucleic Acid Structure and Chemistry 23 S r. RNA PDB 1 FFZ 602 bases Haloarcula marismortui p. 64 of 68

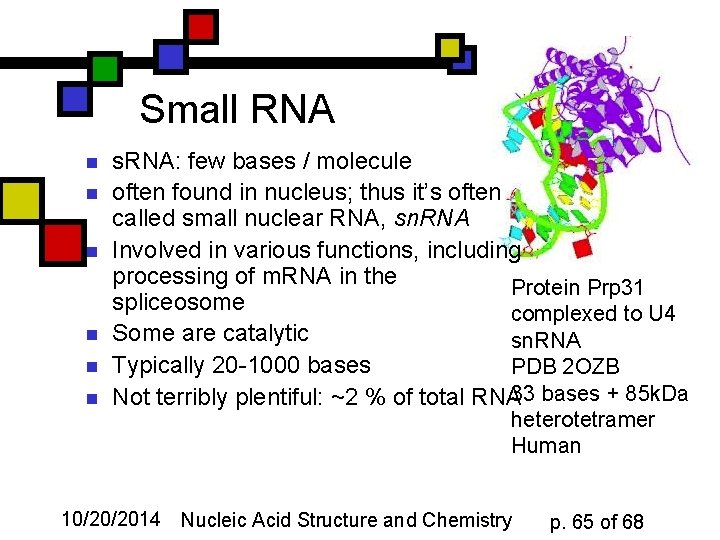
Small RNA n n n s. RNA: few bases / molecule often found in nucleus; thus it’s often called small nuclear RNA, sn. RNA Involved in various functions, including processing of m. RNA in the Protein Prp 31 spliceosome complexed to U 4 Some are catalytic sn. RNA Typically 20 -1000 bases PDB 2 OZB Not terribly plentiful: ~2 % of total RNA 33 bases + 85 k. Da heterotetramer Human 10/20/2014 Nucleic Acid Structure and Chemistry p. 65 of 68

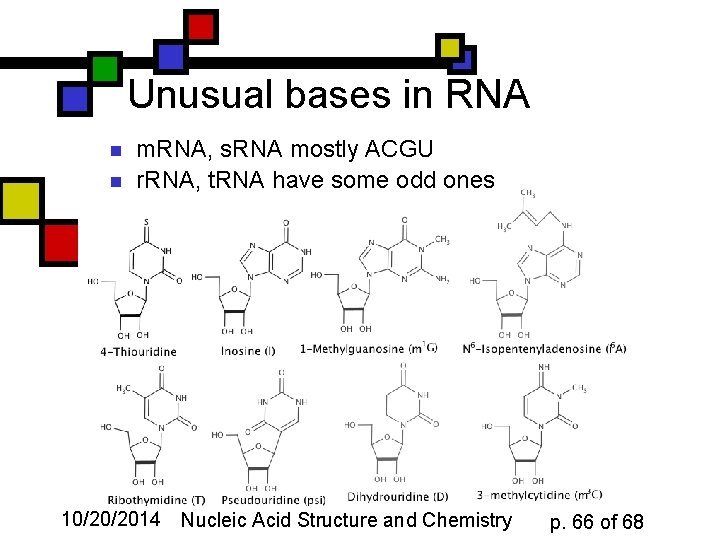
Unusual bases in RNA n n m. RNA, s. RNA mostly ACGU r. RNA, t. RNA have some odd ones 10/20/2014 Nucleic Acid Structure and Chemistry p. 66 of 68

Other small RNAs n n n 21 -28 nucleotides Target RNA or DNA through complementary base-pairing Several types, based on function: n n n Small interfering RNAs (q. v. ) micro. RNA: control developmental timing Small nucleolar RNA: catalysts that (among other things) create the oddball bases sno. RNA 77 courtesy Wikipedia 10/20/2014 Nucleic Acid Structure and Chemistry p. 67 of 68

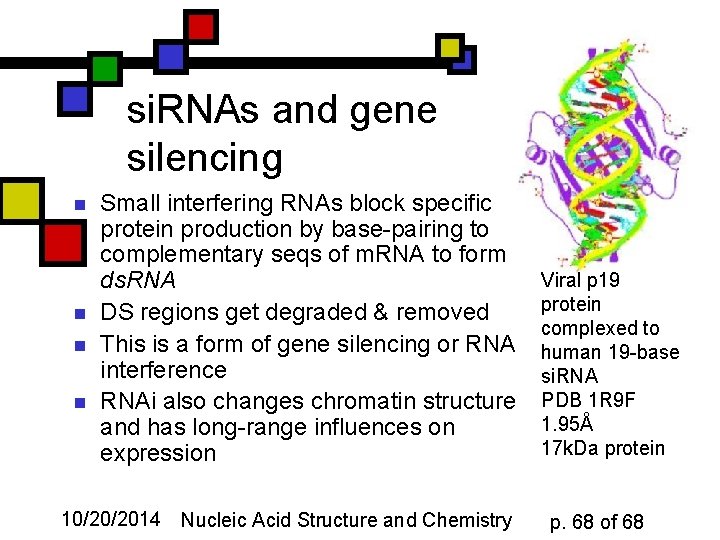
si. RNAs and gene silencing n n Small interfering RNAs block specific protein production by base-pairing to complementary seqs of m. RNA to form ds. RNA DS regions get degraded & removed This is a form of gene silencing or RNA interference RNAi also changes chromatin structure and has long-range influences on expression 10/20/2014 Nucleic Acid Structure and Chemistry Viral p 19 protein complexed to human 19 -base si. RNA PDB 1 R 9 F 1. 95Å 17 k. Da protein p. 68 of 68