Nucleic Acid Hybridization Nucleic acid hybridization is a
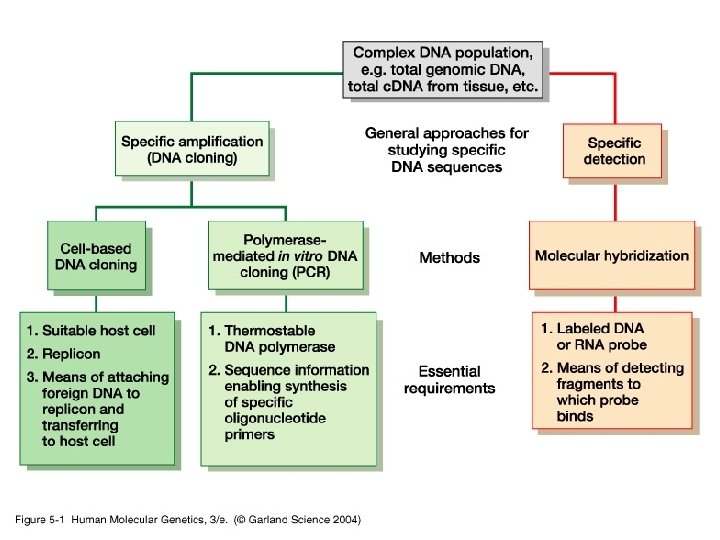

Nucleic Acid Hybridization • Nucleic acid hybridization is a fundamental tool in molecular genetics which takes advantage of the ability of individual single -stranded nucleic acid molecules to form double stranded molecules (that is, to hybridize to each other)
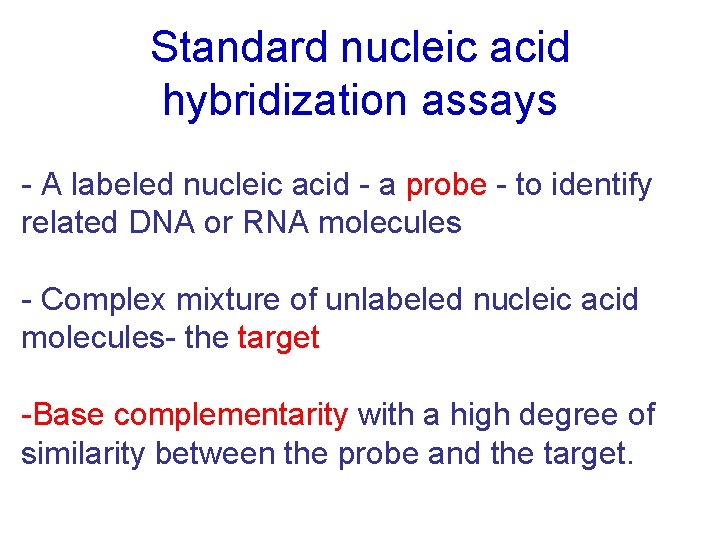
Standard nucleic acid hybridization assays - A labeled nucleic acid - a probe - to identify related DNA or RNA molecules - Complex mixture of unlabeled nucleic acid molecules- the target -Base complementarity with a high degree of similarity between the probe and the target.
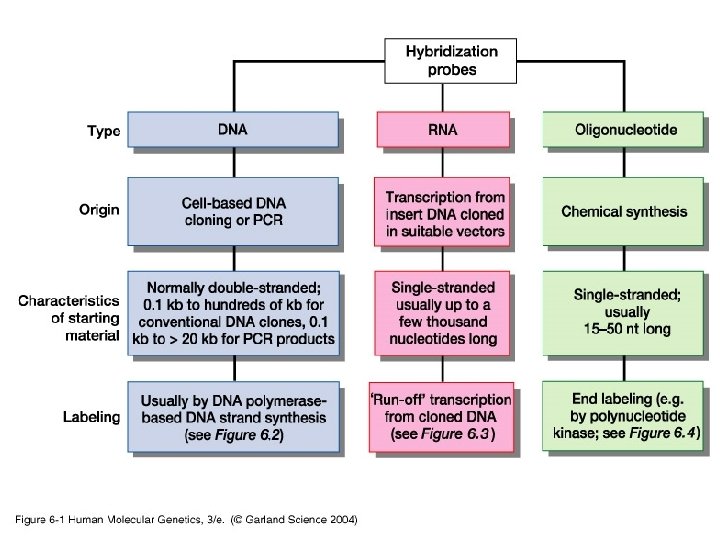
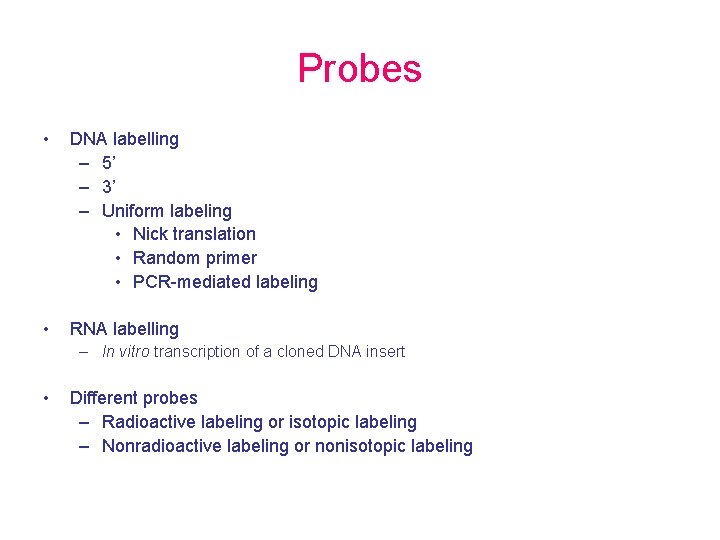
Probes • DNA labelling – 5’ – 3’ – Uniform labeling • Nick translation • Random primer • PCR-mediated labeling • RNA labelling – In vitro transcription of a cloned DNA insert • Different probes – Radioactive labeling or isotopic labeling – Nonradioactive labeling or nonisotopic labeling
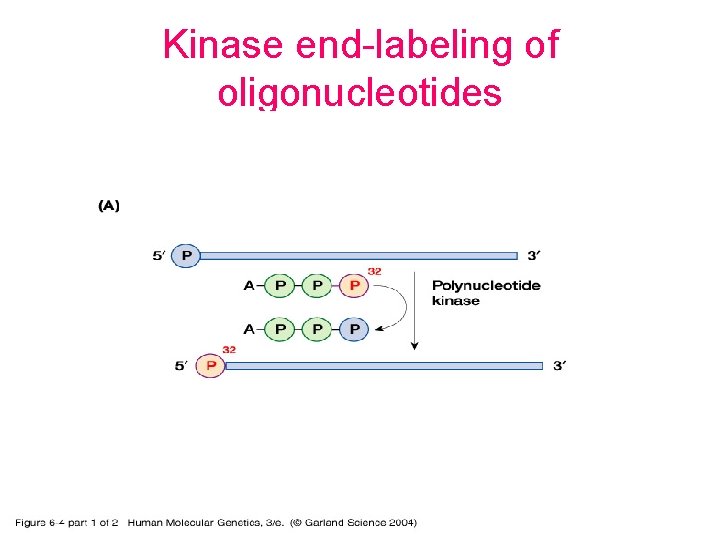
Kinase end-labeling of oligonucleotides
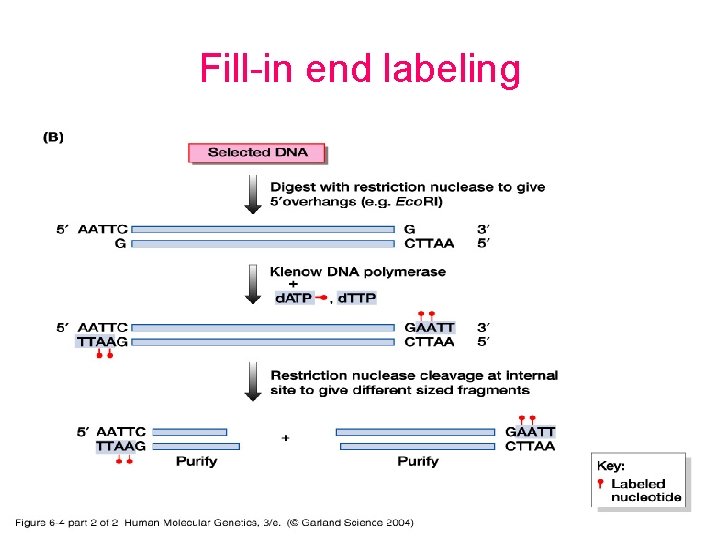
Fill-in end labeling
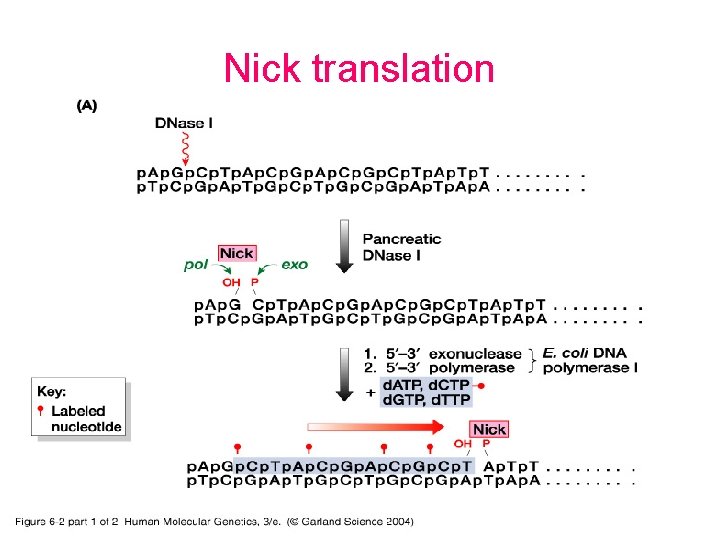
Nick translation
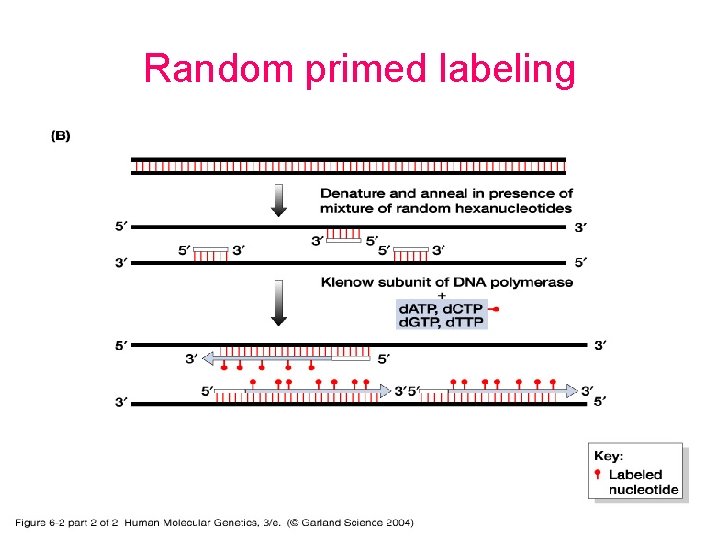
Random primed labeling
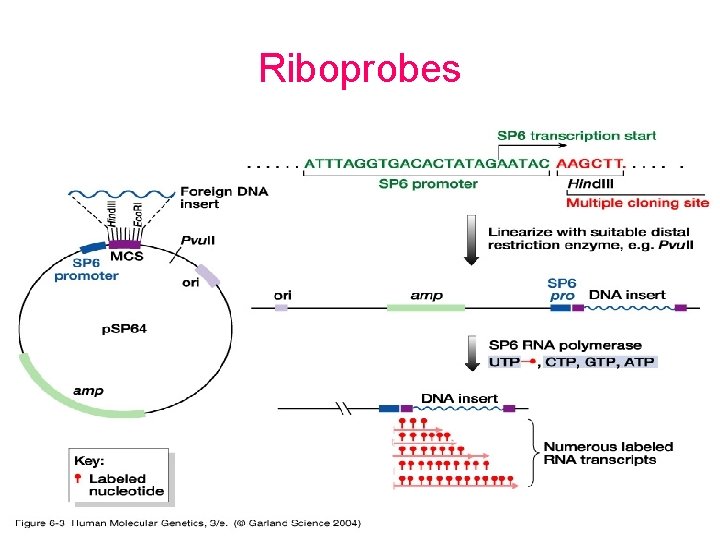
Riboprobes
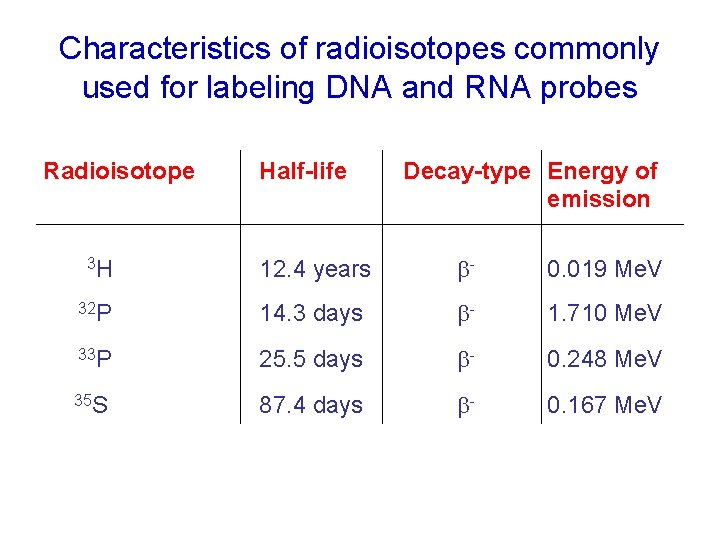
Characteristics of radioisotopes commonly used for labeling DNA and RNA probes Radioisotope Half-life Decay-type Energy of emission 12. 4 years b- 0. 019 Me. V 32 P 14. 3 days b- 1. 710 Me. V 33 P 25. 5 days b- 0. 248 Me. V 35 S 87. 4 days b- 0. 167 Me. V 3 H

Nonisotopic labeling and detection • The use of nonradioactive labels has several advantages: – – – safety higher stability of a probe efficiency of the labeling reaction detection in situ less time taken to detect signal • Major types – Direct nonisotopic labeling (ex. nt labeled with a fluorophore) – Indirect nonisotopic labeling (ex. biotin. -streptavidin system)
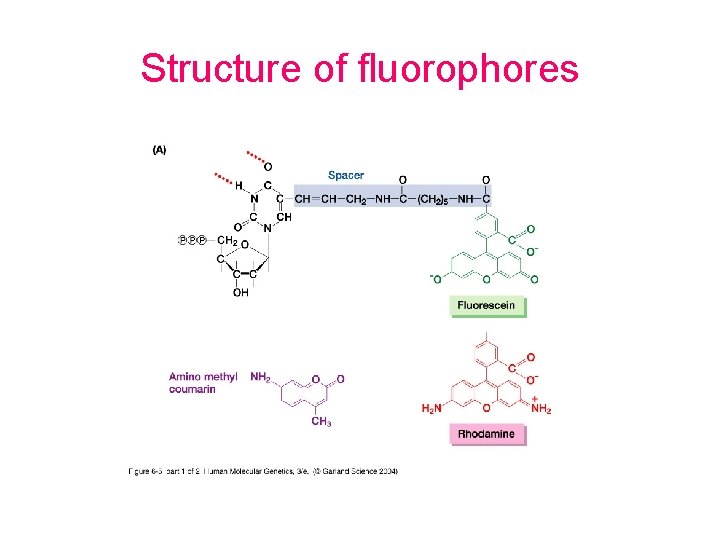
Structure of fluorophores
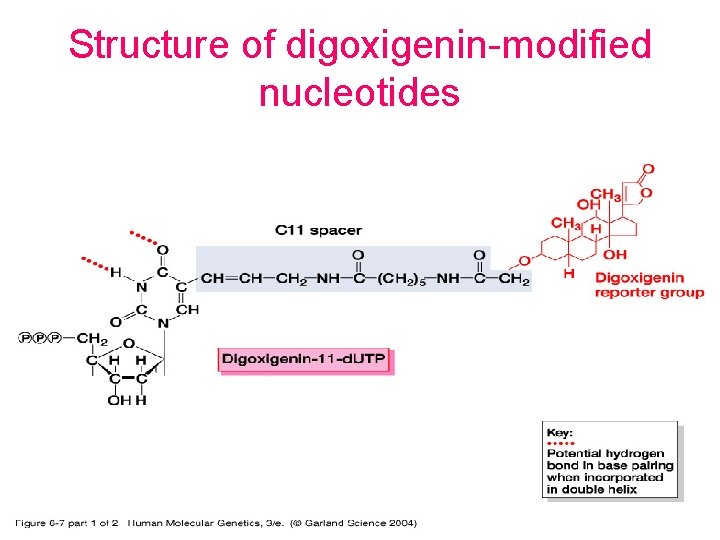
Structure of digoxigenin-modified nucleotides
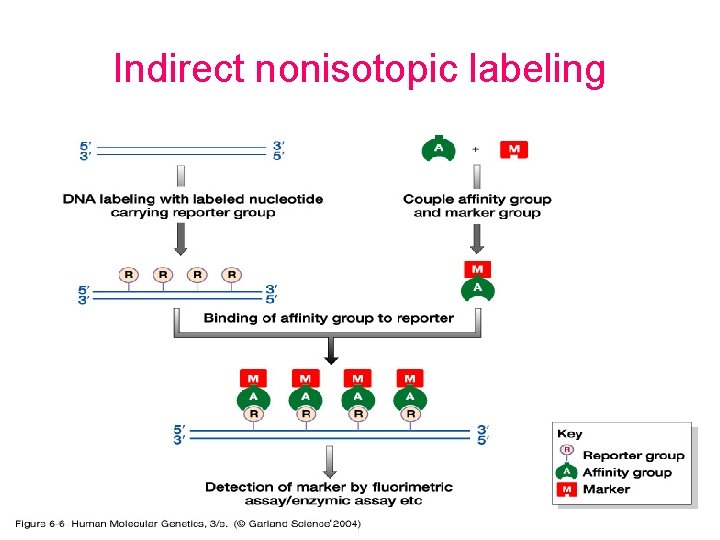
Indirect nonisotopic labeling
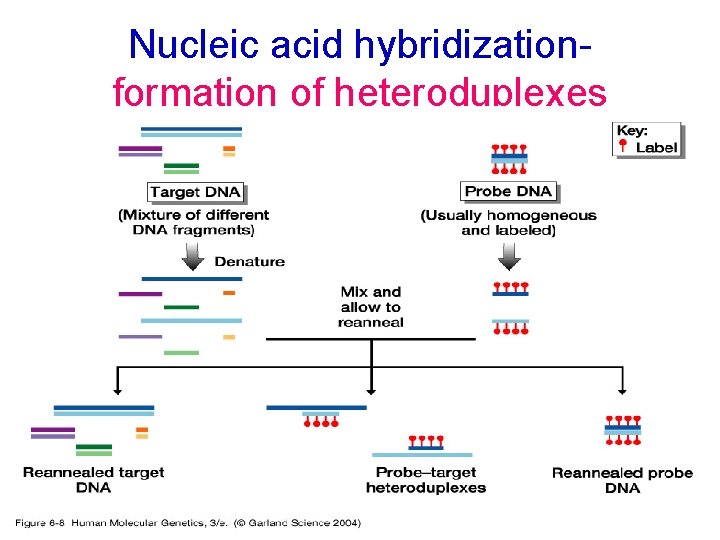
Nucleic acid hybridizationformation of heteroduplexes
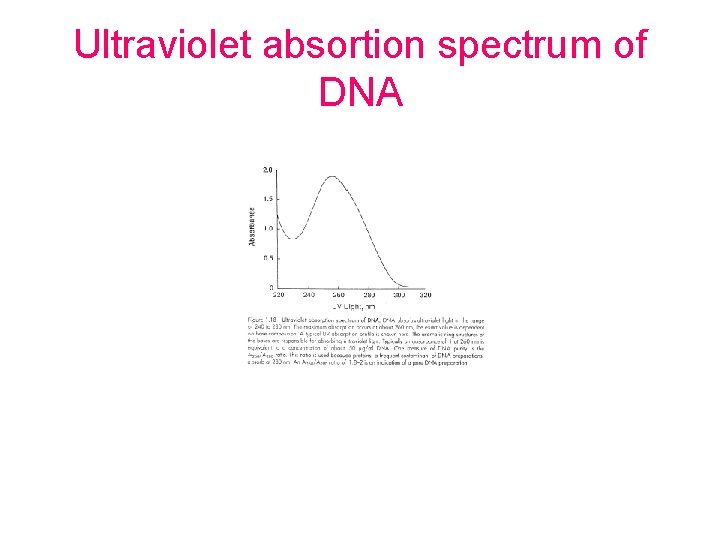
Ultraviolet absortion spectrum of DNA
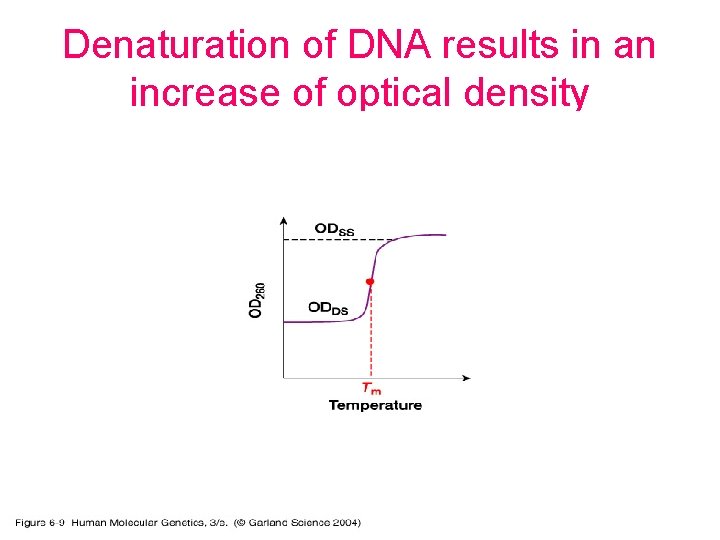
Denaturation of DNA results in an increase of optical density
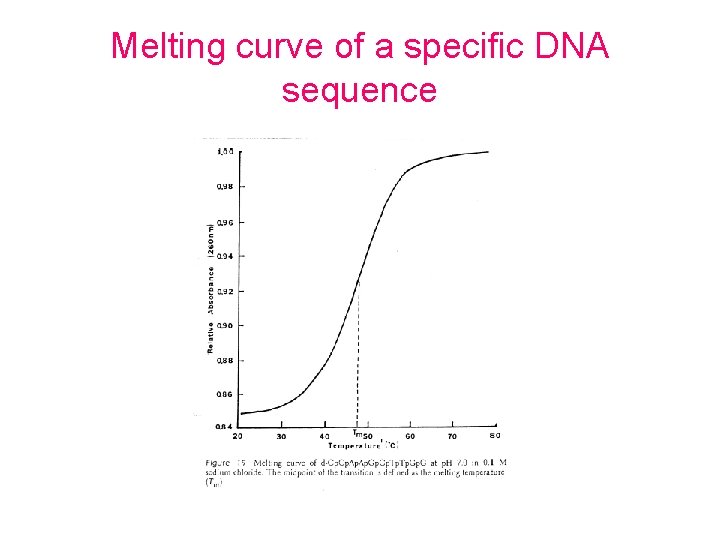
Melting curve of a specific DNA sequence
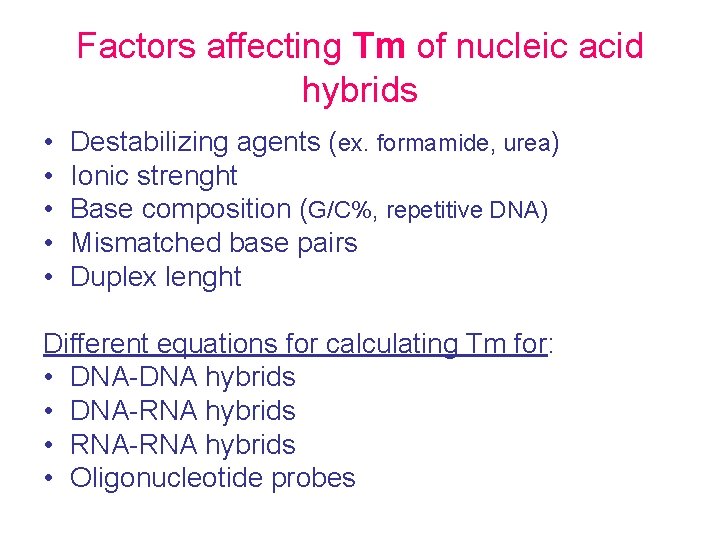
Factors affecting Tm of nucleic acid hybrids • • • Destabilizing agents (ex. formamide, urea) Ionic strenght Base composition (G/C%, repetitive DNA) Mismatched base pairs Duplex lenght Different equations for calculating Tm for: • DNA-DNA hybrids • DNA-RNA hybrids • RNA-RNA hybrids • Oligonucleotide probes

Factors affecting the hybridization for nucleic acids in solution (annealing) • • • Temperature Ionic strenght Destabilizing agents Mismatched base pairs Duplex lenght Viscosity Probe complexity Base composition p. H
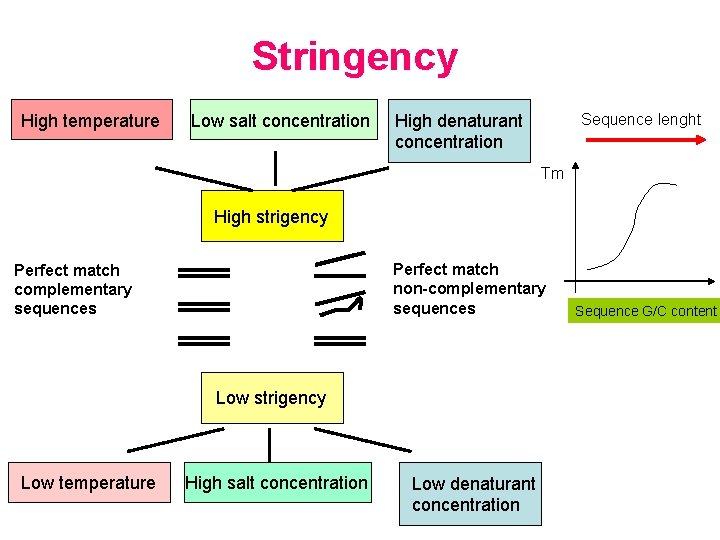
Stringency High temperature Low salt concentration Sequence lenght High denaturant concentration Tm High strigency Perfect match non-complementary sequences Perfect match complementary sequences Low strigency Low temperature High salt concentration Low denaturant concentration Sequence G/C content
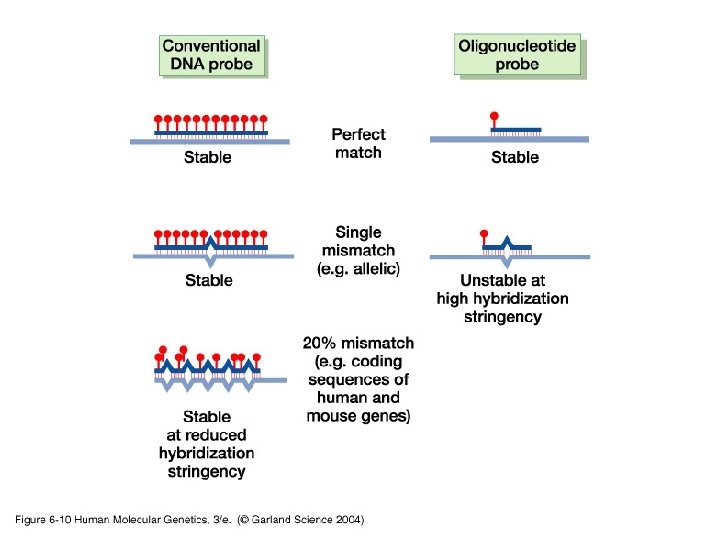
06_10. jpg
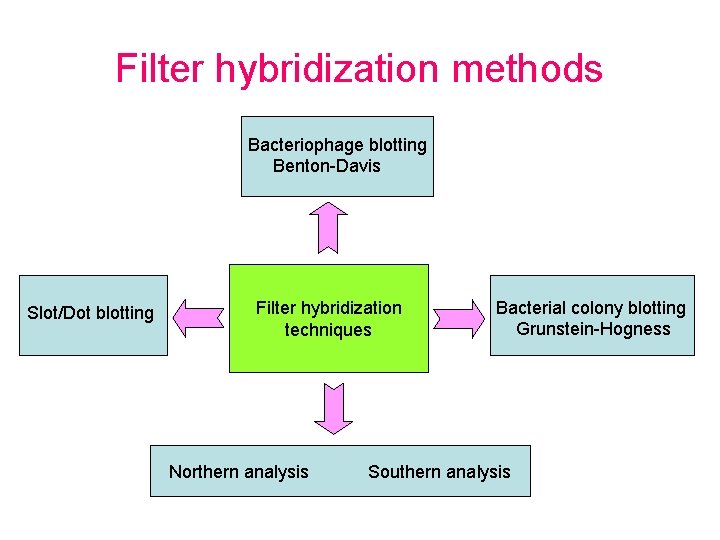
Filter hybridization methods Bacteriophage blotting Benton-Davis Slot/Dot blotting Filter hybridization techniques Northern analysis Bacterial colony blotting Grunstein-Hogness Southern analysis
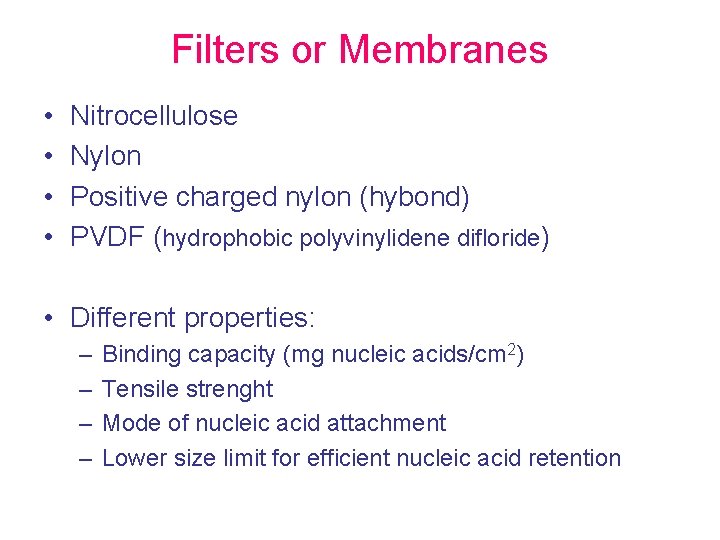
Filters or Membranes • • Nitrocellulose Nylon Positive charged nylon (hybond) PVDF (hydrophobic polyvinylidene difloride) • Different properties: – – Binding capacity (mg nucleic acids/cm 2) Tensile strenght Mode of nucleic acid attachment Lower size limit for efficient nucleic acid retention
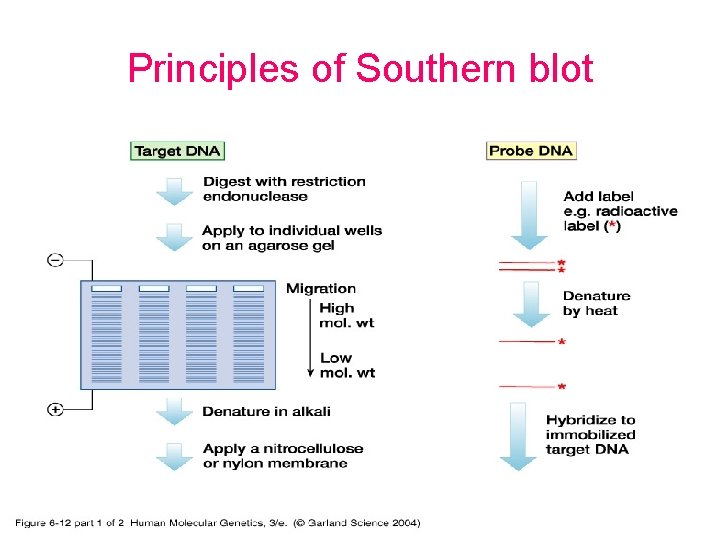
Principles of Southern blot
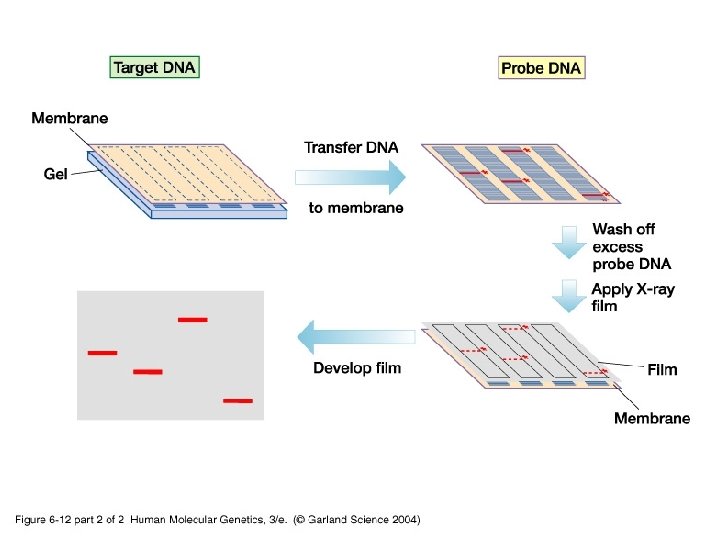
06_12_2. jpg
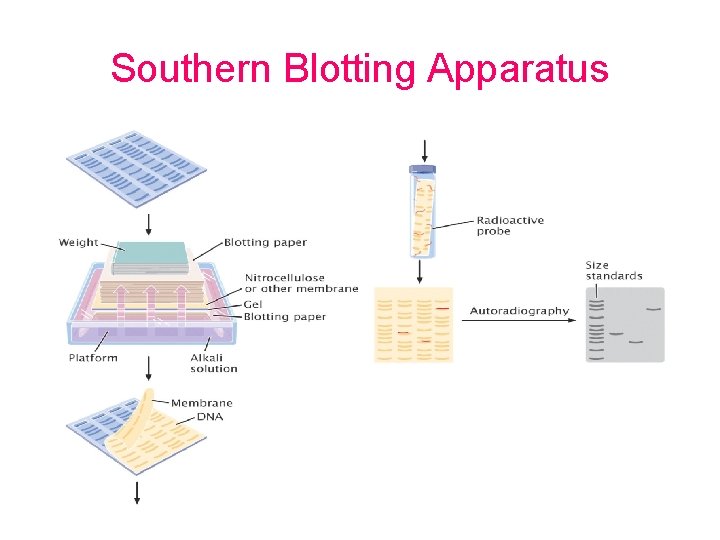
Southern Blotting Apparatus

Depurination/Denaturation of DNA

Typical hybridization solution • High salt solution (SSC or SSPE) • Blocking agent (Denhardts, salmon sperm DNA, yeast t. RNA) • SDS
Southern Applications • Detection of DNA rearrangements and deletions found in several diseases • Identification of structural genes (related in the same species (paralogs) or in different species (orthologs)) • Construction of restriction maps
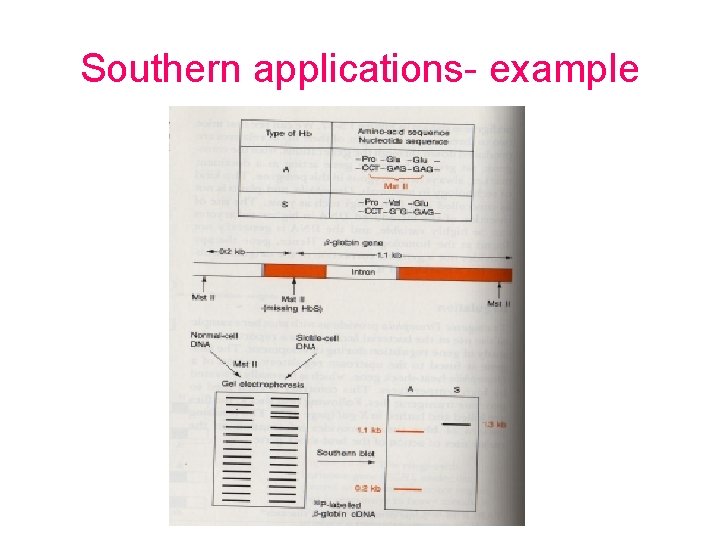
Southern applications- example
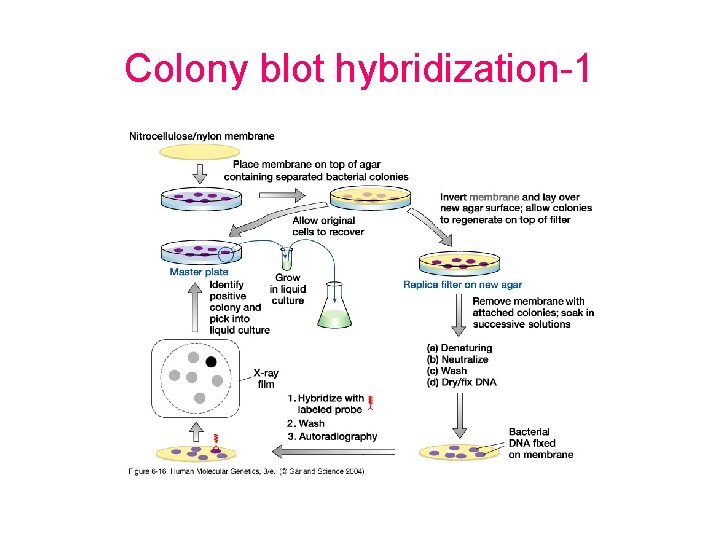
Colony blot hybridization-1
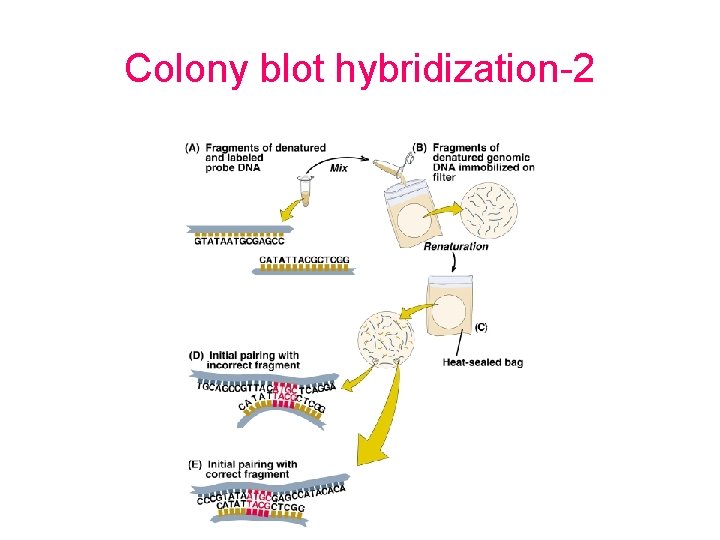
Colony blot hybridization-2
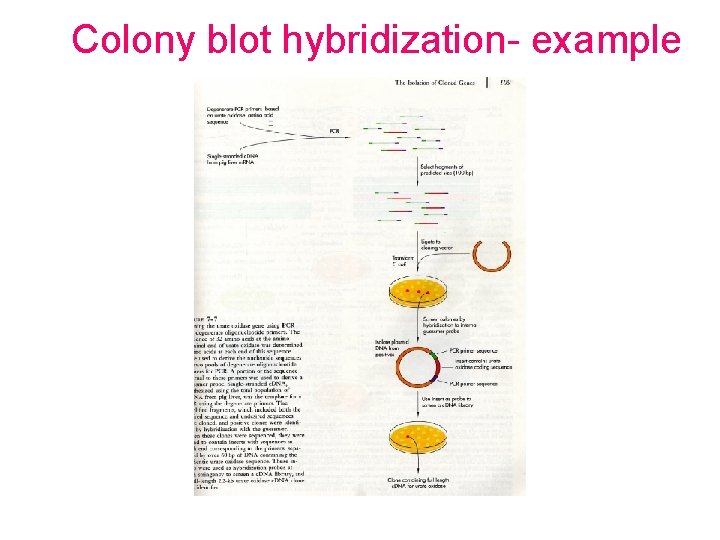
Colony blot hybridization- example
- Slides: 35